Abstract
Indole-3-acetic acid (IAA), the main naturally occurring auxin, is essential for almost every aspect of plant growth and development. However, only recently have studies finally established the first complete auxin biosynthesis pathway that converts tryptophan (Trp) to IAA in plants. Trp is first converted to indole-3-pyruvate (IPA) by the TAA family of amino transferases and subsequently IAA is produced from IPA by the YUC family of flavin monooxygenases. The two-step conversion of Trp to IAA is the main auxin biosynthesis pathway that plays an essential role in many developmental processes.
Keywords: hormonal regulation, hormone biology, genetics, development, auxin
INTRODUCTION
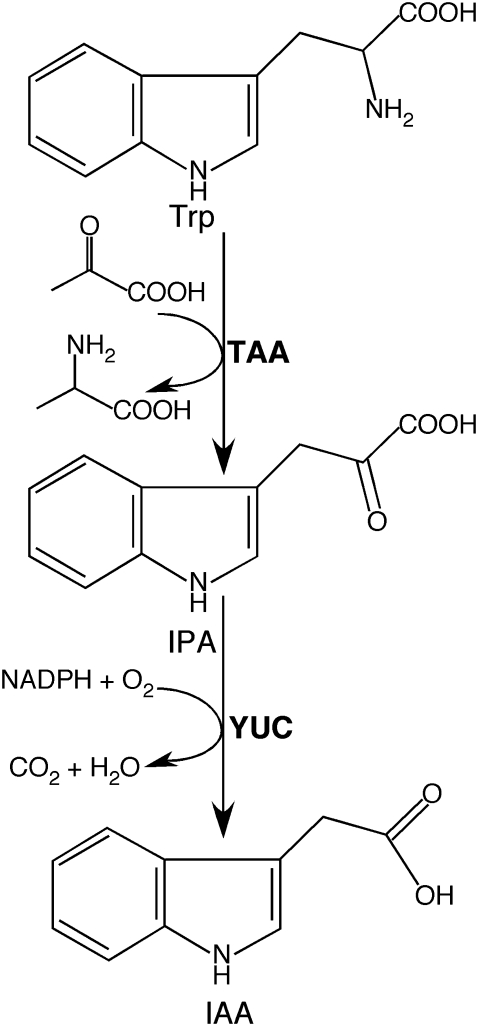
Although auxin has been studied extensively for decades, its main biosynthetic route in plants has only been revealed recently (Mashiguchi et al., 2011; Won et al., 2011) (Figure 1). Indole-3-acetic acid (IAA), the main auxin produced by plants, is known to be synthesized de novo using tryptophan (Trp) as a precursor or using a Trp-independent pathway (reviewed in Zhao, 2010). It has been clearly demonstrated that Trp-dependent auxin biosynthesis is essential for embryogenesis, seedling growth, flower development, vascular pattern formation, and other developmental processes (Cheng et al., 2006, 2007a; Stepanova et al., 2008; Tao et al., 2008). In contrast, the molecular components and the physiological functions of the proposed Trp-independent pathway are not known. In this paper, I review the recent genetic and biochemical results that led to the discovery of the first complete auxin biosynthesis pathway in plants starting at Trp and ending at IAA (Figure 1).
Figure 1.
The Tryptophan-Dependent Auxin Biosynthesis Pathway.
The first step is catalyzed by TAAs that transfer the amino group from Trp to an alpha keto acid such as pyruvate to generate IPA and another amino acid. The second step is an oxygen and NADPH-dependent reaction catalyzed by the YUC flavin-containing monooxygenases.
YUCs AND TAAs ARE ESSENTIAL FOR AUXIN BIOSYNTHESIS
Genetic approaches can be powerful for evaluating whether a proposed route is important for auxin biosynthesis and plant development. If a gene or a gene family plays an important role in auxin biosynthesis, we would expect that inactivation of the gene/gene family leads to dramatic developmental defects similar to those observed in auxin signaling mutants or in polar auxin transport mutants. Otherwise, the gene/gene family may not be a main contributor to auxin biosynthesis.
The YUCCA (YUC) flavin monooxygenase was first identified as a key auxin biosynthesis enzyme because overexpression of YUC in Arabidopsis leads to auxin overproduction (Zhao et al., 2001). Genetic and physiological analyses demonstrated that YUC catalyzes a rate-limiting step in a Trp-dependent auxin biosynthesis pathway (Zhao et al., 2001). In Arabidopsis, there are 11 YUC genes, and members of the YUC family have overlapping functions during development (Cheng et al., 2006, 2007a). Overexpression of each YUC gene in Arabidopsis and in other plants causes the characteristic auxin overproduction phenotypes (Cheng et al., 2006, 2007a). Inactivation of a single YUC gene in Arabidopsis does not cause obvious developmental defects, but simultaneous disruption of several YUC genes in Arabidopsis leads to defects in embryogenesis, seedling growth, flower development, and vascular pattern formation (Cheng et al., 2006, 2007a). The phenotypes of yuc mutant combinations are similar to those of known auxin transport and signaling mutants (Galweiler et al., 1998; Dharmasiri et al., 2005). The developmental defects of the loss-of-function yuc mutants are rescued when the bacterial auxin biosynthesis gene iaaM under the control of a YUC promoter is introduced into the yuc mutants, demonstrating that YUC genes play essential roles in auxin biosynthesis and plant development (Cheng et al., 2006). YUC genes have been identified in all of the sequenced plant genomes and YUC genes have been shown to be important for auxin biosynthesis in other plant species as well (Tobena-Santamaria et al., 2002; Gallavotti et al., 2008).
The TAA family of amino transferases was isolated independently from three genetic screens for mutants with altered responses to shade, ethylene, and the auxin transport inhibitor NPA, respectively (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). Inactivation of TAA1 and its close homologs TAR1 and TAR2 leads to partial auxin deficiency and defects in several developmental processes (Stepanova et al., 2008). The defects of taa mutants are partially rescued by auxin in growth media (Stepanova et al., 2008; Tao et al., 2008). Therefore, the TAA family genes are also considered essential for auxin biosynthesis.
Several other enzymes including CYP79B2/CYP79B3 (Zhao et al., 2002), nitrilase (Bartel and Fink, 1994; Bartling et al., 1994), aldehyde oxidase (Seo et al., 1998), and IPA decarboxylase (Vande Broek et al., 1999) have been proposed to participate in auxin biosynthesis. However, genetic analyses indicate that these enzymes are probably not major players in auxin biosynthesis. CYP79B2/B3 converts Trp to indole-3-acetaldoxime (IAOx), and inactivation of both CYP79B2 and CYP79B3 completely eliminates the production of IAOx (Zhao et al., 2002; Sugawara et al., 2009). However, the cyp79b2 cyp79b3 double mutants only have subtle growth defects, suggesting that the CYP79B2/B3 are not the main contributor to auxin biosynthesis (Zhao et al., 2002). In addition, CYP79B2/B3 appear only to exist in a small number of plant species, making it unlikely that they serve as universal auxin biosynthesis genes. Nitrilases, which hydrolyze indole-3-acetonitrile (IAN) to IAA, were initially isolated in a genetic screen for mutants insensitive to exogenous IAN (Bartel and Fink, 1994). The Arabidopsis genome has four nitrilase genes and it is generally believed that nitrilases play a more important role in glucosinolate metabolism, camalexin homeostasis, and cyanide detoxification than in auxin biosynthesis (Rauhut and Glawischnig, 2009; Su et al., 2011). The IAN levels are more than 100-fold higher than IAA levels in Arabidopsis (Sugawara et al., 2009), but overexpression of nitrilases does not lead to auxin overproduction phenotypes (Normanly et al., 1997), suggesting that IAN probably is not a main intermediate for auxin biosynthesis. Aldehyde oxidases, which can oxidize indole-3-acetaldehyde (IAAld) to IAA, also belong to a family in Arabidopsis and require the molybdenum-cofactor (Moco) for activities. The aba3 mutants, which fail to produce the Moco cofactor and do not have any aldehyde oxidase activities, do not show any obvious auxin-related developmental defects and do not accumulate IAAld, suggesting that aldehyde oxidases probably do not play a major role in auxin biosynthesis (Mashiguchi et al., 2011). IPA decarboxylases are proposed to catalyze the decarboxylation of IPA or other alpha-keto acids. In bacteria, the IPA decarboxylase was shown to convert IPA to IAAld (Vande Broek et al., 2005), but the Arabidopsis counterparts appear not to play a key role in auxin biosynthesis and plant development.
Overall, genetic analyses have so far unambiguously demonstrated that the YUCs and TAAs are two families essential for auxin biosynthesis and plant development. In contrast, analyses on CYP79B2/B3, nitrilases, aldehyde oxidases, and pyruvate decarboxylases indicate that they probably are not the main contributors to auxin biosynthesis.
THE YUCs AND TAAs PARTICIPATE IN THE SAME AUXIN BIOSYNTHESIS PATHWAY
Both YUCs and TAAs are essential for auxin biosynthesis, but they were previously placed in two independent pathways. It was proposed that the YUCs catalyze the conversion of tryptamine to N-hydroxyl tryptamine, which may be converted to IAOx (Zhao et al., 2001; Expósito-Rodríguez et al., 2007; Kim et al., 2007). The TAAs were placed in the step from Trp to IPA (Stepanova et al., 2008; Tao et al., 2008).
Interestingly, some taa mutant combinations and some yuc combinations display similar developmental defects. For example, both the yuc1 yuc4 yuc10 yuc11 quadruple mutants and the taa1 tar1 tar2 triple mutants fail to make the basal parts of Arabidopsis embryos (Cheng et al., 2007a; Stepanova et al., 2008). The phenotypic similarities between yucs and taas prompted speculations that YUCs and TAAs may participate in the same auxin biosynthesis pathway (Strader and Bartel, 2008; Zhao, 2010). Studies on the combinations of yuc and taa mutants in maize are also consistent with the hypothesis that YUC and TAA may participate in the same auxin biosynthesis pathway (Phillips et al., 2011). But there are obvious differences between yuc and taa mutants. For example, the taa mutants were defective in shade avoidance and in responses to ethylene and NPA in roots (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009), but YUCs were never isolated from the genetic screens that initially identified taa mutants. The failure to identify YUCs from the three genetic screens may be due to the redundant nature of YUC genes.
Recent genetic studies have resolved the apparent discrepancy between yuc mutants and taa mutants. All of the taa mutant phenotypes are phenocopied by yuc mutants when the right yuc combinations are used (Won et al., 2011). For example, roots of the yuc3 yuc5 yuc7 yuc8 yuc9 quintuple mutants (yuc Q) are resistant to both ethylene and NPA, which are characteristic phenotypes that were observed previously in taa mutants (Won et al., 2011). The yuc1 yuc4 double mutants behave similarly to taa mutants in shade avoidance assays (Won et al., 2011). All of the characteristic phenotypes of taa mutants can be phenocopied by yuc mutants.
On the other hand, inactivating TAA genes mimics the characteristic yuc mutant phenotypes. Disruption of YUC1 and YUC4 in the pid mutant background completely abolished the initiation of cotyledons (Cheng et al., 2007b, 2008), a phenotype that is perfectly mimicked in the taa1 tar2 pid triple mutants (Won et al., 2011). The synergistic genetic interaction between yuc1 yuc4 double mutants and npy1 is also phenocopied by taa1 tar2 npy1 triple mutants (Cheng et al., 2007b, 2008; Won et al., 2011). Because taa mutants and yuc mutants phenocopy each other in all major developmental processes analyzed, it is concluded that TAAs and YUCs participate in the same auxin biosynthesis pathway (Won et al., 2011).
A TWO-STEP AUXIN BIOSYNTHESIS PATHWAY
Genetic evidence indicates that TAAs and YUCs probably participate in the same auxin biosynthesis pathway (Won et al., 2011). Gain-of-function genetic studies put YUC downstream of TAA genes (Won et al., 2011). Overexpression of YUC genes leads to long hypocotyls and epinastic cotyledons (Zhao et al., 2001), which are the typical auxin overproduction phenotypes. The phenotypes of YUC overexpression lines are further enhanced when both YUC and TAA are overexpressed simultaneously in the same plants (Mashiguchi et al., 2011). On the other hand, the auxin overproduction phenotypes are completely suppressed in the strong taa mutant backgrounds, indicating that YUC-mediated auxin biosynthesis is dependent on TAA activities (Won et al., 2011). Direct measurement of IPA levels in yuc mutants and taa mutants reveals that yuc mutants accumulate IPA whereas taa mutants are partially IPA deficient, suggesting that TAAs participate in the production of IPA and YUCs are involved in metabolizing IPA (Mashiguchi et al., 2011; Won et al., 2011). Finally, direct in vitro biochemical assays have demonstrated that TAA can convert Trp to IPA and that YUCs produce IAA using IPA as a substrate (Mashiguchi et al., 2011). The genetic and biochemical data demonstrate that the TAAs and YUCs catalyze two consecutive reactions that convert Trp to IAA (Figure 1).
Molecular genetic studies in Arabidopsis in combination with analytic biochemistry contributed to the elucidation of the first complete auxin biosynthesis pathways in plants. It is quite surprising that plants use a simple two-step pathway as the main mechanism for de novo synthesis of the essential hormone auxin, given that auxin biosynthesis was previously considered very complicated (Zhao, 2010). Although the chemical reactions of the two-step pathway do not appear very sophisticated, genetic dissection of the pathway is complicated because each step uses a gene family. The involvement of gene families offers an effective way for plants to temporally and spatially control auxin biosynthesis. Tissue/cell-specific auxin biosynthesis can be achieved by selectively expressing particular family members. Indeed, Arabidopsis plants appear to use two separate sets of YUC genes for auxin biosynthesis in roots and shoots. YUC1, YUC2, YUC4, YUC6 are the main YUCs for auxin biosynthesis in shoots and YUC3, YUC5, YUC7, YUC8, YUC9 are responsible for producing auxin in roots (Won et al., 2011). The expression of TAA genes is also temporally and spatially regulated, but Arabidopsis seedlings appear to use the same set of TAA genes for both root and shoot development (Stepanova et al., 2008; Tao et al., 2008).
Both plant pathogenic bacteria and plants use a two-step pathway for auxin biosynthesis, with Trp as the starting point. In bacteria, Trp is oxidized into IAM by the tryptophan-2-monooxygenase iaaM, and IAM is subsequently hydrolyzed by iaaH (Comai and Kosuge, 1982). It appears that the rate-limiting step of the bacterial pathway is the first step that is catalyzed by iaaM. In contrast, the rate-limiting step for auxin biosynthesis in plants is the second step catalyzed by YUCs (Figure 1). Interestingly, the rate-limiting step in both pathways is catalyzed by a flavin-containing enzyme. The differences between the two pathways may reflect different regulatory mechanisms. IAM is destined to IAA by a simple hydrolysis whereas IPA, an alpha keto acid, may be used in other pathways in addition to IAA biosynthesis. Because the step from IPA to IAA is the rate-liming step for auxin biosynthesis in plants, diverting IPA to other pathways can provide an additional mechanism to control auxin biosynthesis. The reaction from Trp to IPA is coupled with the conversion of another alpha keto acid such as pyruvate or alpha-ketoglutarate into another amino acid. Therefore, the TAA-catalyzed step may affect other pathways as well, but YUCs appear to be specific for auxin biosynthesis.
There are still many unanswered questions in auxin biosynthesis. The molecular mechanisms that regulate auxin biosynthesis at transcriptional level and protein level are virtually unknown. In addition to IPA, plants also produce other indolic compounds including IAM, TAM, IAOx, IAN, and IAAld (Sugawara et al., 2009). It is still not clear whether the other indolic intermediates play some roles in auxin biosynthesis or not. Nevertheless, elucidation of the two-step auxin biosynthesis pathway provides a foundation for further dissection of the mechanisms by which auxin controls plant growth and development. We now have the ability to manipulate auxin levels in plants with temporal and spatial precision, providing tools for both basic research and crop improvement.
ADDENDUM
While this paper was in press, another study also suggested that TAAs and YUCs participate in the same auxin biosynthesis pathway (Stepanova et al. (2011), The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 22 November (Epub ahead of print)).
FUNDING
Research in my group is supported by the NIH R01GM068631 and the NSF Plant Genome DBI-0820729.
Acknowledgments
I would like to thank Drs H. Kasahara, K. Mishigushi, Q. Chen, H. De-Paoli and Mr Y. Gao, and Ms A. Zhao and X. Dai for their comments on the manuscript. No conflict of interest declared.
References
- Bartel B, Fink GR. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Schmidt RC, Weiler EW. Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc. Natl Acad. Sci. U S A. 1994;91:6021–6025. doi: 10.1073/pnas.91.13.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007a;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2007b;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Kosuge T. Cloning characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J. Bacteriol. 1982;149:40–46. doi: 10.1128/jb.149.1.40-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez AB, Hernández M, Pérez JA. Cloning and biochemical characterization of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan-dependent auxin biosynthesis pathway. Journal of Plant Growth Regulation. 2007;26:329–340. [Google Scholar]
- Gallavotti A, et al. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl Acad. Sci. U S A. 2008;105:15196–15201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Kim JI, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007;145:722–735. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–1790. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, et al. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell. 2011;23:550–566. doi: 10.1105/tpc.110.075267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut T, Glawischnig E. Evolution of camalexin and structurally related indolic compounds. Phytochemistry. 2009;70:1638–1644. doi: 10.1016/j.phytochem.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Seo M, et al. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 1998;116:687–693. doi: 10.1104/pp.116.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Strader LC, Bartel B. A new path to auxin. Nat. Chem. Biol. 2008;4:337–339. doi: 10.1038/nchembio0608-337. [DOI] [PubMed] [Google Scholar]
- Su T, et al. Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell. 2011;23:364–380. doi: 10.1105/tpc.110.079145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, et al. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2009;106:5430–5435. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobena-Santamaria R, et al. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002;16:753–763. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Broek A, et al. Transcriptional analysis of the Azospirillum brasilense indole-3-pyruvate decarboxylase gene and identification of a cis-acting sequence involved in auxin responsive expression. Mol. Plant–Microbe Interact. 2005;18:311–323. doi: 10.1094/MPMI-18-0311. [DOI] [PubMed] [Google Scholar]
- Vande Broek A, Lambrecht M, Eggermont K, Vanderleyden J. Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J. Bacteriol. 1999;181:1338–1342. doi: 10.1128/jb.181.4.1338-1342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zhao Y, et al. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002;16:3100–3112. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]