Abstract
I reflect on some of our studies on the hyperthermophilic archaeon, Thermococcus kodakarensis KOD1 and its enzymes. The strain can grow at temperatures up to 100 ℃, and also represents one of the simplest forms of life. As expected, all enzymes, DNA, RNA, cytoplasmic membrane, and cytoplasmic solute displayed remarkable thermostability, and we have determined some of the basic principles that govern this feature. To our delight, many of the enzymes exhibited unique biochemical properties and novel structures not found in mesophilic proteins. Here, I will focus on some enzymes whose three-dimensional structures are characteristic of thermostable enzymes. I will also add some examples on the stabilization of DNA, RNA, cytoplasmic membrane, and cytoplasmic solute.
Keywords: thermostability, hyperthermophile, protein, DNA, RNA, cytoplasmic membrane
1. Background of research
Thermophiles are now classified by their growth temperature as follows: thermophiles above 55 ℃, moderate thermophiles above 65 ℃, extreme thermophiles above 75 ℃, and hyperthermophiles above 90 ℃. In the 1970’s, I was focused on moderate thermophiles, namely Bacillus stearothermophilus, and one of the highlights of this period was that our group succeeded in the development of a transformation system in this strain.1) This recombinant technology led to many exciting finding in the following years. In the 1980’s, I was one of many who were taken by surprise with the discovery of hyperthermophiles, first isolated by Stetter and his collagues.2) I was fascinated that these hyperthermophiles could grow, or even preferred to grow, in boiling water. From then on, I have dedicated a major portion of my scientific activities to these “hot bugs,” and this will most likely be the case in the years to come.
2. Isolation of hyperthermophilic archaeon
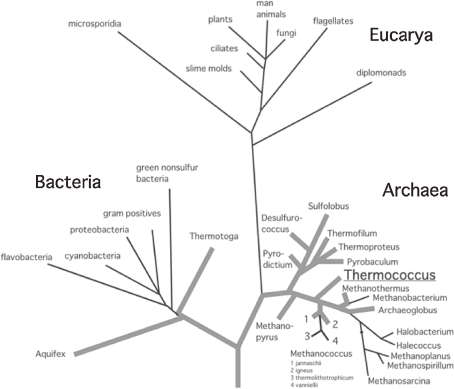
Hyperthermophiles are referred to as microorganisms that optimally grow at temperatures above 80 ℃,3) or as microorganisms that can grow at temperatures above 90 ℃.4) When a phylogenetic tree of all living organisms is constructed based on 16S or 18S ribosomal RNA sequences, three domains are exhibited; Eucarya, Bacteria, and Archaea (Fig. 1).5,6) Hyperthermophiles occupy branches nearest to the root of the tree, suggesting that hyperthermophiles may be the nearest organisms to the last common ancestor of life.
Figure 1.
Universal phylogenetic tree of living organisms based on 16S or 18S ribosomal RNA sequences. The three domains, Archaea, Bacteria, and Eucarya are shown. Boldfaces indicate hyper thermophiles.
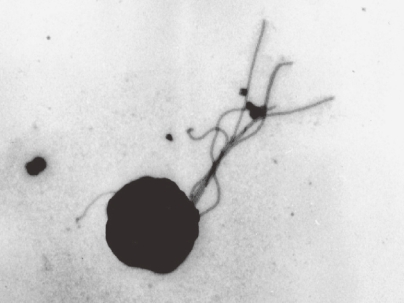
In 1993, we isolated a hyperthermophilic archaeon from a solfatara at Kodakara Island, Kagoshima, Japan.7) The strain exhibited rapid growth on amino acids with elemental sulfur as a terminal electron acceptor, and was later designated Thermococcus kodakarensis KOD1 (Fig. 2).8)
Figure 2.
Electron microscopic observation of Thermococcus kodakarensis KOD1.
T. kodakaraensis KOD1 exhibits cell growth from 60 to 100 ℃. The optimum pH and NaCl concentrations for growth are 7.0 and 3%, respectively. While the strain does grow by fermentation of yeast extract and tryptone, elemental sulfur significantly enhances the growth rate. Excess reducing equivalents are used to reduce the sulfur to hydrogen sulfide, and released in the gas phase. KOD1 cells are irregular cocci with a diameter of approximately 1 µm. The cells are highly motile and harbor several polar flagella. The genome of strain KOD1 turned out to have a length of 2088737 bp.9) The small size of the chromosome, and the high growth rate at high temperatures brought us to examine the strain and its proteins in detail. The 16S rRNA gene sequence indicated that it belongs to genus Thermococcus and named as T. kodakarensis KOD1. This strain is relatively close to the origin of life (Fig. 1). We have exploited the transformation system on this strain and developed gene disruption and/or replacement method on the chromosome.10–12) Now that genome sequence is known, transcriptome analysis by using DNA microarray is made possible. Based on these specific techniques, we have discovered many new enzymes/proteins and new metabolic pathways. Fortunately, the knowledge that we have obtained from this strain has been well above our initial expectations.
In this review, I would like to share some of what we have learned in the past years mainly from strain KOD1.
3. Molecular basis of protein thermostability
When enhancement of protein (thermo) stability is desired, there are a number of strategies available, taking into account four major interactions within a protein; covalent bonds via disulfide bridges, ionic interactions, hydrogen bonds, and hydrophobic interaction.13) Addition of any of these four types of interactions may be considered in order to enhance the thermostability of a protein. Another alternative may be to introduce proline residue at β-turn structures (the proline rule). Here, I will show some examples of protein thermostability.
3.1. Significance of hydrophobic interaction to the thermostability.
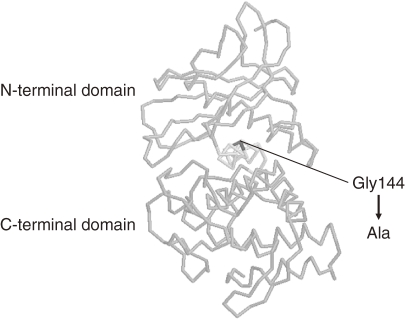
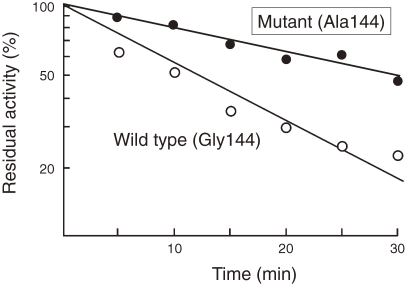
An increase in thermostability of a neutral protease from Bacillus stearothermophilus (NprT) was achieved from a rational approach by comparing its sequence with the thermostable thermolysin from B. thermoproteolyticus. The enzymes were 85% identical, while the thermostability of NprT at 75 ℃ was significantly lower than that of thermolysin. Taking into account the three-dimensional structure of thermolysin, a single mutation G144A was chosen as a candidate to increase the thermostability of NprT. The glycine residue was supposed to be located in an α-helix that connected the N- and C-terminal domains of the enzyme (Fig. 3).14) The mutation was expected to stabilize the α-helix, and increase internal hydrophobicity of the enzyme. Furthermore, the G144A mutation introduces only a small methyl group, minimizing any structural or functional interruption that may be caused by introduction of a new side chain. Indeed, this single mutation led to a significant increase in the thermostability of NprT (Fig. 4).15) This is a good example of the fact that an increase in internal hydrophobicity of an enzyme and stabilization of a secondary structure α-helix leads to an increase in the thermostability of a protein.
Figure 3.
Structure of thermolysin from Bacillus stearothermophilus used for site-directed mutagenesis of neutral protease from B. stearothermophilus.
Figure 4.
G144A mutant displays higher thermotolerance than the wild-type enzyme.
3.2. Significance of ion pairs to the thermostability.
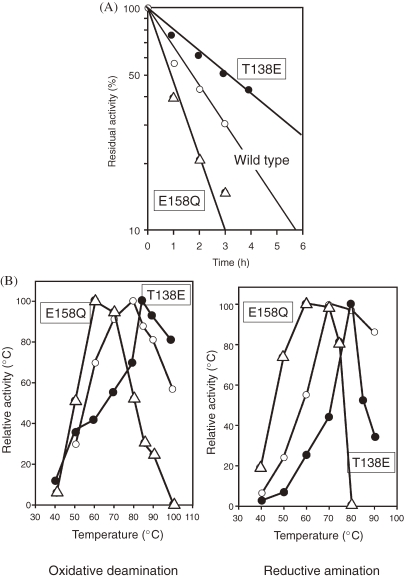
As the importance of ion pairs toward protein thermostability has been stressed in many cases, addition or removal of an ion pair should have significant effects. A clear example is provided by mutagenesis studies of glutamate dehydrogenase from T. kodakarensis KOD1 (Tk-GDH). The GDH from Pyrococcus furiosus (Pf-GDH) and Tk-GDH are 83% identical in terms of primary structure. However, while Pf-GDH displays a half-life of 12 h at 100 ℃, that of Tk-GDH is 4 h. The three-dimensional structure of Pf-GDH has been determined, and exists in a stable hexameric form. A structural model of Tk-GDH was constructed based on the structure of Pf-GDH. A difference was observed between two structures at the monomer–monomer interface. In Pf-GDH, there is a large ion pair network comprised of six residues, Arg35, Asp132, Glu138, Arg164, Arg165, and Lys166. Glu138 is located at the center of the network, interacting with Arg165 and Lys166. In the case of Tk-GDH, Glu138 was replaced by threonine residue. When a T138E mutation was introduced into Tk-GDH, an increase in both thermostability and optimal temperature was observed (Figs. 5A and 5B).16) At one of the two-fold axes of the proteins, Glu158 is at the center of another ion pair network, interacting with Arg124 and Arg128. An E158Q mutation would interrupt this network, and is presumed to destabilize the protein. As expected, the E158Q mutant protein of Tk-GDH displayed a lower optimal temperature for activity and decreased thermostability (Figs. 5A and 5B).16)
Figure 5.
(A) Thermotolerance of wild-type and mutant GDH from T. kodakarensis. (B) Optimum temperature of wild-type and mutant GDH from T. kodakarensis.
3.3. A structural basis for protein thermostability: O6-methylguanine-DNA methyltransferase.
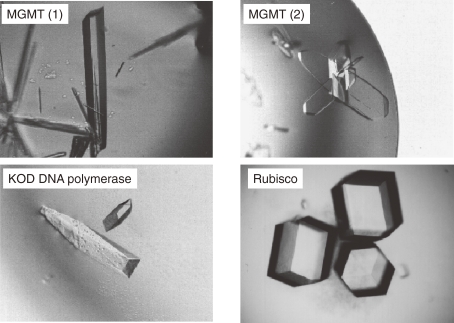
We have pursued attempts to elucidate the three-dimensional structures of various proteins from the hyperthermophilic archaeon T. kodakarensis KOD1. These include DNA polymerase,17,18) homing endonuclease II, O6-methylguanine-DNA methyltransferase (MGMT),19–22) aspartyl-tRNA synthetase,23–25) and ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco)26,27) (Fig. 6).
Figure 6.
Crystals of various proteins from T. kodakarensis KOD1.
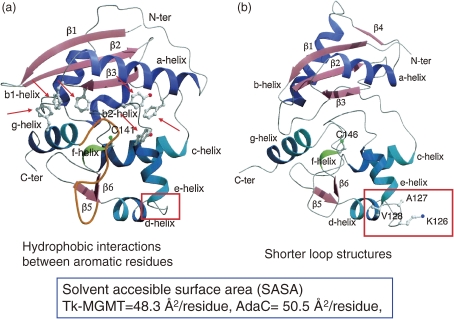
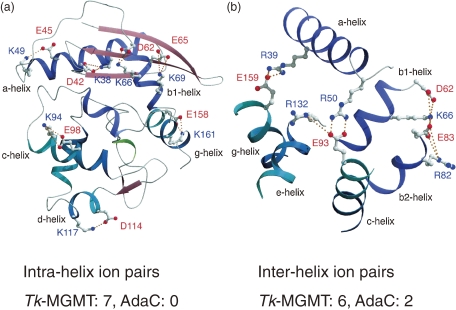
MGMT repairs DNA lesions in a single-step reaction by transferring methyl groups from O6-methylguanine of DNA to the cysteine residue of its own molecule. The enzyme from strain KOD1, Tk-MGMT, is a monomeric protein composed of 174 amino acid residues.28) Recombinant Tk-MGMT exhibits methyltransferase activity, and is stable at 90 ℃ for 30 min. We crystallized the protein, and its three-dimensional structure was elucidated at 1.8 Å resolution.21) This structure was compared with its counterpart from Escherichia coli (AdaC, C-terminal fragment of Ada protein)29) (Fig. 7). It should be noted that Tk-MGMT contains more aromatic residues (17 residues) than AdaC (8 residues), generating aromatic clusters within the protein to stabilize the internal packing of the protein (Fig. 7). We observed seven intra-helix ion pairs in Tk-MGMT, while none was detected in AdaC. It is presumed that these intra-helix ion pairs contribute to reinforcement of the stability of the alpha-helices. Furthermore, the number of inter-helix ion pairs was also higher in Tk-MGMT (six pairs) compared to AdaC (two pairs), stabilizing the internal packing of the tertiary structure (Fig. 8).20,21)
Figure 7.
Structural comparison between TK-MGMT (a) and AdaC (b).
Figure 8.
Intra- and inter-helices in Tk-MGMT.
Differences in the content of hydrophobic, polar, and charged residues were less than expected, as shown in Table 1. The only alteration observed was a 6% increase in charged residues in Tk-MGMT compared to AdaC. However, discrepancies became more evident when we focused on the solvent-accesible surface area (SASA) of the two proteins. Although the SASA of Tk-MGMT (8160 Å2) and AdaC (8339 Å2) were comparable, the percentage of charged residues in the SASA of Tk-MGMT (54%) was considerably higher than that of AdaC (35%). This was also accompanied by a decrease in hydrophobic and polar residues in the SASA from 32% to 24%. These comparisons clearly indicate that in the thermostable Tk-MGMT, there is a shift of hydrophobic residues to the interior of the molecule, and charged residues to the surface of the molecule.21) These observations suggest that the presence of both ion pairs and hydrophobic interaction can stabilize internal packing of its tertiary structure, leading to thermostability of the protein.
Table 1.
Comparison of solvent-accessible surface areas (SASAs)
| Tk-MGMT | AdaC | |
|---|---|---|
| No. of residues in crystal structure | 169 | 165 |
| No. of hydrophobic residues | 75 (44%) | 78 (47%) |
| No. of polar residues | 46 (27%) | 50 (30%) |
| No. of charged residues | 48 (28%) | 37 (22%) |
| Total SASA | 8160 Å2 | 8339 Å2 |
| SASA of hydrophobic residues | 1935 Å2 (24%) | 2638 Å2 (32%) |
| SASA of polar residues | 1797 Å2 (22%) | 2752 Å2 (33%) |
| SASA of charged residues | 4428 Å2 (54%) | 2949 Å2 (35%) |
3.4. Significance of quaternary structure to thermostability: ribulose 1,5-bisphosphate carboxylase/oxygenase.
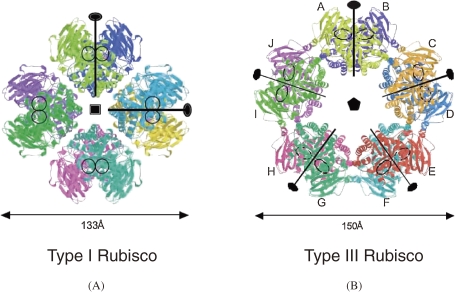
Rubisco is found predominantly in higher plants, algae, cyanobacteria, and photosynthetic bacteria.30,31) It is the most abundant enzyme on our planet, and plays one of the most important roles in our ecosystem. Rubisco catalyzes the covalent addition of carbon dioxide to ribulose 1,5-bisphosphate, producing two molecules of 3-phosphoglycerate.32) The function and the abundance of the enzyme provide a major link between inorganic and organic carbon in our biosphere. Through the Calvin cycle, the fixed carbon is then converted into sugars and other cell material, which will ultimately be utilized as the carbon and energy source of virtually all heterotrophic organisms. The significance of Rubisco has attracted scientists for decades, consequently leading to an extraordinary accumulation of knowledge on the enzyme. In almost all organisms, Rubisco is a hexadecamer consisting of eight large (L) and eight small (S) subunits, known as the Type I enzyme. Another type of Rubisco, found in some β-purple bacteria, is a homodimer of L subunits only and referred to as the Type II enzyme. While the primary structures of Rubisco L subunits of the same type are highly similar (>70%), those of Type I and Type II enzymes display less than 50% similarity.
Through genome analysis of strain KOD1, we came upon an open reading frame that displayed similarity to previously known Rubiscos. At first we were skeptical as to whether the ORF encoded a genuine Rubisco. The deduced amino acid sequence showed only 50% similarity to the previously known types of Rubisco, and moreover, why would a heterotrophic hyperthermophilic archaeon need a Rubisco?
Phylogenetic analysis of Rubiscos indicated that Tk-Rubisco clustered with the other archaeal sequences, and was distinct from Type I and Type II enzymes in terms of primary structure. We expressed the gene in Escherichia coli, and purified the recombinant enzyme. Activity measurements revealed that Tk-Rubisco could catalyze the carboxylation and oxygenation of ribulose 1,5-bisphosphate, indicating that the protein was a bona fide Rubisco.33,34) In fact, as Tk-Rubisco displayed activity at temperatures as high as 100 ℃, its specific activity was higher than any previously characterized Rubisco. We also found that Tk-Rubisco exhibited an unprecedentedly high carboxylase specificity. The native Tk-Rubisco consisted solely of large subunits,35) and allowed us to perform further structural analysis with the recombinant protein.
Observation of the purified enzyme by electron microscopy indicated a pentagonal structure, indicating that Tk-Rubisco was a decamer composed only of large subunits, with a pentagonal ring-like structure.35) We further crystallized the protein using ammonium sulfate as a precipitant, and determined the structure of Tk-Rubisco at 2.8 Å resolution.
As indicated by the above results, Tk-Rubisco is a novel (L2)5 decameric structure (Type III) (Fig. 9).27) Compared to previously known Type I enzymes, each L2 dimer was inclined approximately 16° to form a toroid-shaped decamer with unique L2–L2 interfaces. Differential scanning calorimetry (DSC), circular dichroism (DC), and gel permeation chromatography (GPC) showed that Tk-Rubisco maintained its secondary structure and decameric assembly even at high temperatures. Despite the low sequence homology, monomer (L) and dimer (L2) structures of Tk-Rubisco were well conserved compared with those of Type I and Type II enzymes. In its unique L2–L2 interface which includes rims of the L–L dimerization surface, an intensive ionic network was observed. This ionic network was considered to stabilize not only the pentagonal (L2)5 assembly, but also the L–L association in each dimer itself. It might contribute especially to the structural thermostability of the active sites, which are also located at the L–L dimerization surface. When the ionic network was broken, the thermostability decreased, though the L2 structure was maintained. These results clearly indicate that the quaternary structure is important to increase thermostability of the enzyme.35)
Figure 9.
Type I Rubisco (L8 structure, small subunits are not shown) from spinach and Type III Rubisco from T. kodakarensis show octamer and decamer structures, respectively. Ribbon diagram is shown. Each monomer is shown in different colors and labeled from A to J.
In spite of the accumulation of knowledge on the biochemical and structural features of Tk-Rubisco, little progress has been made in elucidating the physiological role of the enzyme. Recently, we found that Tk-Rubisco is involved in a new metabolic pathway, AMP degradation.36)
3.5. Significance of S–S bond: DNA polymerase.
Recombinant KOD DNA polymerase was purified, and its characteristics were studied and compared with those of DNA polymerases from various (hyper)thermophiles.37) The frequency of deoxyribonucleotide misincorporation was very low (0.35%), indicating that KOD DNA polymerase was an enzyme with high fidelity. Processivity, or the persistence of sequential nucleotide polymerization, of the enzyme was 10- to 15-fold greater than those for Pfu DNA polymerase and Deep Vent DNA polymerase. The extension rate of KOD DNA polymerase was 106 to 138 bases s−1, which is much higher than those of Pfu (25 bases s−1), Deep Vent (23 bases s−1), and Taq (61 bases s−1) DNA polymerases. It turned out that KOD DNA polymerase was a superb enzyme in terms of fidelity (low mutation frequency), processivity, and elongation speed for PCR (polymerase chain reaction).
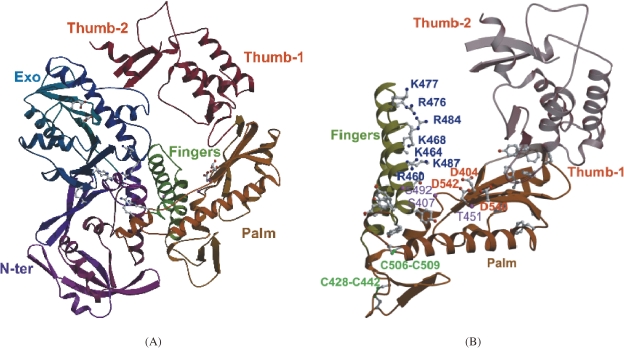
The excellent performance of KOD DNA polymerase tempted us to further determine its three-dimensional structure.17,18) The enzyme had a disk-like shape with dimensions of 60 Å × 80 Å × 100 Å (Fig. 10A). The enzyme was comprised of four distinct domains with subdomains; N-terminal domain, Exonuclease domain, Polymerase domain (including Palm and Fingers subdomains), and the Thumb domain (including Thumb-1 and Thumb-2 subdomains). Two disulfide bonds were found in the polymerase domain (Fig. 10B), and these bonds may contribute to the thermostability of the enzyme. Based on our recent results of tertiary structural studies, we are now clarifying the mechanisms that lead to the high fidelity and elongation rate of KOD DNA polymerase. Further elucidation of the structure-function relationship will provide better understanding of DNA polymerases and will suggest strategies to engineer better PCR enzymes.
Figure 10.
Three-dimensional structure of KOD DNA polymerase. (A) Overall structure of KOD DNA polymerase. The structure is composed of domains and subdomains, which are indicated as follows. N-Terminal domain (N-ter, purple), Exonuclease domain (Exo, blue), Polymerase domain including Palm (brown) and Fingers (green) subdomains, and the Thumb domain (red) including Thumb-1 and Thumb-2 subdomains. (B) The polymerase domain of KOD DNA polymerase. Conserved acidic residues (D404, D540, and D542) are represented by ball-and-stick models and indicated in red. Basic residues in the Fingers domain are also represented by ball-and stick models and indicated in blue.
3.6. Significance of chaperonin.
It is well known that chaperonin is important for protein folding. Strain KOD1 has two chaperonins CpkA and CpkB, and they are mainly expressed and functioned at lower and higher temperatures, respectively.38–40) When chaperonin is added into a protein solution in vitro, it prevents the inactivation and aggregation of protein at high temperature. Some other proteins such as prefoldin are also induced by heat shock and exhibit the same function as chaperonin. These proteins can support the growth of hyperthermophiles at high temperatures.
4. The importance of high temperature for maturation of thermostable proteins: glutamate dehydrogenase
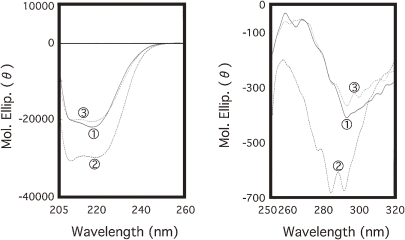
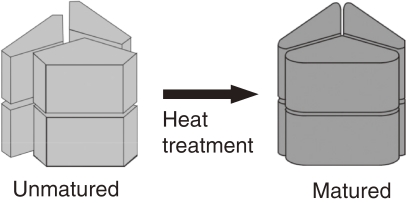
An intriguing characteristic of thermostable proteins was revealed during studies on glutamate dehydrogenase of T. kodakarensis KOD1 (Tk-GDH). The native enzyme purified from KOD1 cells was obtained solely in a hexameric form and displayed high specific activity.41) In contrast, when we expressed the gene in Escherichia coli and purified the recombinant protein, we obtained a mixture of hexamers and monomers. Only the recombinant hexamers exhibited GDH activity, but the specific activity was significantly lower than the native hexameric enzyme. Comparison of CD spectra between the native and recombinant hexamers revealed that their structures were markedly different (Fig. 11). The results suggested that the temperature of protein production might be important for proper assembly of thermostable proteins.42) We pursued this possibility by incubating the recombinant protein at high temperature (80 ℃, 15 min), and found that the CD spectra of the heat-treated protein displayed a similar profile to that of the native Tk-GDH. The specific activity of the protein, although still lower than the native enzyme, also increased 2.6-fold. Likewise, heat treatment of the inactive monomeric form of the recombinant enzyme converted a portion of the monomers to a hexameric form. From these studies, we realized that high temperature plays a pivotal role (heat maturation) in the proper folding and oligomerization of thermostable enzymes (Fig. 12).
Figure 11.
CD spectra of native Tk-GDH (1), and hexameric forms of recombinant Tk-GDH prior to (2) and following (3) heat treatment (80 ℃, 15 min). Far UV-spectra are shown on the left panel, and near UV-spectra on the right panel.
Figure 12.
A model describing the structural change of Tk-GDH induced by heat treatment.
5. Stabilization mechanism of DNA at high temperature
5.1. Significance of histone and polyamine.
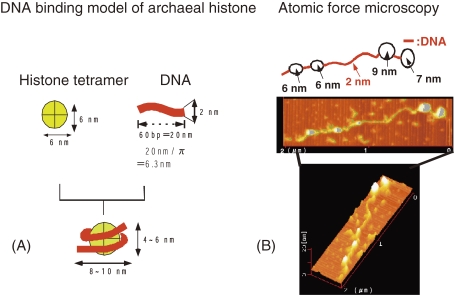
When a solution of linear DNA is heated, double-stranded DNA is melted and two single-stranded DNA molecules appear. How is the chromosomal DNA of hyperthermophile stabilized at high temperatures such as boiling point? T. kodakarensis KOD1 possesses two histones (basic proteins, HpkA & HpkB) that are essential for DNA compaction and nucleosome formation (Fig. 13), leading to higher melting temperature by 20 ℃ or more.43,44) In addition, high concentration of potassium ion and polyamines (positively charged aliphatic compounds) such as spermine and spermidine can stabilize the DNA.
Figure 13.
(A) DNA binding model of archaeal histone. (B) Observation of nucleosome-like structure of T. Kodakarensis by atomic force microscopy.
5.2. Significance of reverse gyrase.
Reverse gyrase is worthy of remark from the standpoint of both DNA stabilization and evolution of life. The enzyme is specific for hyperthermophiles, because the enzyme gene (including homologues) has been found in every hyperthermophile ever analyzed without exception and none is found in thermophiles or mesophiles. Reverse gyrase is a kind of topoisomerase and gives a positive superhelix structure, giving high thermostability to DNA. When the reverse gyrase gene was deleted from the chromosome of T. kodakarensis, the Δrgy mutant strain grew slowly at 80 ℃ or higher and could not grow at 93 ℃, while the wild-type strain could grow at 100 ℃.45) These results clearly show that reverse gyrase is essential to stabilize chromosomal DNA at high temperatures.
6. Stabilization mechanism of RNA at high temperature
6.1. Modification of RNA.
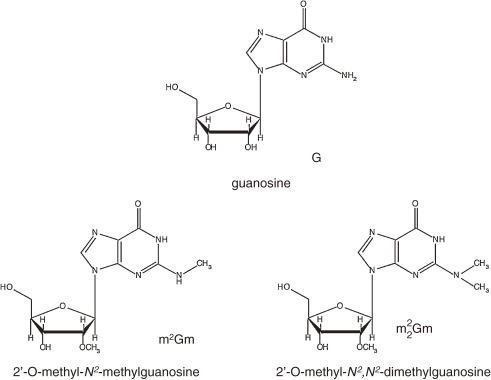
Posttranscriptional modification of tRNA and rRNA is well known. In archaeal tRNA, various modified nucleosides such as archaeosine (7-formamidino-7 deazaguanosine), 1-methylpseudouridine (m1Ψ), N2,2′-O-dimethylguanosine (m2Gm), and N2,N2-O-trimethylguanosine (m22Gm) have been reported46) (Fig. 14). In relation to high growth temperature, m2Gm and m22Gm deserves our attention. These material are found in hyperthermophiles (Archaeoglobus, Methanothermus, Thermoproteus, Thermococcus, Pyrobaculum, Pyrodictium), but not found in thermophiles (Methanobacterium, Thermoplasma). In addition, Pyrococcus furiosus cells grown at 100 ℃ contained three times more m2Gm and m22G than those cells at 70 ℃.47) This phenomenon strongly suggests that modified nucleosides contribute to stabilization of tRNA.
Figure 14.
Posttranscriptional modification of nucleoside.
6.2. Significance of branched-chain polyamine.
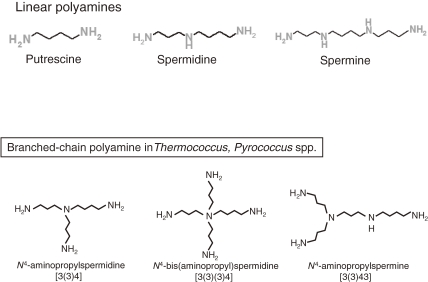
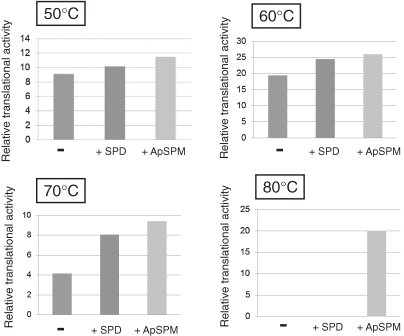
Extreme thermophiles produce two types of unusual polyamine: long linear polyamines such as caldopentamine and caldohexamine, and branched polyamines (Fig. 15). Linear polyamines such as spermidine, spermine and longer polyamines can stabilize DNA. In contrast, tRNA can be stabilized by branched polyamines.48) Since tRNA has tertiary structure, branched polyamines may tightly interact with tRNA ant stabilize it. Cytoplasmic polyamines were analyzed for cells cultivated at various growth temperatures in the hyperthermophilic archaeon Thermococcus kodakarensis. Spermidine (linear) and N4-aminopropylspermine (branched) were identified as major polyamines at 60 ℃, and the amount of N4-aminopropylspermine increased as the growth temperature rose.49) We have also examined the effect of N4-aminopropylspermine on in vitro protein synthesis by using S30 fraction of T. kodakarensis. When spermidine was added at a concentration of 0.2 mM, protein was not synthesized at 80 ℃, while addition of N4-aminopropylspermine (0.2 mM) supported protein synthesis at 80 ℃ (Fig. 16) (unpublished data). These data suggest the physiological importance of branched polyamine at high temperature.
Figure 15.
Linear polyamines and branched-chain polyamines.
Figure 16.
Effect of linear and branched-chain polyamines on in vitro protein synthesis by using S30 fraction of T. kodakarensis.
7. Stabilization of cytoplasmic membrane at high temperature
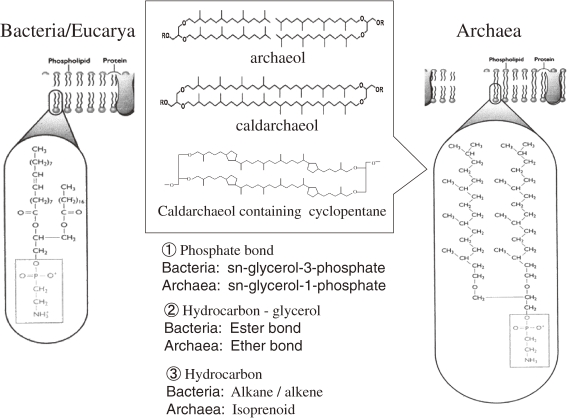
Generally speaking, archaea have unique membrane lipids typified by ether linkages of the glycerol-to-isoprenoid chains with sn-2,3 stereochemistry that runs against the naturally occurring sn-1,2 stereochemistry of the glycerophospholipids of bacteria and eucarya. The ether linkage in archaea is thought to contribute to greater chemical stability at extreme pH levels, unlike the ester linkage in bacteria and eucarya, and the isoprenoid chains are more stable against oxidation than fatty acids. The fluidity of cytoplasmic membrane to environmental change was maintained by the following features; (1) the length of the hydrocarbon chains, (2) side-chain saturation such as double-bond hydrogenation, or (3) side-chain modification such as cyclopentane ring formation. The cytoplasmic membrane of hyperthermophile has generally been stabilized by longer hydrocarbon chains with side-chain modification. For example, in Thermococcales strains, the ratio of caldarchaeol (C40 isoprenoid units, dibiphytaqnyl diglycerol tetraethers) to archaeol (C20 isoprenoid units, diphytanyl glycerol diethers) increased with increasing growth temperature. In Methanocaldococcus jannaschii, macrocyclic archaeol polar lipids increase as the temperature rises. Core lipid from Thermoplasmatales strains was caldarchaeol, having different numbers of cyclopentane rings, and the degree of cyclization increased with increasing growth temperature. These complex characteristics are important to stabilize membrane at high temperature (Fig. 17).50–53)
Figure 17.
Comparison of membrane lipid.
8. General effect of cytoplasmic solute
Hyperthermophiles accumulate unusual organic solutes in response to osmotic as well as heat stress. Mannosyl glycerate, mannosyl glyceramide, di-myo-inositol phosphate (DIP), mannosyl-di-myo-inositol phosphate, diglycerolphosphate, and glycerol-phospho-myo-inositol are examples of compatible solutes highly restricted to thermophiles and hyperthermophiles. It is shown that the solute plays a role of thermoprotection.54)
9. Protection of thermolabile metabolic intermediate
Some metabolic intermediate are thermolabile, and those substances are quickly transferred to the next enzyme for the protection as in the tryptophan biosynthetic pathway.55) Another example is that heat-labile triosephosphates are quickly removed and trapped in stable fructose 6-phosphate in the reactions of the bifunctional FBP aldolase/phosphatase in thermophilic Archaea such as Ignicoccus hospitaqlis, Metallosphaera sedula and Thermoproteus neutrophillus.56) This kind of protection is also important in hyperthermophiles.
10. Prevention of thermal inactivation and aggregation of enzymes
Proteins tend to form inactive aggregate at high temperatures. We have found that polyamines effectively prevent thermal inactivation and aggregation of hen egg lysozyme. These results imply that polyamines are the candidates as molecular additives for preventing the thermal aggregation and inactivation of relatively heat labile proteins.57) Intracellular polyamines may have the same kind of functions in thermophiles.
11. Future perspectives
The examples mentioned above describe some interesting aspects for stabilization of enzymes, DNA, RNA, cytoplasmic membrane, and cytoplasmic solute in hyperthermophilic archaea. Until now, our studies have been mainly biased towards biochemical and structural studies on the enzymes of strain KOD1. In future studies, we are planning to focus a little more on the cell itself, with its diverse metabolic and regulatory pathways. Elucidating the functions of hypothetical genes will be one of the main tasks to address. We have determined the complete genome sequence of strain KOD1,9) and 2306 genes at most are present on the genome, and in some way, contribute to the activities in the cell. The relatively small number of genes implies that the mechanisms supporting life in strain KOD1 should also be relatively simple. We have also exploited an excellent gene disruption (replacement and deletion) method in this hyperthermophile10) the physiological function of the target gene (and gene product) can be directly analyzed. We hope this will lead us to better understand some of the basic principles of life. There is still a long ride ahead, but we are certain that it will be a “hot” one.
Acknowledgements
I would like to express my sincere gratitude to all members who contributed to the research mentioned here. Much of the research was performed with H. Atomi, T. Fukui, and T. Kanai at the Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, and with M. Takagi, M. Morikawa, and S. Fujiwara at Department of Biotechnology, Graduate School of Engineering, Osaka University. I would also like to extend my sincere thanks to Prof. Y. Kai, Osaka University, Prof. K. Miki, Kyoto University, with whom fruitful collaborations have led to so many beautiful protein structures.
Profile
Tadayuki Imanaka, male, biotechnologist, graduated from Osaka University, receiving the degree of Bachelor of Engineering in 1967, finished the post-graduate course at the same university, receiving the degree of Master of Engineering in 1969, and awarded the degree of Doctor of Engineering from Osaka University in 1973. Postdoctoral research associate at Massachusetts Institute of Technology (USA) from 1973 to 1974 (an official leave of absence), Associate Professor of Biotechnology at Osaka University since 1981, and Professor of Biotechnology at Osaka University since 1989. Professor at Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University since 1996, and Professor at Department of Biotechnology, Ritsumeikan University since April, 2008. He was awarded Biotechnology award of the Society for Bioscience and Bioengineering, Japan, in 2001, Arima Prize of Japanese Biotechnology Association, in 2001, Fellow in American Academy of Microbiology, in 2003, The Chemical Society of Japan Award, in 2005, Japan Society for Environmental Biotechnology Award, in 2008, and Fellow in The Chemical Society of Japan, in 1009. He was selected as a member, Science Council of Japan, since 2005. He received the Medal with Purple Ribbon from Japanese Emperor in 2010. He has been working on extremophiles including hyperthermophiles.
References
- 1).Fujii M., Takagi M., Imanaka T., Aiba S. (1983) Molecular cloning of a thermostable neutral protease gene from Bacillus stearothermophilus in a vector plasmid and its expression in Bacillus stearothermophilus and Bacillus subtilis. J. Bacteriol. 154, 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Fischer F., Zillig W., Stetter K.O., Schreiber G. (1983) Chemolithoautotrophic metabolism of anaerobic extremely thermophilic archaebacteria. Nature 301, 511–513 [DOI] [PubMed] [Google Scholar]
- 3).Stetter K.O. (1999) Extremophiles and their adaptation to hot environments. FEBS Lett. 452, 22–25 [DOI] [PubMed] [Google Scholar]
- 4).Adams M.W., Kelly R.M. (1998) Finding and using hyperthermophilic enzymes. Trends Biotechnol. 16, 329–332 [DOI] [PubMed] [Google Scholar]
- 5).Woese C.R., Kandler O., Wheelis M.L. (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Barns S.M., Delwiche C.F., Palmer J.D., Pace N.R. (1996) Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. U.S.A. 93, 9188–9193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. (1994) Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60, 4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Atomi H., Fukui T., Kanai T., Morikawa M., Imanaka T. (2004) Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1, 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T. (2005) Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Sato T., Fukui T., Atomi H., Imanaka T. (2003) Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Sato T., Fukui T., Atomi H., Imanaka T. (2005) An improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Matsumi R., Manabe K., Fukui T., Atomi H., Imanaka T. (2007) Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host/marker system based on antibiotic resistance. J. Bacteriol. 189, 2683–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Imanaka, T. and Atomi, H. (2002) Innzyme Catalysis in Organic Synthesis (eds. Drauz, K. and Waldmann, E.). Wiley-VCH Verlag, Weinheim, pp. 67–93. [Google Scholar]
- 14).Takagi M., Imanaka T., Aiba S. (1985) Nucleotide sequence and promoter region for the neutral protease gene from Bacillus stearothermophilus. J. Bacteriol. 163, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Imanaka T., Shibazaki M., Takagi M. (1986) A new way of enhancing the thermostability of proteases. Nature 324, 695–697 [DOI] [PubMed] [Google Scholar]
- 16).Rahman R.N.Z.A., Fujiwara S., Nakamura H., Takagi M., Imanaka T. (1998) Ion pairs involved for maintaining a thermastable structure of glutamate dehydrogenase (GDH) from a hyperthermophilic archaeon. Biochem. Biophys. Res. Commun. 248, 920–926 [DOI] [PubMed] [Google Scholar]
- 17).Hashimoto H., Matsumoto T., Nishioka M., Yuasa T., Takeuchi S., Inoue T., Fujiwara S., Takagi M., Imanaka T., Kai Y. (1999) Crystallographic studies on family B DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis strain KOD1. J. Biochem. 125, 983–986 [DOI] [PubMed] [Google Scholar]
- 18).Hashimoto H., Nishioka M., Fujiwara S., Takagi M., Imanaka T., Inoue T., Kai Y. (2001) Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 306, 469–477 [DOI] [PubMed] [Google Scholar]
- 19).Leclere M.M., Nishioka M., Yuasa T., Fujiwara S., Takagi M., Imanaka T. (1998) The O6-methylguanine-DNA methyltransferase from a hyperthermophilic archaeon Pyrococcus sp. KOD1: A thermostable repair enzyme. Mol. Gen. Genet. 258, 69–77 [DOI] [PubMed] [Google Scholar]
- 20).Hashimoto H., Nishioka M., Inoue T., Fujiwara S., Takagi M., Imanaka T., Kai Y. (1998) Crystallization and preliminary X-ray crystallographic analysis of archaeal O6-methyl-guanine-DNA methyltransferase. Acta Crystallogr. D, Biol. Crystallogr. 54, 1395–1396 [DOI] [PubMed] [Google Scholar]
- 21).Hashimoto H., Inoue T., Nishioka M., Fujiwara S., Takagi M., Imanaka T., Kai Y. (1999) Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase. J. Mol. Biol. 292, 707–716 [DOI] [PubMed] [Google Scholar]
- 22).Shiraki K., Nishikori S., Fujiwara S., Hashimoto H., Kai Y., Takagi M., Imanaka T. (2001) Comparative analyses of the conformational stability of a hyperthermophilic protein and its mesophilic counterpart. Eur. J. Biochem. 268, 4144–4150 [DOI] [PubMed] [Google Scholar]
- 23).Imanaka T., Lee S., Takagi M., Fujiwara S. (1995) Aspartyl-tRNA synthetase of the hyperthermophilic archaeon Pyrococcus sp. KOD1 has a chimerical structure of eukaryotic and bacterial enzymes. Gene 164, 153–156 [DOI] [PubMed] [Google Scholar]
- 24).Fujiwara S., Lee S., Haruki M., Kanaya S., Takagi M., Imanaka T. (1996) Unusual enzyme characteristics of aspartyl-tRNA synthetase from hyperthermophilic archaeon Pyrococcus sp. KOD1. FEBS Lett. 394, 66–70 [DOI] [PubMed] [Google Scholar]
- 25).Schmitt E., Moulinier L., Fujiwara S., Imanaka T., Thierry J.C., Moras D. (1998) Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD1; archaeon specificity and catalytic mechanism of adenylate formation. EMBO J. 17, 5227–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Maeda N., Kitano K., Fukui T., Ezaki S., Atomi H., Miki K., Imanaka T. (1999) Ribulose bisphosphate carboxylase/oxygenase from the hyperthermopholic archaeon Pyrococcus kodakaraensis KOD1 is composed solely of large subunits and forms a pentagonal strucuture. J. Mol. Biol. 293, 57–66 [DOI] [PubMed] [Google Scholar]
- 27).Kitano K., Maeda N., Fukui T., Atomi H., Imanaka T., Miki K. (2001) Crystal structure of a novel-type archaeal Rubisco with pentagonal symmetry. Structure 9, 473–481 [DOI] [PubMed] [Google Scholar]
- 28).Takagi M., Kai Y., Imanaka T. (2001) Methylguanine methyltransferase from Thermococcus kodakaraensis KOD1. Methods Enzymol. 334, 239–248 [DOI] [PubMed] [Google Scholar]
- 29).Moore M.H., Gulbis J.M., Dodson E.J., Demple B., Moody P.C. (1994) Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 13, 1495–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Shively J.M., van Keulen G., Meijer W.G. (1998) Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52, 191–230 [DOI] [PubMed] [Google Scholar]
- 31).Watson G.M., Tabita F.R. (1997) Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol. Lett. 146, 13–22 [DOI] [PubMed] [Google Scholar]
- 32).Hartman F.C., Harpel M.R. (1994) Structure, function, regulation, and assembly of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 63, 197–234 [DOI] [PubMed] [Google Scholar]
- 33).Ezaki S., Maeda N., Kishimoto T., Atomi H., Imanaka T. (1999) Presence of a structurally novel type Rubisco in the hyperthermophilic archaeon, Pyrococcus kodakaraensis KOD1. J. Biol. Chem. 274, 5078–5082 [DOI] [PubMed] [Google Scholar]
- 34).Atomi H., Ezaki S., Imanaka T. (2001) Ribulose 1,5-bisphosphate carboxylase/oxygenase from Thermococcus kodakaraensis KOD1. Methods Enzymol. 331, 353–365 [DOI] [PubMed] [Google Scholar]
- 35).Maeda N., Kanai T., Atomi H., Imanaka T. (2002) The unique pentagonal structure of an archaeal Rubisco is essential for its high thermostability. J. Biol. Chem. 277, 31656–31662 [DOI] [PubMed] [Google Scholar]
- 36).Sato T., Atomi H., Imanaka T. (2007) Archaeal Type III Rubiscos function in a pathway for AMP metabolism. Science 315, 1003–1006 [DOI] [PubMed] [Google Scholar]
- 37).Takagi M., Nishioka M., Kakihara H., Kitabayashi M., Inoue H., Kawakami B., Oka M., Imanaka T. (1997) Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 63, 4504–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Yan Z., Fujiwara S., Kohda K., Takagi M., Imanaka T. (1997) In vitro stabilization and in vivo solubilization of foreign proteins by the β-subunit of a chaperonin from the hyperthermophilic archaeon Pyrococcus sp. strain KOD1. Appl. Environ. Microbiol. 63, 785–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Izumi M., Fujiwara S., Takagi M., Kanaya S., Imanaka T. (1999) Isolation and characterization of a second subunit of molecular chaperonin from Pyrococcus kodakaraensis KOD1: Analysis of ATPase deficient mutant. Appl. Environ. Microbiol. 65, 1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Izumi M., Fujiwara S., Takagi M., Fukui K., Imanaka T. (2001) Two kinds of archaeal chaperonin with different temperature dependency from a hyperthermophile. Biochem. Biophys. Res. Commun. 280, 581–587 [DOI] [PubMed] [Google Scholar]
- 41).Rahman R.N.Z.A., Fujiwara S., Takagi M., Kanaya S., Imanaka T. (1997) Effect of heat treatment on proper oligomeric structure formation of thermostable glutamate dehydrogenase from a hyperthermophilic archaeon. Biochem. Biophys. Res. Commun. 241, 646–652 [DOI] [PubMed] [Google Scholar]
- 42).Rahman R.N.Z.A., Fujiwara S., Takagi M., Imanaka T. (1998) Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and comparison of enzyme characteristics of native and recombinant GDHs. Mol. Gen. Genet. 257, 338–347 [DOI] [PubMed] [Google Scholar]
- 43).Higashibata H., Fujiwara S., Takagi M., Imanaka T. (1999) Analysis of DNA compaction profile and intracellular contents of archaeal histones from Pyrococcus kodakaraensis KOD1. Biochem. Biophys. Res. Commun. 258, 416–424 [DOI] [PubMed] [Google Scholar]
- 44).Higashibata H., Fujiwara S., Ezaki S., Takagi M., Fukui K., Imanaka T. (2000) Effect of polyamines on histone-induced DNA compaction of hyperthermophilic archaea. J. Biosci. Bioeng. 89, 103–106 [DOI] [PubMed] [Google Scholar]
- 45).Atomi H., Matsumi R., Imanaka T. (2004) Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 186, 4829–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).McCloskey J.A., Graham D.E., Zhou S., Crain P.F., Ibba M., Konisky J., Soll D., Olsen G.J. (2001) Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 29, 4699–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Kowalak J.A., Dalluge J.J., McCloskey J.A., Stetter K.O. (1994) The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33, 7869–7876 [DOI] [PubMed] [Google Scholar]
- 48).Oshima T., Moriya T., Terui Y. (2011) Identification, chemical synthesis, and biological functions of unusual polyamines produced by extreme thermophiles. Methods Mol. Biol. 720, 81–111 [DOI] [PubMed] [Google Scholar]
- 49).Morimoto N., Fukuda W., Nakajima N., Masuda T., Terui Y., Kanai T., Oshima T., Imanaka T., Fujiwara S. (2010) Dual biosynthesis pathway for longer-chain polyamines in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 192, 4991–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Albers S.V., van de Vossenberg J.L., Driessen A.J., Konings W.N. (2000) Adaptations of the archaeal cell membrane to heat stress. Front. Biosci. 5, D813–D820 [DOI] [PubMed] [Google Scholar]
- 51).Hanford M.J., Peeples T.L. (2002) Archaeal tetraether lipids: unique structures and applications. Appl. Biochem. Biotechnol. 97, 45–62 [DOI] [PubMed] [Google Scholar]
- 52).Carballeira N.M., Reyes M., Sostre A., Huang H., Verhagen M.F., Adams M.W. (1997) Unusual fatty acid compositions of the hyperthermophilic archaeon Pyrococcus furiosus and the bacterium Thermotoga maritime. J. Bacteriol. 179, 2766–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Matsuno Y., Sugai A., Higashibata H., Fukuda W., Ueda K., Uda I., Sato I., Itoh T., Imanaka T., Fujiwara S. (2009) Effect of growth temperature and growth phase on the lipid composition of the archaeal membrane from Thermococcus kodakaraensis. Biosci. Biotechnol. Biochem. 73, 104–108 [DOI] [PubMed] [Google Scholar]
- 54).Borges N., Matsumi R., Imanaka T., Atomi H., Santos H. (2010) Thermococcus kodakaraensis mutants deficient in di-myo-inositol phosphate use aspartate to cope with heat stress. J. Bacteriol. 192, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Tang X.-F., Ezaki S., Atomi H., Imanaka T. (2000) Biochemical analysis of a thermostable tryptophan synthase from a hyperthermophilic archaeon. Eur. J. Biochem. 267, 6369–6377 [DOI] [PubMed] [Google Scholar]
- 56).Say R.F., Fuchs G. (2010) Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464, 1077–1081 [DOI] [PubMed] [Google Scholar]
- 57).Kudou M., Shiraki K., Fujiwara S., Imanaka T., Takagi M. (2003) Prevention of thermal inactivation and aggregation of lysozyme by polyamines. Eur. J. Biochem. 270, 4547–4554 [DOI] [PubMed] [Google Scholar]