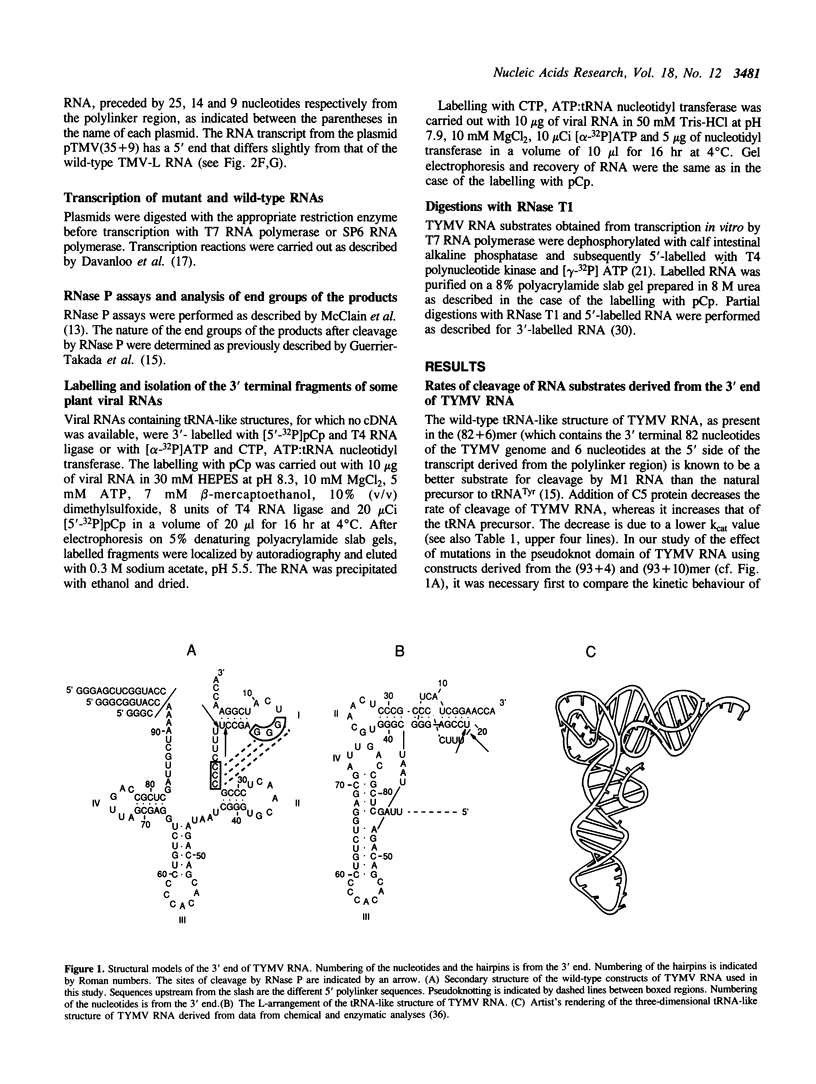
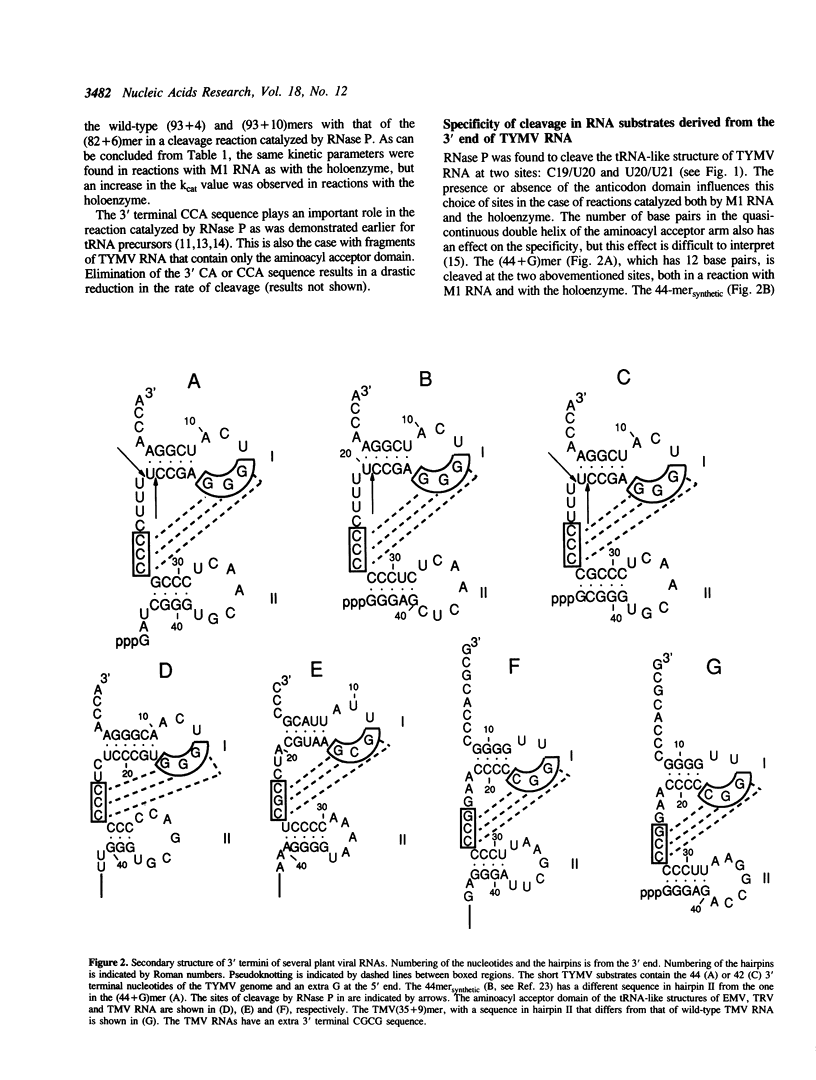
Abstract
In a previous study it was shown that RNase P from E. coli cleaves the tRNA-like structure of turnip yellow mosaic virus (TYMV) RNA in vitro (Guerrier-Takada et al. (1988) Cell, 53, 267-272). Cleavage takes place at the 3' side of the loop that crosses the deep groove of the pseudoknot structure present in the aminoacyl acceptor domain. In the present study fragments of TYMV RNA with mutations in the pseudoknot, generated by transcription in vitro, were tested for susceptibility to cleavage by RNase P. Changes in the specificity with respect to the site of cleavage and decreases in the rate of cleavage were observed with most of these substrates. The behaviour of various mutants in the reaction catalyzed by RNase P is in agreement with the present model of the TYMV RNA pseudoknot (Dumas et al. (1987), J. Biomol. Struct. Dyn. 263, 652-657). Base substitutions in the loop that crosses the shallow groove of the pseudoknot structure resulted, however, in an unexpected decrease in the rate of cleavage, probably due to conformational changes in the substrates. Studies on other tRNA-like structures revealed an important role in the reaction with RNase P for both the nucleotide at the 3' side of the loop that spans the deep groove and the nucleotide at position 4, which correspond to positions--1 and 73, respectively, in tRNA precursors.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angenent G. C., Posthumus E., Bol J. F. Biological activity of transcripts synthesized in vitro from full-length and mutated DNA copies of tobacco rattle virus RNA 2. Virology. 1989 Nov;173(1):68–76. doi: 10.1016/0042-6822(89)90222-5. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard U., Söll D. The unusually long amino acid acceptor stem of Escherichia coli selenocysteine tRNA results from abnormal cleavage by RNase P. Nucleic Acids Res. 1988 Dec 23;16(24):11617–11624. doi: 10.1093/nar/16.24.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard U., Willis I., Söll D. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J Biol Chem. 1988 Feb 15;263(5):2447–2451. [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T. W., Florentz C., Giege R. Valylation of tRNA-like transcripts from cloned cDNA of turnip yellow mosaic virus RNA demonstrate that the L-shaped region at the 3' end of the viral RNA is not sufficient for optimal aminoacylation. Biochimie. 1988 Dec;70(12):1719–1727. doi: 10.1016/0300-9084(88)90030-2. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Hall T. C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988 May 5;201(1):31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- Dumas P., Moras D., Florentz C., Giegé R., Verlaan P., Van Belkum A., Pleij C. W. 3-D graphics modelling of the tRNA-like 3'-end of turnip yellow mosaic virus RNA: structural and functional implications. J Biomol Struct Dyn. 1987 Apr;4(5):707–728. doi: 10.1080/07391102.1987.10507674. [DOI] [PubMed] [Google Scholar]
- Furdon P. J., Guerrier-Takada C., Altman S. A G43 to U43 mutation in E. coli tRNAtyrsu3+ which affects processing by RNase P. Nucleic Acids Res. 1983 Mar 11;11(5):1491–1505. doi: 10.1093/nar/11.5.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Vold B. S., Morch M. D., Joshi R. L., Haenni A. L. Ionic conditions for the cleavage of the tRNA-like structure of turnip yellow mosaic virus by the catalytic RNA of RNase P. J Biol Chem. 1988 Aug 25;263(24):11617–11620. [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Structural requirements for processing of synthetic tRNAHis precursors by the catalytic RNA component of RNase P. J Biol Chem. 1988 Jan 15;263(2):652–657. [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984 Jan 20;223(4633):285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. M1 RNA with large terminal deletions retains its catalytic activity. Cell. 1986 Apr 25;45(2):177–183. doi: 10.1016/0092-8674(86)90381-8. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Structure in solution of M1 RNA, the catalytic subunit of ribonuclease P from Escherichia coli. Biochemistry. 1984 Dec 18;23(26):6327–6334. doi: 10.1021/bi00321a006. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., McClain W. H., Altman S. Cleavage of tRNA precursors by the RNA subunit of E. coli ribonuclease P (M1 RNA) is influenced by 3'-proximal CCA in the substrates. Cell. 1984 Aug;38(1):219–224. doi: 10.1016/0092-8674(84)90543-9. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., van Belkum A., Pleij C. W., Altman S. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988 Apr 22;53(2):267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Joshi R. L., Joshi S., Chapeville F., Haenni A. L. tRNA-like structures of plant viral RNAs: conformational requirements for adenylation and aminoacylation. EMBO J. 1983;2(7):1123–1127. doi: 10.1002/j.1460-2075.1983.tb01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumelsky N., Altman S. Selection and characterization of randomly produced mutants in the gene coding for M1 RNA. J Mol Biol. 1988 Aug 5;202(3):443–454. doi: 10.1016/0022-2836(88)90277-x. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- McPheeters D. S., Stormo G. D., Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J Mol Biol. 1988 Jun 5;201(3):517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Reich C., James B. D., Olsen G. J., Pace B., Waugh D. S. Structure and catalytic function in ribonuclease P. Cold Spring Harb Symp Quant Biol. 1987;52:239–248. doi: 10.1101/sqb.1987.052.01.029. [DOI] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly R. M., RajBhandary U. L. A single mutation in loop IV of Escherichia coli SuIII tRNA blocks processing at both 5'- and 3'-ends of the precursor tRNA. J Biol Chem. 1986 Feb 25;261(6):2928–2935. [PubMed] [Google Scholar]
- Rether B., Bonnet J., Ebel J. P. Studies on tRNA nucleotidyltransferase from baker's yeast. 1. Purification of the enzyme. Protection against thermal inactivation and inhibition by several substrates. Eur J Biochem. 1974 Dec 16;50(1):281–288. doi: 10.1111/j.1432-1033.1974.tb03896.x. [DOI] [PubMed] [Google Scholar]
- Rietveld K., Linschooten K., Pleij C. W., Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984 Nov;3(11):2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Pleij C. W., Bosch L. Three-dimensional models of the tRNA-like 3' termini of some plant viral RNAs. EMBO J. 1983;2(7):1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H., Shimura Y. Functional domains of the RNA component of ribonuclease P revealed by chemical probing of mutant RNAs. EMBO J. 1988 Dec 1;7(12):3817–3821. doi: 10.1002/j.1460-2075.1988.tb03266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque A., Arnez J., Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J Mol Biol. 1988 Aug 20;202(4):835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Belkum A., Cornelissen B., Linthorst H., Bol J., Pley C., Bosch L. tRNA-like properties of tobacco rattle virus RNA. Nucleic Acids Res. 1987 Apr 10;15(7):2837–2850. doi: 10.1093/nar/15.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Verlaan P., Kun J. B., Pleij C., Bosch L. Temperature dependent chemical and enzymatic probing of the tRNA-like structure of TYMV RNA. Nucleic Acids Res. 1988 Mar 25;16(5):1931–1950. doi: 10.1093/nar/16.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Wiersema P. J., Joordens J., Pleij C., Hilbers C. W., Bosch L. Biochemical and biophysical analysis of pseudoknot-containing RNA fragments. Melting studies and NMR spectroscopy. Eur J Biochem. 1989 Aug 15;183(3):591–601. doi: 10.1111/j.1432-1033.1989.tb21088.x. [DOI] [PubMed] [Google Scholar]