Background: Sister chromatid arm cohesion is resolved at prophase/prometaphase, but its mechanism remains unclear.
Results: The ST159P motif on Sororin is phosphorylated by Cdk1/cyclin B, and its subsequent interaction with the polo box domain of Plk1 facilitates the resolution of arm cohesion.
Conclusion: Sororin mediates Plk1-mediated phosphorylation of SA2 to resolve sister chromatid arm cohesion.
Significance: The mechanism of chromatid resolution is important to understand chromosomal missegregation often seen in cancer.
Keywords: CDK (Cyclin-dependent Kinase), Chromosomes, Cohesin, Mitosis, Phosphorylation, Arm Cohesion, Plk1, SA2, Sororin
Abstract
Unlike in budding yeast, sister chromatid cohesion in vertebrate cells is resolved in two steps: cohesin complexes are removed from sister chromatid arms during prophase via phosphorylation, whereas centromeric cohesins are removed at anaphase by Separase. Phosphorylation of cohesin subunit SA2 by polo-like kinase 1 (Plk1) is required for the removal of cohesins at prophase, but how Plk1 is recruited to phosphorylate SA2 during prophase is currently not known. Here we report that Sororin, a cohesin-interacting protein essential for sister chromatid cohesion, plays a novel role in the resolution of sister chromatid arms by direct interaction with Plk1. We identified an evolutionarily conserved motif (ST159P) on Sororin, which was phosphorylated by Cdk1/cyclin B and bound to the polo box domain of Plk1. Mutating Thr159 into alanine prevented the interaction of Plk1 and Sororin and inhibited the resolution of chromosomal arm cohesion. We propose that Sororin is phosphorylated by Cdk1/cyclin B at prophase and acts as a docking protein to bring Plk1 into proximity with SA2, resulting in the phosphorylation of SA2 and the removal of cohesin complexes from chromosomal arms.
Introduction
The accurate separation of sister chromatids into two daughter cells relies on an evolutionarily conserved protein complex called cohesin. Cohesion along the length of the sister chromatid is formed by cohesins during DNA replication in S phase, and sister chromatids are kept together until the transition from metaphase to anaphase during mitosis. After the degradation of the inhibitory chaperones securin and cyclin B, Separase, an endopeptidase that cleaves the cohesin subunit Scc1/Mcd1 (Rad21 in humans), is activated (1–6). In Saccharomyces cerevisiae all cohesin complexes dissociate from the chromosomes simultaneously when the cohesin subunit Scc1/Mcd1 is cleaved by Separase before the sister chromatids are separated into daughter cells. In metazoa, however, cohesins are removed from the chromatids in two steps (7). At prophase, after Plk13 phosphorylates SA2, the bulk of the cohesin complexes dissociates from the chromosome arms (8–10). This step is independent of Separase activation. However, it relies on Plk1 and Aurora B to phosphorylate cohesin SA2 (9–11) and Wapl to remove the modified cohesins (12–14). The centromeric cohesins and leftover cohesins along the chromosome arms, which are protected by Shugoshin, which prevents phosphorylation of SA2 by Plk1 (15, 16), are removed by Separase at the transition from metaphase to anaphase (17, 18). Although it has been shown that Shugoshin prevents the phosphorylation of SA2 by Plk1, the molecular mechanisms of Plk1-mediated phosphorylation of SA2 remain to be determined.
In addition to the cohesin complex, a number of other cohesin-associated proteins, including Pds5A, Pds5B, Wapl, and Sororin, play important roles in sister chromatid cohesion (13, 19–22). Sororin is a nuclear protein conserved in vertebrates. A Sororin ortholog has recently been reported in Drosophila (14), but no apparent ortholog has been found in S. cerevisiae. Sororin has a “KEN box” that is ubiquitinated by Cdh1-activated anaphase-promoting complex (APCCdh1) and degraded during the G1 phase of the cell cycle (22). Sororin interacts with the cohesin complex and is required for sister chromatid cohesion (21, 22). Co-expression analysis indicates that the Sororin gene CDCA5 is co-regulated with known cell cycle genes, such as CDK1, CYCLIN B, BUB 1, and CDC20 (23). Immunoblotting data suggest that Sororin is phosphorylated during mitosis (22), and high throughput protein phosphorylation analysis confirms Sororin phosphorylation (24–27). Recent studies indicate that Sororin is phosphorylated by Cdk1/cyclin B (28) and that mitotic phosphorylation of Sororin impairs its ability to inhibit the binding of cohesin release factor Wap1 and cohesin-interacting protein Pds5 (14).
Here we provide evidence that Sororin interacts with Plk1 and functions in the resolution of sister chromatid arm cohesion. We used cell biology and biochemical approaches to study the phosphorylation of Sororin, the interaction of Sororin and Plk1, and the resolution of arm cohesion. We identified and confirmed three Cdk1/cyclin B phosphorylation sites in an in vitro kinase assay and mass spectrometry. We also found that Plk1 could not phosphorylate Sororin directly but could do so if Sororin was phosphorylated a priori by Ckd1/cyclin B. Moreover, we identified an evolutionarily conserved motif on Sororin, ST159P, which was phosphorylated by Cdk1/cyclin B and interacted with Plk1. Mutation at ST159P not only prevented Plk1 binding but also inhibited the resolution of arm cohesion. Based on these observations, we propose a model describing the role of Sororin and Plk1 in the removal of cohesins from chromosomal arms.
EXPERIMENTAL PROCEDURES
Antibodies
The sources of the antibodies used in this study were as follows: rabbit polyclonal antibodies FLAG (Sigma), human Rad21 (29), and GFP (Abcam, Cambridge, MA); goat polyclonal antibody SA2 (Novus Biologicals, Littleton, CO); and mouse monoclonal antibodies: FLAG (Sigma), Myc (Calbiochem), Cdk1 (Millipore, Billerica, MA), and Plk1 (Zymed Laboratories Inc.). Rabbit anti-Sororin polyclonal antibody was developed using peptide 212EKQKRKKKKMPEILK227.
Stably Transfected HeLa Cell Lines Expressing Tagged Sororin
To develop stable cell lines expressing GFP-tagged wild type or mutant Sororin, Sororin cDNA was cloned into pCruz GFP vector (Santa Cruz Biotechnology, Santa Cruz, CA) and transfected into HeLa cells. Individual clones were selected and maintained in DMEM containing 10% FBS, 300 μg/ml G418, 100 units/ml penicillin, and 100 μg/ml streptomycin. To develop a HeLa Myc-Sororin stably transfected Tet-On cell line, Sororin cDNA was cloned into pTRE2hyg2-Myc vector (Clontech) and transfected into HeLa Tet-On cells. Positive clones were selected and cultured in DMEM with 10% FBS, 300 μg/ml G418, 100 μg/ml hygromycin, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Mutation and Cloning of Sororin
Mutation of serine and threonine residues into alanine was performed using site-specific PCR. Sororin cDNA was cloned into EcoRI and EcoRV cloning sites of pFLAG-CMV2 for expression in mammalian cells or cloned into BamHI and XhoI sites of pET21a(+) vector for expression as a His6 epitope-tagged protein in bacteria.
Recombinant Protein
Sororin cDNA was cloned into pGEX4T3 or pET21a(+) vector (Novagen, Madison, WI) with GST or His6 tagged to Sororin N terminus or C terminus, respectively. Recombinant Sororin was expressed in Escherichia coli BL21-CodonPlus. GST-Sororin was purified with glutathione-Sepharose 4B and eluted with 10 mm glutathione in 50 mm Tris buffer, pH 8.0. Sororin-His was purified with nickel-nitrilotriacetic acid beads and eluted with 250 mm imidazole.
Cdk1 Phosphorylation Assay
The Cdk1/cyclin B phosphorylation assay was performed by mixing 1 μl of 2 μg/μl substrate, 4 μl of 5× reaction buffer (100 mm Tris-HCl, pH 7.5, 500 mm NaCl, 60 mm MgCl2, 5 mm DTT), 1 μl of 0.25 mm ATP, 1 μl of [γ-32P]ATP (4 μCi; PerkinElmer Life Sciences BLU002A), 2 μl of 10 ng/μl Cdk1 kinase (Millipore), and H2O up to final volume of 20 μl. The mixture was incubated for 30 min at 30 °C with constant agitation. The sample was mixed with 10 μl of 3× SDS sample buffer and resolved with SDS-PAGE. The gel was dried and placed on a phosphorscreen imager cassette overnight. The phosphorylated protein bands were visualized by a GE Healthcare Storm 860 phosphorimaging scanner.
Plk1 Kinase Assay
The substrate, reaction buffer, ATP, and [γ-32P]ATP were mixed as in the Cdk1 kinase assay described above. One μl of 125 ng/μl Plk1 kinase (Millipore) was used, and H2O was added up to a final volume of 20 μl. The mixture was incubated for 30 min at 30 °C. To minimize the interference of Cdk1/cyclin B in the sequential phosphorylation of Sororin by Cdk1/cyclin B followed by Plk1, Cdk1/cyclin B was depleted using Cdk1 mAb. In addition, Cdk1 inhibitor CGP74514A (Calbiochem) was added to the reaction mixture at a final concentration of 1 μm.
siRNA and Transfection
Sororin siRNA duplex targeting the coding region (Dharmacon, Lafayette, CO) or 3′-UTR (Qiagen, Valencia, CA) was transfected into cells using DharmaFECTTM 1 (Dharmacon) according to the protocol provided by the manufacturer. Silencer® Negative Control siRNA 1 (Applied Biosystems, Foster City, CA) was used as the negative control.
Real Time RT-PCR
Two days after siRNA transfection, total RNA in HeLa cells was extracted using an RNeasy Mini kit (Qiagen). cDNA was synthesized using oligo(dT)12–18 primer (Invitrogen) and SuperScript II reverse transcriptase (Invitrogen). Primers for the gene were designed close to the 3′-coding region of Cdca5: 5′-AGTCTCGCCAGTGGTGTGCT-3′ (forward) and 5′-TTCAACCAGGAGATCAAACTGC-3′ (reverse). SYBR Green quantitative RT-PCR was performed with the Eppendorf Mastercycler EP Realplex System. The ΔΔCt method was used for data analysis, and GAPDH was used as the internal control for normalization. The primer sequences for GAPDH have been described previously (30).
Plasmid Transfection
The calcium phosphate method (31) was used to transfect appropriate plasmids into 293T cells. EffecteneTM transfection reagent from Qiagen was used to transfect HeLa cells.
Metaphase Chromosome Spread
Cells were trypsinized and spun down at 100 × g for 6 min. The cells were mixed with 10 ml of prewarmed (37 °C) hypotonic solution (0.075 m KCl), which was added dropwise with gentle agitation. The samples were incubated at 37 °C for 15 min. One milliliter of fixative (freshly prepared 3:1 methanol:acetic acid) was added to each sample followed by centrifugation at 100 × g for 6 min. Ten milliliters of fixative was added to the cells, which were incubated at room temperature for at least 30 min. The fixative was changed twice, and the cells were resuspended in 0.5 ml of fixative. Two to three drops of cell suspension were dropped from a height of 12 inches onto angled slides. After air drying, the slides were stained with Giemsa solution for 5 min and rinsed in running water for 10 min.
Protein Isolation, Immunoprecipitation, and Immunoblotting
Unless otherwise noted, cells were harvested 40–48 h after transfection, and lysates were made as described previously (32). The protocol for immunoprecipitation (IP) and immunoblot analysis has been previously described (32).
Immunofluorescence Microscopy
HeLa cells were grown on four-chamber slides. The cells were fixed with 4% paraformaldehyde in PEM buffer (80 mm potassium PIPES, pH 6.8, 5 mm EGTA, 2 mm MgCl2) for 15 min followed by washing three times with PEM buffer. Next, the samples were incubated in 50 mm NH4Cl, PEM buffer for 5 min; washed with PEM buffer; and incubated in 1% Triton X-100, PEM buffer for 10 min followed by washing three times with PEM buffer. Cells were incubated in 1% BSA, PEM buffer for 1 h. The primary and secondary antibodies were diluted in 1% BSA, PEM buffer and applied to slides followed by washing three times with 0.1% Tween 20, PEM buffer. The cells were mounted with Vectashield® mounting medium containing DAPI (Vector Laboratories, Burlingame, CA). Images were obtained with a microscope (E800, Nikon) equipped with Quips imaging software (Applied Imaging) and a 100×/1.4 objective lens (Nikon). For immunostaining of chromosome spread, mitotic cells were centrifuged onto slides using Cytospin. The samples were extracted with 0.1% Triton X-100 for 5 min. The rest steps were the same as described above (for details, also see Ref. 32).
Mass Spectrometry
Recombinant Sororin was phosphorylated by Cdk1/cyclin in vitro. The protein was resolved with SDS-PAGE and stained with Coomassie Blue. The Sororin gel band was excised and sequentially washed with 25 mm ammonium bicarbonate, acetonitrile, and 10 mm dithiothreitol at 60 °C and 50 mm iodoacetamide at room temperature. Trypsin and elastase digestion was performed at 37 °C for 4 h. After quenching with formic acid, the supernatant was analyzed. Each gel digest was analyzed by nano-LC/MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Orbitrap Velos Pro by MS Bioworks, LLC (Ann Arbor, MI). Peptides were loaded on a trapping column and eluted over a 75-μm analytical column at 300 nl/min; both columns were packed with Jupiter Proteo resin (Phenomenex). The mass spectrometer was operated in data-dependent mode with MS performed in the Orbitrap at 60,000 full-width half-maximal resolution and MS/MS performed in Thermo Scientific LTQ Orbitrap mass spectrometer. The 15 most abundant ions were selected for MS/MS. Data were searched using a local copy of Mascot with the Swiss-Prot Human Database. Mascot DAT files were parsed into Scaffold software for validation and filtering. A nonredundant list for each sample was created. Data were filtered using a minimum protein value of 99%, a minimum peptide value of 50% (Prophet scores), and at least two unique peptides per protein.
RESULTS
Sororin Is Post-translationally Modified with Phosphorylation
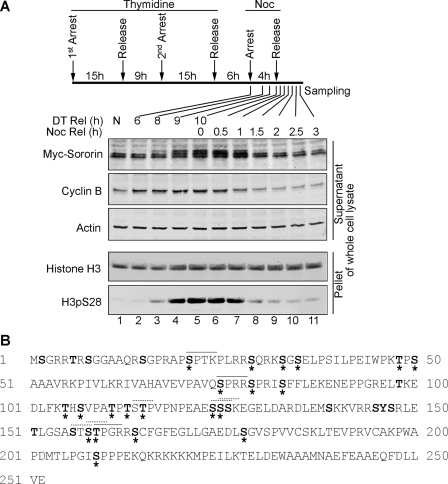
During the cell cycle, the level of Sororin protein not only oscillates but also undergoes modifications. This is evident from its higher molecular weight shift and slower migration in mitosis (M) phase (Fig. 1A, lanes 4–7). This shift is likely due to phosphorylation. There are 33 putative phosphorylation sites in Sororin, including 23 serines, 9 threonines, and 1 tyrosine (Fig. 1B). All the predicted phosphorylation sites are in the N terminus and middle part of Sororin. Among the 33 putative phosphorylation sites, 21 sites have been confirmed to be phosphorylated by high throughput mass spectrometry analysis (Fig. 1B and Table 1) (22, 24–27, 33, 34). Although Sororin is phosphorylated by Cdk1 (28), the exact phosphorylation sites and the physiological significance of phosphorylation remain to be characterized.
FIGURE 1.
Sororin protein is post-translationally modified. A, Sororin protein profile during mitotic stages of the cell cycle. HeLa Myc-Sororin stably transfected Tet-On cells were induced with doxycycline. The cells were synchronized using double thymidine blocks. Six hours after release from the second thymidine block, nocodazole (Noc) was applied. Four hours later, cells were released from nocodazole arrest. Cell samples were harvested at 6, 8, 9, and 10 h after double thymidine release (DT Rel) and 0, 0.5, 1, 1.5 2, 2.5, and 3 h after nocodazole release (Noc Rel). Non-synchronized (NS) cells were used as a control. Whole cell lysate was prepared. The supernatant and pellet were immunoblotted. Cyclin B indicates the G2/M phase, and the H3pS28 level represents M phase. B, phosphorylation of Sororin was predicted using NetPhos and Phosphonet. The letters in bold are predicted to be phosphorylated by kinases, and the phosphorylation sites that have been confirmed are labeled with *. The solid line (—) over the letters shows the putative Cdk1/cyclin B phosphorylation site, whereas the dashed line (· · ·) indicates the putative PBD binding motifs.
TABLE 1.
Putative and confirmed phosphorylation sites in Sororin
| Phosphosites | Predicted by NetPhos | Predicted by Phosphonet | Peptide sequence | Experimentally confirmed |
|---|---|---|---|---|
| 1 | Ser2 | MSGRRTRSG | ||
| 2 | Thr6 | MSGRRTRSGGAAQ | ||
| 3 | Ser8 | MSGRRTRSGGAAQRS | ||
| 4 | Ser15 | SGGAAQRSGPRAPSP | ||
| 5 | Ser21 | Ser21 | RSGPRAPSPTKPLRR | This study, Refs. 26, 33, 44 |
| 6 | Ser29 | Ser29 | PTKPLRRSQRKSGSE | 26 |
| 7 | Ser33 | Ser33 | LRRSQRKSGSELPSI | 26, 34 |
| 8 | Ser35 | RSQRKSGSELPSILP | 26 | |
| 9 | Thr48 | Thr48 | LPEIWPKTPSAAAVR | 25 |
| 10 | Ser50 | EIWPKTPSAAAVRKP | 25 | |
| 11 | Ser75 | Ser75 | VEVPAVQSPRRSPRI | This study, Refs. 24, 26, 27, 34 |
| 12 | Ser79 | Ser79 | AVQSPRRSPRISFFL | 24, 26, 44 |
| 13 | Ser83 | Ser83 | PRRSPRISFFLEKEN | 26 |
| 14 | Thr98 | EPPGRELTKEDLFKT | ||
| 15 | Thr105 | TKEDLFKTHSVPATP | 26 | |
| 16 | Ser107 | EDLFKTHSVPATPTS | 26 | |
| 17 | Thr111 | Thr111 | KTHSVPATPTSTPVP | 26 |
| 18 | Thr113 | Thr113 | HSVPATPTSTPVPNP | 26 |
| 19 | Thr115 | Thr115 | VPATPTSTPVPNPEA | 26 |
| 20 | Ser124 | VPNPEAESSSKEGEL | 26 | |
| 21 | Ser125 | Ser125 | PNPEAESSSKEGELD | 26 |
| 22 | Ser126 | NPEAESSSKEGELDA | ||
| 23 | Ser139 | DARDLEMSKKVRRSY | ||
| 24 | Ser145 | Ser145 | MSKKVRRSYSRLETL | |
| 25 | Tyr146 | SKKVRRSYSRLETLG | ||
| 26 | Ser147 | Ser147 | KKVRRSYSRLETLGS | |
| 27 | Thr151 | RSYSRLETLGSASTS | ||
| 28 | Ser156 | LETLGSASTSTPGRR | ||
| 29 | Ser158 | Ser158 | TLGSASTSTPGRRSC | 34 |
| 30 | Thr159 | Thr159 | LGSASTSTPGRRSCF | This study, Ref. 27 |
| 31 | Ser164 | Ser164 | TSTPGRRSCFGFEGL | 45 |
| 32 | Ser178 | Ser178 | LLGAEDLSGVSPVVC | 45 |
| 33 | Ser209 | Ser209 | DMTLPGISPPPEKQK | 34, 44 |
Sororin Is Phosphorylated by Ckd1/Cyclin B
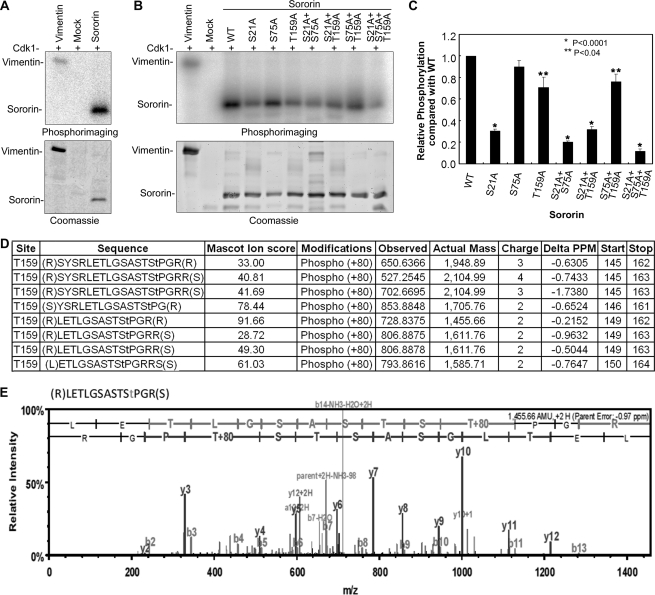
Cdk1/cyclin B phosphorylates Ser or Thr residues in the consensus sequence (S/T)PX(K/R). We examined the Sororin protein primary structure and identified three putative Cdk1/cyclin B phosphorylation sites, S21PTK, S75PRR, and T159PGR (Fig. 1B). To confirm whether Sororin is phosphorylated by Cdk1/cyclin B, we performed an in vitro kinase assay using recombinant human Sororin with a C-terminal His6 tag. In this assay, Cdk1/cyclin B phosphorylated vimentin (the positive control) and Sororin (Fig. 2A). To confirm the putative Cdk1/cyclin B phosphorylation sites, seven Sororin mutants, including three single mutations (S21A, S75A, and T159A), three double mutations (S21A/S75A, S21A/T159A, and S75A/T159A), and one triple mutation (S21A/S75A/T159A) were constructed by replacing Ser21, Ser75, and Thr159 residues with alanine. In vitro Cdk1 kinase assays indicated that, except for Sororin S75A mutant, phosphorylation of the other six Sororin mutants was significantly reduced (p < 0.001 and p < 0.04, respectively) in comparison with wild type (WT) (Fig. 2, B and C). The data suggested that Ser21 and Thr159 are phosphorylated by Cdk1/cyclin B with Ser75 phosphorylated to a lesser degree. The major Cdk1/cyclin B phosphorylation site on Sororin is Ser21, which accounted for 70% of the total Sororin phosphorylation, whereas Thr159 accounted for 20% (Fig. 2C). We conclude that Ser21 and Thr159 are Cdk1/cyclin B phosphorylation sites.
FIGURE 2.
Sororin is phosphorylated by Ckd1/cyclin B. A, recombinant Sororin was phosphorylated by Cdk1/cyclin B in vitro. Vimentin was used as a positive control, and Mock (no Sororin) was a negative control. After phosphorylation, the samples were resolved using SDS-PAGE. The phosphorylation bands with 32P signals were visualized using phosphorimaging (upper panel), whereas protein bands were detected with Coomassie staining (lower panel) of the same gel. B, Sororin WT and mutants with single, double, and triple mutations of putative Cdk1/cyclin B phosphorylation sites (S21A, T75A, and T159A) were used for in vitro Cdk1/cyclin B kinase assays. Vimentin and Mock served as positive and negative controls, respectively. The upper panel shows the phosphorimage, and the lower panel shows the Coomassie staining of the same gel. C, densitometric quantification of the phosphorylation shown in B. The ratio of the density of the phosphorylated band over that of the Coomassie-stained band was used to calculate the relative phosphorylation of Sororin mutant over WT. The relative phosphorylation of Sororin WT and mutants was compared using Student's t test. The asterisks (*) indicate that the phosphorylation of a mutant is significantly different from that of WT (*, p < 0.0001; **, p < 0.04). Data are the means ± S.E. from six experiments. D and E, validation of Thr159 phosphorylation status by mass spectrometry analysis of recombinant Sororin phosphorylated in vitro by Cdk1/cyclinB. The peptides containing phosphorylated Thr159 identified and manually validated are shown in D. A sample mass spectrum of a peptide containing phosphorylated Thr159 is shown in E.
To confirm the Cdk1/cyclin B phosphorylation sites on Sororin, we analyzed the in vitro phosphorylation of Sororin by Cdk1/cyclin B using mass spectrometry analysis. Mass spectrometry confirmed that Ser21, Ser75, and Thr159 were phosphorylated. The data confirming phosphorylation of Thr159 are shown in Fig. 2, D and E. Based on these studies, we conclude that Sororin protein is phosphorylated by Cdk1/cyclin B at Ser21, Ser75, and Thr159.
Phosphorylation of Sororin by Plk1
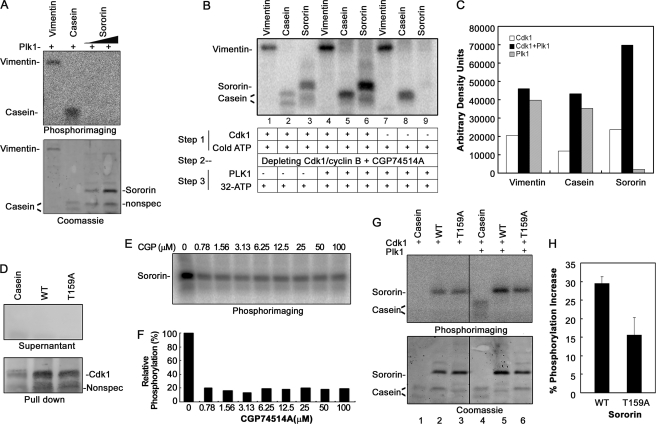
Further bioinformatics analysis of Sororin protein sequence revealed that several stretches of amino acids had sequence similarity to the consensus Plk1 phosphorylation site ((D/E)X(S/T)ΦX(D/E) where Φ is a hydrophobic amino acid). To investigate whether Sororin can be phosphorylated by Plk1, we performed an in vitro Plk1 kinase assay. In this assay, recombinant Sororin protein could not be phosphorylated by Plk1, whereas the two positive controls, vimentin and casein, were phosphorylated as expected (Fig. 3A).
FIGURE 3.
Phosphorylation of Sororin by Cdk1 is prerequisite for Plk1 to phosphorylate Sororin.
A, Plk1 cannot phosphorylate Sororin in an in vitro kinase assay. The gel was stained with Coomassie Blue after it was visualized using phosphorimaging. Vimentin and casein were used as positive controls. B, Sororin can be phosphorylated by Plk1 only after it is phosphorylated by Cdk1/cyclin B. Sororin was first phosphorylated by Cdk1/cyclin B with cold ATP (Step 1), and then Cdk1/cyclin B was depleted by anti-cyclin B mAb and inhibited by Cdk1 inhibitor CGP74514A (1 μm) (Step 2) before Plk1 and [32P]ATP were added to phosphorylate Sororin (Step 3). Lanes 1–3 represent Cdk1/cyclin B phosphorylation alone, lanes 4–6 represent additional phosphorylation of Sororin by Plk1 after Sororin was phosphorylated by Cdk1/cyclin B, and lanes 7–9 represent phosphorylation of Sororin by Plk1 alone. C, bar graph showing the densitometric quantification of the phosphorylation by Cdk1/cyclin B alone (□), Cdk1 plus Plk1 (■), or Plk1 alone ( ), which correspond to lanes 1–3, lanes 4–6, and lanes 7–9 in B, respectively. D, efficiency of Cdk1/cyclin B immunodepletion. Cdk1/cyclin B was depleted using anti-cyclin B mAb after Sororin was phosphorylated with Cdk1/cyclin B and before Plk1 was added to the reaction mixture. To check the efficiency of depletion, the reaction mixture after Cdk1/cyclin B depletion (Supernatant) and Cdk1/cyclin B-bound agarose beads (Pull down) were immunoblotted with anti-Cdk1 mAb. Nonspec, nonspecific band. E, efficiency of CGP74514A (CGP) in inhibiting Cdk1/cyclin B activity. Various concentrations of CGP74514A were added to the reaction mixture of the in vitro assay of Sororin phosphorylation by Cdk1/cyclin B. After SDS-PAGE, the phosphorylation signal was detected using phosphorimaging. F, densitometric quantification of Sororin phosphorylation shown in E. G, phosphorylation is reduced in Sororin mutant T159A. Sororin WT and mutant (T159A) were sequentially phosphorylated by Ckd1/cyclin B and Plk1 as described in B above. 32P phosphorimaging (upper panel) and Coomassie staining (lower panel) of the same gel are shown. Lanes 1–3, Cdk1/cyclin B only; lanes 4–6, Cdk1/cyclin B and Plk1. H, densitometric quantification of Sororin WT and T159A mutant phosphorylated by Cdk1/cyclin B (Pcdk1) or Cdk1/cyclin B plus Plk1 (Pcdk1+plk1), which was normalized with the amount of protein loaded. Changes in phosphorylation were calculated using the equation (Pcdk1+plk1 − Pcdk1)/Pcdk1+plk1. Data are the means ± S.E. from three experiments.
), which correspond to lanes 1–3, lanes 4–6, and lanes 7–9 in B, respectively. D, efficiency of Cdk1/cyclin B immunodepletion. Cdk1/cyclin B was depleted using anti-cyclin B mAb after Sororin was phosphorylated with Cdk1/cyclin B and before Plk1 was added to the reaction mixture. To check the efficiency of depletion, the reaction mixture after Cdk1/cyclin B depletion (Supernatant) and Cdk1/cyclin B-bound agarose beads (Pull down) were immunoblotted with anti-Cdk1 mAb. Nonspec, nonspecific band. E, efficiency of CGP74514A (CGP) in inhibiting Cdk1/cyclin B activity. Various concentrations of CGP74514A were added to the reaction mixture of the in vitro assay of Sororin phosphorylation by Cdk1/cyclin B. After SDS-PAGE, the phosphorylation signal was detected using phosphorimaging. F, densitometric quantification of Sororin phosphorylation shown in E. G, phosphorylation is reduced in Sororin mutant T159A. Sororin WT and mutant (T159A) were sequentially phosphorylated by Ckd1/cyclin B and Plk1 as described in B above. 32P phosphorimaging (upper panel) and Coomassie staining (lower panel) of the same gel are shown. Lanes 1–3, Cdk1/cyclin B only; lanes 4–6, Cdk1/cyclin B and Plk1. H, densitometric quantification of Sororin WT and T159A mutant phosphorylated by Cdk1/cyclin B (Pcdk1) or Cdk1/cyclin B plus Plk1 (Pcdk1+plk1), which was normalized with the amount of protein loaded. Changes in phosphorylation were calculated using the equation (Pcdk1+plk1 − Pcdk1)/Pcdk1+plk1. Data are the means ± S.E. from three experiments.
The mechanism of Plk1 phosphorylation has been elucidated (35–37). The kinase domain of Plk1 needs to be localized to the phosphorylating site on the substrate protein by PBD via binding to a motif that is presented either on the substrate itself or on an adjacent docking protein. To investigate the possibility that phosphorylation of Sororin by Plk1 requires a putative PBD binding motif on Sororin to be phosphorylated first, we performed sequential kinase assays by first using Cdk1/cyclin B to phosphorylate Sororin (Fig. 3B, Step 1), then immunodepleting and inhibiting Cdk1/cyclin B with CGP74514A to minimize the potential incorporation of [32P]ATP by Cdk1/cylin B (Fig. 3B, Step 2), and finally adding Plk1 to phosphorylate Sororin (Fig. 3B, Step 3). The immunodepletion of Cdk1/cyclin B was confirmed by Western blot, which suggests that no Cdk1 could be detected in the reaction mixture (Fig. 3D), and the activity of Cdk1/cyclin B could be inhibited up to 80% by CGP75414A (Fig. 3, E and F). The results indicate that Cdk1 phosphorylates Sororin (Fig. 3B, lane 3) but that Plk1 phosphorylates Sororin only after Sororin is phosphorylated by Cdk1 (Fig. 3B, lane 6), whereas Plk1 alone fails to do so (Fig. 3B, lane 9). Fig. 3C quantitatively shows the additional phosphorylation of vimentin and casein (both positive controls) as well as Sororin by Plk1 after they were phosphorylated by Cdk1. Collectively, these results suggest that Sororin can be sequentially phosphorylated by Cdk1/cyclin B and then Plk1.
The consensus motif that PBD binds is S(pS/pT)(P/X) where pS is phosphoserine and pT is phosphothreonine (36). There are five putative PBD binding sites on Sororin: ST115P, SS125S, SS126K, ST157S, and ST159P (Fig. 1B). Among these, ST159P is also a Cdk1/cyclin B phosphorylation site. We tested the possibility that after ST159P is phosphorylated by Cdk1/cyclin B SpT159P functions as a PBD binding motif to facilitate Plk1 to phosphorylate Sororin. We used the same assay described above in Fig. 3B. Indeed, when the threonine 159 site was mutated into alanine, Sororin phosphorylation by Plk1 was significantly reduced compared with the wild type Sororin (Fig. 3, G and H). A protein-protein interaction assay showed that ST159P is a bona fide PBD binding motif, whereas the other four putative PBD binding motifs are not (see below). We also examined several putative Plk1 phosphorylation sites (ELT98KED, ESS125SKE, EMS139KKV, SYS147RLE, DLS178GVS, and DMT204LPG) on Sororin by mutating Ser/Thr residues into alanine individually or in combinations. However, we were unable to identify specific Plk1 phosphorylation site(s) on Sororin using the in vitro kinase assay (data not shown).
ST159P Motif of Sororin Interacts with Plk1
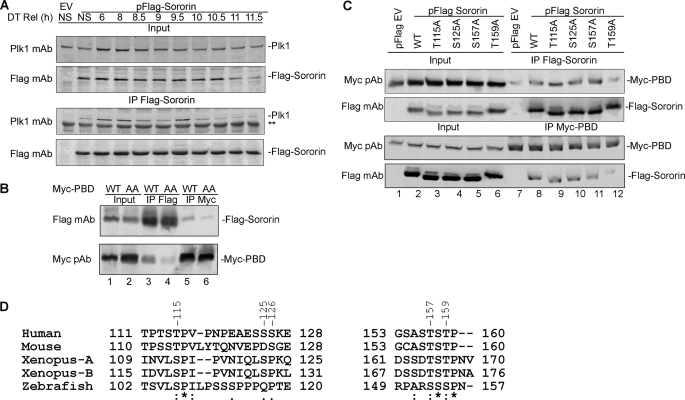
The sequence ST159PGR on Sororin is not only a Cdk1/cyclin B phosphorylation consensus motif but also a putative consensus motif to which the PBD of Plk1 can bind. To examine whether there is a stable physical interaction between Sororin and Plk1 in vivo, we expressed FLAG-Sororin in HeLa cells. The cells were then synchronized using double thymidine, and samples were harvested at different time points after release from double thymidine. Co-IP of endogenous Plk1 by FLAG-Sororin was performed. Immunoblot results show that endogenous Plk1 was co-immunoprecipitated by Sororin (Fig. 4A).
FIGURE 4.
Interaction of Plk1 and Sororin. A, HeLa cells were transfected with pFLAG-Sororin or pFLAG empty vector (EV). Twenty-four hours after transfection, the cells were synchronized with double thymidine (DT). Non-synchronized (NS) cells were used as a control. Cells were harvested 6–11.5 h after release from double thymidine. Endogenous Plk1 was co-immunoprecipitated by FLAG-Sororin and visualized with immunoblotting. ** indicates a nonspecific band. B, WT, but not mutant (AA) PBD, interacts with Sororin. FLAG-Sororin was expressed in HeLa S3 Myc-PBD WT and Myc-PBD(AA) Tet-On cells for 40 h before Myc-PBD WT and mutant Myc-PBD(AA) were induced with doxycycline for 16 h. Co-IP of FLAG-Sororin and Myc-PBD was performed. C, Sororin T159A mutation significantly reduces its interaction with PBD. HeLa S3 Tet-On cells stably expressing Myc-tagged PBD were transfected with FLAG-tagged Sororin WT and four putative PBD binding motif mutants (T115A, S125A, S157A, and T159A). Empty vector (EV)-transfected cells were used as a control. Reciprocal IP and immunoblotting were performed. D, sequence comparison for the five putative PBD binding motifs at ST115P, SS125S, SS126K, ST157S, and ST159P of human Sororin with other vertebrate species. The numbers on the top line show the site of human Sororin. Invariant, conserved, and semiconserved residues are indicated by an asterisk (*), colon (:), and period (.), respectively. pAb, polyclonal antibody.
Plk1 function relies on its C-terminal PBD to bind to either a docking protein or a target protein and its N-terminal kinase domain to phosphorylate the target protein. To confirm the interaction between the PBD of Plk1 and the PBD binding motif of Sororin, we utilized two HeLa S3 Tet-On cell lines that were stably transfected with either Myc-PBD WT or Myc-PBD mutant (AA) (38). Myc-PBD(AA) contains two mutations (H538A and K540A) that disrupt PBD binding to its partner. We expressed FLAG-Sororin in these two cell lines and performed reciprocal IP. Immunoblot results show that PBD WT and Sororin could co-immunoprecipitate each other (Fig. 4B, lanes 3 and 5), whereas PBD(AA)-Sororin interaction was significantly reduced (Fig. 4B, lanes 4 and 6). This suggests that Plk1 interacts with Sororin via its PBD.
To identify the physiologically relevant PBD binding sites on Sororin, we made several constructs by mutating the second Ser or Thr residue in the putative PBD binding motifs (ST115P, SS125S, ST157S, and ST159P) into alanine. Because SS125S126K contains two putative PBD binding sites, mutation of Ser125 is likely to disrupt both sites. We expressed WT and mutant FLAG-Sororin in HeLa S3 Myc-PBD Tet-On cells. Co-IP was performed, and immunoblot results demonstrated that mutation of Thr115, Ser125, and Thr157 did not affect the interaction of Sororin with the PBD (Fig. 4C, lanes 9–11). Only the T159A mutant considerably reduced the interaction of Sororin with the PBD (Fig. 4C, lane 12). These results are supported by bioinformatics data that only ST159P is conserved in vertebrates (Fig. 4D). Based on the above results, we conclude that Sororin interacts with the PBD of Plk1 via its ST159P motif.
Sororin Co-localizes with Plk1
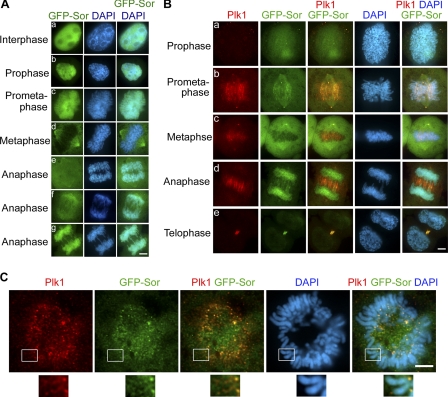
It is known that Plk1 is essential for the removal of sister chromatid arm cohesion (9), and our co-IP results shown above demonstrate that Sororin and Plk1 interact with each other. These data prompted us to hypothesize that Sororin collaborates in the Plk1-mediated removal of sister chromatid arm cohesion. Due to the lack of specific Sororin antibody to investigate the localization of endogenous Sororin during the cell cycle, we established a stably transfected HeLa cell line expressing GFP-Sororin. GFP-Sororin is functional because it not only can interact with cohesin (see Fig. 6B, lane 5), but it also can rescue the Sororin depletion by siRNA (see below). Fluorescence microscopy showed that localization of GFP-Sororin is dynamic during mitosis (Fig. 5). Sororin was localized in the nucleus in interphase (Fig. 5A, panel a). At prophase, GFP-Sororin was predominantly localized on chromatin (Fig. 5A, panel b). At prometaphase, it was found on chromatids (Fig. 5A, panel c) and the spindle and centrosomes (Fig. 5B, panel b). At metaphase, although GFP-Sororin was still on the spindle and centrosomes, a majority of GFP-Sororin had dissociated from the chromatids (Fig. 5A, panel d). Interestingly, after sister chromatids were separated, Sororin gradually reassociated with the segregated chromosomes with the progression of anaphase (Fig. 5A, panels e, f, and g), but the Sororin signal disappeared at telophase (Fig. 5B, panel e). The association of Sororin with chromosomes in late anaphase has also been observed in time lapse live cell imaging (14, 28).
FIGURE 6.
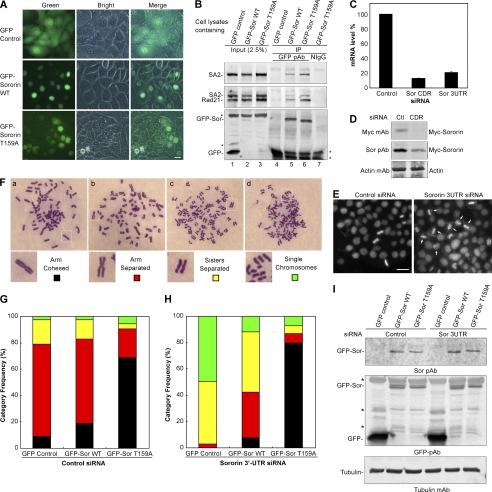
Sororin T159A mutant inhibits resolution of chromosomal arm cohesion. A, fluorescence microscopy showing the expression of GFP, GFP-Sororin WT, and GFP-Sororin T159A mutant in HeLa stable cell lines. B, co-IP of GFP-Sororin (Sor) and cohesin. GFP and GFP-Sororin were immunoprecipitated with rabbit anti-GFP polyclonal antibody (pAb) (lanes 4–6). Normal rabbit IgG (NIgG) was used as a control (lane 7). 2.5% of the total input protein used for co-IP was loaded in lanes 1–3. Co-IP of cohesin by GFP-Sororin was analyzed with immunoblotting using SA2 polyclonal antibody and Rad21 mAb. The middle panel shows both SA2 and Rad21 bands, which were probed with Rad21 antibody after SA2 was blotted. * indicates non specific bands. C–E, efficiency of endogenous Sororin knockdown by siRNA. C, exponentially growing HeLa cells were transfected with Sororin coding region siRNA (Sor CDR) or Sororin 3′-UTR (Sor 3UTR). Silencer Negative Control 1 siRNA was used as a control. Forty-eight hours after transfection, total RNA in HeLa cells was extracted, and RT-PCR was performed. The mRNA level in cells transfected with Sororin 3′-UTR or coding region siRNA was significantly reduced compared with that in cells transfected with control siRNA. Data are the means ± S.E. from three samples. D, Myc-Sororin in HeLa Tet-On Sororin stably transfected cells induced with 2 μg/ml doxycycline. The cells were transfected with control or Sororin coding region siRNA for 48 h. Whole cell lysates were used for immunoblotting with anti-Myc mAb, Sororin polyclonal antibody (Sor pAb), and Actin mAb. E, HeLa cells were transfected with control siRNA or Sororin 3′-UTR siRNA for 24 h. The cells were fixed and stained with DAPI. Cells treated with Sororin 3′-UTR siRNA arrested at prometaphase (arrow). F–I, HeLa stable cell lines shown in A were treated with control siRNA (G) or Sororin 3′-UTR siRNA (H) before metaphase spread of chromosomes was performed. More than 100 mitotic cells were randomly examined and categorized according to the resolution of sister chromatid cohesion in F (panels a–d). The frequency of each category was plotted in G and H. I, immunoblotting shows the GFP-Sororin level in the cells used for metaphase spread in G and H. * indicates nonspecific bands. Bar size, 25 μm.
FIGURE 5.
Sororin co-localizes with Plk1 on sister chromatids at prometaphase. A, panels a–g, cellular localization of Sororin in HeLa cells stably expressing GFP-Sororin. B, panels a–e, HeLa GFP-Sororin (Sor) stable cells were fixed and stained with Plk1 mAb followed by rhodamine-labeled secondary antibody. Sororin and Plk1 were found co-localized on chromosome at prometaphase. C, immunostaining of chromosome spreads showing the co-localization of Plk1 and GFP-Sororin on chromosome arm. GFP-Sororin is shown in green, Plk1 is shown in red, and the co-localization of GFP-Sororin and Plk1 is shown in yellow. Nuclear material was stained with DAPI (blue). Scale bar, 5 μm.
We investigated the co-localization of GFP-Sororin and Plk1 during mitosis. Sororin and Plk1 were found on sister chromatids in prometaphase (Fig. 5, B, panel b, and C), centrosomes and centromeres in prometaphase and metaphase (Fig. 5B, panels b and c), and the midbody in anaphase (Fig. 5B, panels b and c). However, the frequency of the co-localization of GFP-Sororin and Plk1 on prophase/prometaphase chromosome arms was found to be <1% on the fixed mitotic cells, which is likely due to the transient interactions of Sororin and Pkl1 on chromosome arms. The co-localization of Sororin and Plk1 on chromatids coincided with the dissociation of chromatid arm cohesins, supporting the notion that Sororin and Plk1 cooperate to resolve arm cohesion.
Sororin Mediates Resolution of Arm Cohesion
Previous studies showed that Plk1 is required for the removal of sister chromatid arm cohesion (9, 10). Our data demonstrated that Sororin and Plk1 not only interacted with each other but also co-localized on chromosomes at prometaphase. Based on these data, we reason that Sororin perhaps assists Plk1 in resolving sister chromatid arm cohesion. To test this possibility, we established three HeLa stable cell lines that expressed GFP (as control), GFP-Sororin WT, and GFP-Sororin T159A mutant, respectively. The expression of GFP and GFP-tagged Sororin WT and T159A mutant can be observed using fluorescence microscopy. Although GFP (control) signal was both cytoplasmic and nuclear, GFP-Sororin WT and T159A mutant were exclusively nuclear (Fig. 6A). GFP-tagged Sororin WT and T159A mutant could efficiently co-immunoprecipitate cohesin core subunits, such as Rad21 and SA2 (Fig. 6B), indicating that neither the GFP tag nor the T159A mutation interrupted the interaction of Sororin and cohesin.
To investigate the direct role of Sororin on the resolution of sister chromatid arms, we used Sororin 3′-UTR siRNA to knock down endogenous Sororin while sparing the ectopic expression of GFP-Sororin WT and T159A mutant. Because our Sororin polyclonal antibody recognizes overexpressed Sororin and is not sensitive enough to react with endogenous Sororin, we used real time PCR, immunoblotting, and microscopy to confirm the efficiency of Sororin 3′-UTR siRNA (Fig. 6, C–E). We tested two Sororin siRNAs: one against the Sororin coding region and the other against the 3′-UTR. Real time PCR showed that both siRNAs could efficiently knock down Sororin mRNA (Fig. 6C). Immunoblots indicated that Sororin protein was reduced by at least 75% (Fig. 6D). After HeLa cells were transfected with Sororin 3′-UTR siRNA for 24 h, the cells arrested at prometaphase (Fig. 6E), a typical phenotype seen in cells with Sororin depletion (22). These results show that Sororin 3′-UTR siRNA can efficiently deplete endogenous Sororin.
To investigate the effect of the Sororin PBD binding motif ST159P on the resolution of sister chromatid arms, we treated HeLa GFP, GFP-Sororin WT, and GFP-Sororin T159A cell lines with control siRNA or Sororin 3′-UTR siRNA and performed metaphase chromosome spreads to analyze the status of sister chromatid arm resolution. The protein levels of GFP-Sororin WT and T159A were equivalent in the cells after siRNA treatment (Fig. 6I). Mitotic cells were counted and categorized into four groups according to the chromosomal morphology shown in Fig. 6F: (i) arm-cohesed in which the two sister chromatids are held together with no discernible separation, (ii) arm open in which the arms of sister chromatids are separated but are held together at the centromere, (iii) sisters separated in which both arms and the centromere of the sister chromatids are separated but the sister chromatids are still aligned side by side, and (iv) single chromosomes in which sister chromatids are not only separated but also are scattered randomly with no evident pairing.
In control siRNA-treated cells, the frequency of arm-cohesed chromosomes increased about 10% in Sororin WT cells compared with that in GFP control cells. However, the frequency of arm-cohesed chromosomes in Sororin T159A mutant cells increased by more than 60%, which was significantly higher than that found in both GFP control and Sororin WT cells (Fig. 6G). In contrast, the frequency of arm-separated chromosomes in Sororin T159A cells (20%) was significantly less than that of the GFP control and GFP-Sororin WT cells (70%) (Fig. 6G). The phenotype shown by the T159A mutant was quite interesting. In this experiment, the mutant proteins were expressed in addition to the endogenous WT protein. Sororin T159A mutant, despite its lack of interaction with Plk1, was still proficient in its interaction with cohesin subunits (Fig. 6B) and cohesion function (Fig. 6G). It is also interesting to note that the Sororin T159A mutation appeared to have a dominant negative effect that overrode the function of endogenous Sororin in the resolution of sister chromatid arm cohesion. Collectively, these data indicate that overexpression of WT Sororin stabilizes sister chromatid arm cohesion, whereas the Sororin T159A mutant inhibits the resolution of sister chromatid arm cohesion.
To investigate the effect of GFP-Sororin alone on arm cohesion, the endogenous Sororin was depleted with Sororin 3′-UTR siRNA. Metaphase chromosome spread analysis showed that sister chromatids were separated in more than 95% of GFP control cells (sisters separated plus single chromatids), and no arm-cohesed sister chromatids were found (Fig. 6H), suggesting that the endogenous Sororin was depleted. GFP-Sororin WT could rescue the phenotype of the Sororin 3′-UTR siRNA-treated cells by restoring the frequency of arm-cohesed sister chromatids to the level equivalent to that in GFP control cells with control siRNA treatment (Fig. 6, G and H), implying that GFP-Sororin is fully functional. In contrast, GFP-Sororin T159A mutant not only rescued the phenotype caused by Sororin 3′-UTR siRNA, but also resulted in stronger cohesion, demonstrating more than 80% arm-cohesed sister chromatids (Fig. 6H). Collectively, the above results suggest that a mutation at Thr159 of Sororin does not affect its cohesion function but prevents the resolution of arm cohesion due to the lack of its interaction with Plk1. These data underscore the functional significance of the interaction of the ST159P motif of Sororin with Plk1, which plays a critical role in arm cohesion resolution.
DISCUSSION
How Plk1 is recruited to phosphorylate SA2 during prophase is not currently known. In this study, we report that the interaction of Plk1 with Sororin plays a pivotal role in resolving arm cohesion. Our data suggest that Sororin acts as a docking protein and recruits Plk1 to phosphorylate SA2 during prophase, which facilitates the removal of cohesins from sister chromatid arms. Although Sororin, a cohesin-interacting protein, is known to play an essential role in chromosomal cohesion, our data demonstrate a novel role for Sororin in chromosomal arm resolution.
Analysis of the Sororin protein sequence using various bioinformatics tools indicated that there are three putative Cdk1/cyclin B phosphorylation sites and five putative PBD binding sites. All three putative Cdk1/cyclin B phosphorylation sites (Ser21, Ser75, and Thr159) and one of the five putative PBD binding sites (ST159P) were confirmed in this study. These sequence features suggest that Sororin is a kinase-regulated protein, and the phosphorylation status of Sororin may dictate its cellular functions. For example, non-phosphorylated Sororin can displace Wapl that binds to Pds5A, whereas phosphorylated Sororin cannot (14).
Plk1 contains a highly conserved kinase domain in its N-terminal region and two polo boxes constituting the PBD in the C-terminal region (39). Although the kinase activity of Plk1 is critical for the various cellular functions, the PBD is responsible for proper cellular localization of Plk1 (35–37). It has been reported that the PBD of Plk1 binds to a docking protein to bring its catalytic domain to the vicinity of its substrate (35, 39). We found that one of the five putative PBD binding sites of Sororin overlaps with the Cdk1/cyclin B phosphorylation site, ST159P. Co-IP results demonstrated that the T159A mutation on Sororin abolished its interaction with Plk1 in vivo, confirming that ST159P constitutes both the Cdk1/cyclin B phosphorylation site and PBD binding site. Moreover, ST159P is conserved in vertebrate Sororin, and the phosphorylation of Thr159 has been confirmed by mass spectrometry analysis (27).
Previous studies indicate that both Rad21 and SA2 are heavily phosphorylated but that only Plk1-mediated SA2 phosphorylation is essential for the dissociation of cohesin from chromosome arms during prophase and prometaphase (11). Knockdown of Plk1 or expression of SA2 non-phosphorylatable mutants prevents the removal of cohesin complexes from chromosomal arms (9, 11), whereas overexpression of Plk1 leads to chromosomal instability and chromosomal missegregation apparently due to the premature loss of sister chromatid arm cohesion (40, 41). Although SA2 can be phosphorylated by Plk1, no PBD binding site (S(pS/pT)P) has been found on SA2 or other cohesin core subunits, making it unclear how SA2 is phosphorylated by Plk1 in vivo. It has been reported that the C terminus of Sororin interacts with cohesin (42), and our data show that the Sororin T159A mutant prevented Plk1 binding and inhibited the resolution of sister chromatid arm cohesion, suggesting that Sororin mediates Plk1 to phosphorylate SA2.
It has been reported that Wapl controls the dynamic association of cohesin with chromosomes (12, 13). Depletion of Wapl causes accumulation of chromosomes with unresolved chromosome arms typically found in prometaphase cells. Overexpression of Wapl, however, results in precocious chromosome separation. Direct association of Wapl with cohesin via Pds5 and SA1/2 strengthens the notion that Wapl is responsible for the non-catalytic removal of cohesin. One recent study showed that Wapl function is antagonized by Sororin (14). Both Sororin and Wapl have a common motif that interacts with Pds5. It is believed that Sororin binds to Pds5 to prevent the interaction of Wapl with Pds5 from S phase to prophase so that cohesin stably associates with sister chromatids. When the cell cycle progresses into prophase/prometaphase, Sororin is phosphorylated and loses its ability to inhibit Wapl interaction with Pds5, leading to the removal of cohesin (14). Our data show that Sororin can be sequentially phosphorylated by Cdk1/cyclin B and Plk1 in vitro. The activity of Cdk1/cyclin B and Plk1 reaches the highest level once cells enter mitosis. It is therefore plausible that Cdk1/cyclin B and Plk1 are responsible for phosphorylating Sororin in vivo and destabilizing its interaction with Pds5.
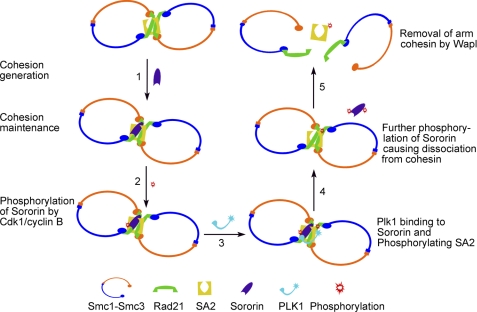
Based on our results, we propose a model to show how Sororin can mediate the dissociation of cohesin complexes from chromosomal arms at prophase (Fig. 7). During DNA replication at S phase, sister chromatid cohesion is generated after the cohesin complexes are loaded to newly synthesized sister chromatids, Smc3 is acetylated, and Sororin associates with Pds5. Sororin binding to Pds5 displaces Wapl, which is critical in generating and maintaining sister chromatid cohesion (Fig. 7, Step 1; for simplicity, Pds5 and Wapl are not shown). When the cell cycle enters prophase, cyclin B activity increases, and Sororin is phosphorylated by Cdk1/cyclin B at the PBD binding site ST159P (Fig. 7, Step 2). Sororin then functions as a docking protein to recruit Plk1 into the vicinity of the cohesin complex via the interaction between the Plk1 PBD and SpT159P of Sororin. This interaction results in SA2 phosphorylation by Plk1 (Fig. 7, step 3). After the completion of SA2 phosphorylation, Plk1 and/or other kinases further phosphorylate Sororin, causing changes in its configuration that lead to its dissociation from Pds5. Sororin dissociation allows Wapl to bind to Pds5 (Fig. 7, step 4). Once Wapl is bound to Pds5, it removes the cohesin complexes with phosphorylated SA2, resulting in the resolution of sister chromatid arms (Fig. 7, step 5). The failure of Plk1 to dissolve centromeric cohesin and some arm cohesin may be due to its inability to bind to Sororin, which is prevented either by the lack of phosphorylation on the PBD binding motif of Sororin or by the protection provided by other proteins, such as Shugoshin. The lines of evidence supporting this model include 1) the requirement of Sororin for the generation and maintenance of cohesion (14, 21, 22, 43), 2) the direct interaction of Sororin with cohesin (22, 42), 3) competition of Sororin and Wapl for binding to Pds5 (14), 4) phosphorylation of Sororin by Cdk1/cyclin B, 5) direct interaction and co-localization of Sororin with Plk1 on the chromosome arms during prophase and prometaphase, and 6) persistent arm cohesion upon Sororin T159A mutation. To directly prove that SA2 phosphorylation by Plk1 is mediated by Sororin in vivo, it is necessary to analyze SA2 phosphorylation status in cells expressing knocked in Sororin T159A mutant.
FIGURE 7.
Model for Sororin-mediated resolution of arm cohesion. During generation of cohesion at S phase, Sororin is incorporated into the cohesin complex by interacting with cohesin and is required for the maintenance of sister chromatid cohesion both on chromosome arms and centromeres (Step 1). Protection of centromeric cohesion by Shugoshin is not shown. To resolve chromosomal arm cohesion during prophase, Sororin is phosphorylated by Cdk1/cyclin B at the PBD binding site ST159P (Step 2), which functions as a docking protein to recruit Plk1 to the vicinity of the cohesin complex and results in SA2 phosphorylation by Plk1 (Step 3). Once SA2 is phosphorylated, Sororin is further phosphorylated and dissociates from cohesin (Step 4). Wapl displaces phosphorylated Sororin and binds with Pds5 to facilitate the removal of cohesin complexes from sister chromatid arms (Step 5).
Acknowledgments
We thank Dr. Herman H. W. Sillje at the Department of Cell Biology, Max Planck Institute for Biochemistry for HeLa S3 Tet-On Myc-PBD cell lines. We thank Dr. Eryong Huang for assistance in RT-PCR and Dr. Terzah Horton for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1RO1 CA109478 and 1RO1 CA109330 from the NCI (to D. P.). This work was also supported by a 2008 Virginia and L. E. Simmons Family Foundation collaborative research award (to D. P.).
- Plk1
- polo-like kinase 1
- IP
- immunoprecipitation
- PBD
- polo box domain.
REFERENCES
- 1. Cohen-Fix O., Peters J. M., Kirschner M. W., Koshland D. (1996) Genes Dev. 10, 3081–3093 [DOI] [PubMed] [Google Scholar]
- 2. Zachariae W., Nasmyth K. (1999) Genes Dev. 13, 2039–2058 [DOI] [PubMed] [Google Scholar]
- 3. Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. (1998) Cell 93, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 4. Stemmann O., Zou H., Gerber S. A., Gygi S. P., Kirschner M. W. (2001) Cell 107, 715–726 [DOI] [PubMed] [Google Scholar]
- 5. Uhlmann F., Lottspeich F., Nasmyth K. (1999) Nature 400, 37–42 [DOI] [PubMed] [Google Scholar]
- 6. Uhlmann F., Wernic D., Poupart M. A., Koonin E. V., Nasmyth K. (2000) Cell 103, 375–386 [DOI] [PubMed] [Google Scholar]
- 7. Waizenegger I. C., Hauf S., Meinke A., Peters J. M. (2000) Cell 103, 399–410 [DOI] [PubMed] [Google Scholar]
- 8. Sumara I., Vorlaufer E., Gieffers C., Peters B. H., Peters J. M. (2000) J. Cell Biol. 151, 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sumara I., Vorlaufer E., Stukenberg P. T., Kelm O., Redemann N., Nigg E. A., Peters J. M. (2002) Mol. Cell 9, 515–525 [DOI] [PubMed] [Google Scholar]
- 10. Giménez-Abián J. F., Sumara I., Hirota T., Hauf S., Gerlich D., de la Torre C., Ellenberg J., Peters J. M. (2004) Curr. Biol. 14, 1187–1193 [DOI] [PubMed] [Google Scholar]
- 11. Hauf S., Roitinger E., Koch B., Dittrich C. M., Mechtler K., Peters J. M. (2005) PLoS Biol. 3, e69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gandhi R., Gillespie P. J., Hirano T. (2006) Curr. Biol. 16, 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kueng S., Hegemann B., Peters B. H., Lipp J. J., Schleiffer A., Mechtler K., Peters J. M. (2006) Cell 127, 955–967 [DOI] [PubMed] [Google Scholar]
- 14. Nishiyama T., Ladurner R., Schmitz J., Kreidl E., Schleiffer A., Bhaskara V., Bando M., Shirahige K., Hyman A. A., Mechtler K., Peters J. M. (2010) Cell 143, 737–749 [DOI] [PubMed] [Google Scholar]
- 15. McGuinness B. E., Hirota T., Kudo N. R., Peters J. M., Nasmyth K. (2005) PLoS Biol. 3, e86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kitajima T. S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S. A., Watanabe Y. (2006) Nature 441, 46–52 [DOI] [PubMed] [Google Scholar]
- 17. Hauf S., Waizenegger I. C., Peters J. M. (2001) Science 293, 1320–1323 [DOI] [PubMed] [Google Scholar]
- 18. Haering C. H., Nasmyth K. (2003) BioEssays 25, 1178–1191 [DOI] [PubMed] [Google Scholar]
- 19. Losada A., Yokochi T., Hirano T. (2005) J. Cell Sci. 118, 2133–2141 [DOI] [PubMed] [Google Scholar]
- 20. Panizza S., Tanaka T., Hochwagen A., Eisenhaber F., Nasmyth K. (2000) Curr. Biol. 10, 1557–1564 [DOI] [PubMed] [Google Scholar]
- 21. Schmitz J., Watrin E., Lénárt P., Mechtler K., Peters J. M. (2007) Curr. Biol. 17, 630–636 [DOI] [PubMed] [Google Scholar]
- 22. Rankin S., Ayad N. G., Kirschner M. W. (2005) Mol. Cell 18, 185–200 [DOI] [PubMed] [Google Scholar]
- 23. Walker M. G. (2001) Curr. Cancer Drug Targets 1, 73–83 [DOI] [PubMed] [Google Scholar]
- 24. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 25. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 26. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cantin G. T., Yi W., Lu B., Park S. K., Xu T., Lee J. D., Yates J. R., 3rd (2008) J. Proteome Res. 7, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 28. Dreier M. R., Bekier M. E., 2nd, Taylor W. R. (2011) J. Cell Sci. 124, 2976–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pati D., Zhang N., Plon S. E. (2002) Mol. Cell. Biol. 22, 8267–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Genome Biol. 3, RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freshney R. I. (2000) Culture of Animal Cells: a Manual of Basic Technique, 4th Ed., pp. 456–457, Wiley-Liss, Inc., New York [Google Scholar]
- 32. Zhang N., Kuznetsov S. G., Sharan S. K., Li K., Rao P. H., Pati D. (2008) J. Cell Biol. 183, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 34. Chen R. Q., Yang Q. K., Lu B. W., Yi W., Cantin G., Chen Y. L., Fearns C., Yates J. R., 3rd, Lee J. D. (2009) Cancer Res. 69, 2663–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lowery D. M., Mohammad D. H., Elia A. E., Yaffe M. B. (2004) Cell Cycle 3, 128–131 [PubMed] [Google Scholar]
- 36. Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 37. Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. (2003) Cell 115, 83–95 [DOI] [PubMed] [Google Scholar]
- 38. Hanisch A., Wehner A., Nigg E. A., Silljé H. H. (2006) Mol. Biol. Cell 17, 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lowery D. M., Lim D., Yaffe M. B. (2005) Oncogene 24, 248–259 [DOI] [PubMed] [Google Scholar]
- 40. Yamamoto Y., Matsuyama H., Kawauchi S., Matsumoto H., Nagao K., Ohmi C., Sakano S., Furuya T., Oga A., Naito K., Sasaki K. (2006) Oncology 70, 231–237 [DOI] [PubMed] [Google Scholar]
- 41. Li J. J., Li S. A. (2006) Pharmacol. Ther. 111, 974–984 [DOI] [PubMed] [Google Scholar]
- 42. Wu F. M., Nguyen J. V., Rankin S. (2011) J. Biol. Chem. 286, 3579–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lafont A. L., Song J., Rankin S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20364–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen M. H., Koinuma J., Ueda K., Ito T., Tsuchiya E., Nakamura Y., Daigo Y. (2010) Cancer Res. 70, 5337–5347 [DOI] [PubMed] [Google Scholar]
- 45. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Sci. Signal. 3, ra3 [DOI] [PubMed] [Google Scholar]