Abstract
We report a food-related outbreak of salmonellosis in humans caused by a nonmotile variant of Salmonella enterica serotype Typhimurium in France in 2009. This nonmotile variant had been circulating in laying hens but was not considered as Typhimurium and consequently escaped European poultry flock regulations.
Keywords: bacteria, foodborne infections, outbreak, nonmotile Salmonella, Salmonella enterica, food poisoning, laying hens, poultry, eggs, tiramisu, France
Identification of Salmonella enterica serotypes is based on flagellar and somatic antigens. Of the 2,610 serotypes distinguished in the Kauffmann-White-Le Minor Scheme (1,2), only Gallinarum are obligatory nonflagellate and hence, nonmotile. Nonmotile variants of diphasic Salmonella spp. are rarely reported in humans, food, or animals (3,4) as opposed to monophasic variants (5), in particular the monophasic variant of serotype Typhimurium (antigenic formula 1,4,[5],12:i:–) that has emerged worldwide (6–8). The French National Reference Center for Salmonella (FNRC-Salm) network is based upon voluntary reporting from ≈1,500 hospital and private clinical laboratories representing two thirds of all clinical Salmonella spp. strains isolated per year. In France, serotype Typhimurium is the most prevalent serotype and remains stable with ≈4,000 human isolates per year; 1,4,[5],12:i:– strains have dramatically increased since 2005 (≈100 isolates) to reach ≈1,000 isolates in 2009.
We report a 2009 outbreak (8 cases) of salmonellosis in humans caused by a nonmotile strain of Salmonella enterica subsp. enterica with the antigenic formula 4,5,12:–:–. To estimate the extent of circulation and to determine the molecular subtypes of these nonmotile Salmonella strains, we performed a comprehensive, molecular epidemiologic study on human and nonhuman 1,4,[5],12:–:– strains isolated during 2000–2009 in France.
The Study
In May 2009, diarrhea and fever developed in 8 persons living in southwestern France 1 day after they ate a homemade tiramisu prepared with raw eggs. Fecal analysis was performed on samples from 5 of the 8 persons. We also cultured a sample from the tiramisu. In medical laboratories, the isolation was performed by using standard procedures (i.e., use of conventional selective media). Isolation from the food sample was performed as required by the current International Organization for Standardization ISO 6579:2002 (i.e., by 2 selective enrichment media) (9). All cultures yielded S. enterica subsp. enterica 4,5,12:–:–.
An investigation at the suspected layer farm was conducted and showed the presence of 11 nonmotile Salmonella spp. isolates (with the same antigenic formula) in dust and feces collected from laying-hen houses. The layer farm, located in northwestern France, is a major farm that produces >32,000,000 eggs per year. All 17 isolates (5 from humans, 1 from the tiramisu, and 11 from the laying hens) were pan-susceptible to all antimicrobial drugs tested (10).
Analysis by Pulsenet (www.cdc.gov/pulsenet/protocols.htm) standardized XbaI pulsed-field gel electrophoresis (PFGE) showed an indistinguishable profile, XTYM-1, associated with the multidrug-resistant S. enterica serotype Typhimurium DT104 clone (10,11) (Figure A1). Multilocus variable number tandem repeat analysis (MLVA) typing (12) showed an unique type 3–14–7–21–311 (loci STTR9–5–6–10–3, respectively) in 16/17 isolates and a single-locus variant (differing by 1 repetition) for the remaining isolate, which was an environmental isolate collected 1 month after the control measures were implemented (slaughter of laying hens) (Figure A1).
The tiramisu isolate 09CEB3100 was also characterized by multilocus sequence typing (MLST) for the fliC and fljB genes encoding the flagellar antigens (13). The isolate belonged to MLST sequence type 19, the main sequence type of serotype Typhimurium (http://mlst.ucc.ie/mlst/dbs/Senterica). PCR and sequencing identified the fliC gene encoding the i antigen and the fljB gene encoding for the 1,2 antigen, confirming that this Salmonella spp. strain of the antigenic formula 4,5,12:–:– was a nonmotile variant of serotype Typhimurium. The investigation and molecular data concluded that a nonmotile variant strain of serotype Typhimurium, genetically distinct from the emerging monophasic variant described worldwide, has been circulating in laying hens, whose contaminated eggs had likely caused food poisoning.
During 2000–2009, a total of 108,362 serotyped Salmonella spp. isolates from humans were registered at the FNRC-Salm, of which 374 (0.3%) were nonmotile. The 1,4,[5],12:–:– strains were the most prevalent (147/374, 39%). Such 1,4,[5],12:–:– strains have been rarely reported in food, environment or animals with 166 registered isolates collected by the French Food Safety Agency Salmonella network during 2001–2009 (compared with 21,214 serotype Typhimurium and 157,885 Salmonella spp. isolates registered).
To determine whether the population of nonmotile Salmonella spp. was circulating, we studied 43 additional S. enterica serotype 1,4,[5],12:–:– strains: 22 human and 21 nonhuman strains (5 from laying hens, 6 from cattle, 2 from sheep, 2 from partridges, 1 from chickens, 3 from the environment, and 3 from foodstuffs) isolated during 2001–2009. These strains were selected on the basis of their diversity (geographic area, year of isolation, and source). All the strains were characterized by antimicrobial drug susceptibility testing, PFGE, MLVA, and by fliC and fljB genes sequencing.
The 60 S. enterica serotype 1,4,[5],12:–:– isolates studied (17 isolates linked to the food poisoning and 43 additional isolates) displayed 22 different PFGE profiles. Only 10 profiles, from 49 isolates (81.6%), matched with profiles from the FNRC-Salm PFGE Typhimurium database (which includes 207 profiles generated in routine surveillance on 632 strains isolated during 1981–2010). All 49 strains were confirmed as nonmotile variants of Typhimurium by the identification of the fliC gene encoding the i antigen and the fljB gene encoding for the 1,2 antigen. The 11 strains whose PFGE pattern did not match any Typhimurium PFGE profile in our database belonged to serotypes Typhimurium (corresponding to 4 new PFGE profiles), Paratyphi B (n = 3), Agona, Derby, Indiana, and Saintpaul (1 each).
Among the 49 strains with a XTYM profile, 37 (75%) belonged to XTYM-1 (Figure A1). The nonmotile XTYM-1 strains were divided into 2 groups regarding their susceptibility to antimicrobial agents: pan-susceptible for those (17) linked to the food poisoning plus 6 strains isolated before 2009 (3 human and 3 nonhuman isolates) and penta-resistant profile (resistant to amoxicillin, streptomycin/spectinomycin, sulfonamide, chloramphenicol, and tetracycline) for the 14 remaining strains. Notably, all 23 nonmotile XTYM-1 pan-susceptible strains had a single amino acid substitution in the fliC gene (Asp251 encoding an asparagine residue) compared with those available in public database, including serotype Typhimurium reference strain LT2. The search was made with BLASTN (www.ncbi.nlm.nih.gov). The 12 nonmotile strains that matched other non–XTYM-1 profiles exhibited 7 PFGE profiles, suggesting the loss of motility was acquired independently by 7 distinct serotype Typhimurium populations.
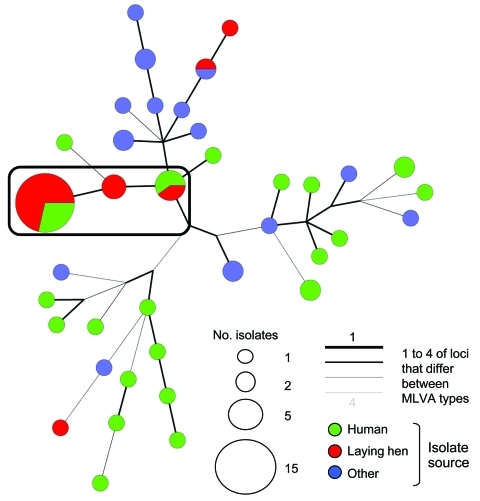
Thirty-eight MLVA types were found after testing the 60 isolates. The MLVA type 3–14–7-21–311 found in the food poisoning strains was not observed in the other strains tested. A minimum-spanning tree based on the MLVA types is shown in the Figure. The strains from the food poisoning clustered at the external extremity of a tree branch, whereas 3 human and 3 laying hen strains (from the same producer) isolated earlier (2005–2007) clustered at the internal extremity of the same branch. The 2 MLVA types differed by only 1 repetition difference at 2 loci. Two laying-hen strains isolated in 2009 (also from the same producer) were grouped in the interconnecting node. This finding, combined with antimicrobial drug susceptibility testing, PFGE, and fliC sequencing data, suggested that the 2009 strain causing the food poisoning is a derivative of the 2005 strain; both strains were isolated from the same egg producer during a 4-year interval.
Conclusions
Regarding the European Directive and the Commission Regulation on the monitoring and reduction of zoonotic agents (14), the French Regulation has extended the target for reduction of prevalence of Salmonella spp. in poultry producers to include notification of monophasic (because of the recent emergence in humans) and nonmotile (because of this food poisoning) variants of Typhimurium after January 2010 (15). This food poisoning outbreak also highlighted the need for a second selective enrichment media for Salmonella spp. detection not based on the motility in complement to the modified semisolid Rappaport-Vassiladis medium recommended as a single medium by the European Directive.
We report a foodborne outbreak caused by a nonmotile S. enterica 4,5,12:–:– strain in France. This strain has been present in laying hens in France for the past decade. Despite continuous advances in food safety and disease surveillance, control, and prevention, atypical pathogenic Salmonella spp. strains that bypass existing procedures do emerge. Foodborne bacterial infections remain a major public health concern.
Acknowledgments
We thank the French microbiological laboratories participating in the human Salmonella network for isolate processing. We thank Angie Bone for helping with the English revision of the manuscript and Erwan Trochu for his technical assistance.
Biography
Dr Le Hello is a medical biologist and deputy director of the French National Reference Center for Salmonella at Institut Pasteur. His research interests include the molecular characterization of Salmonella populations.
Figure A1.
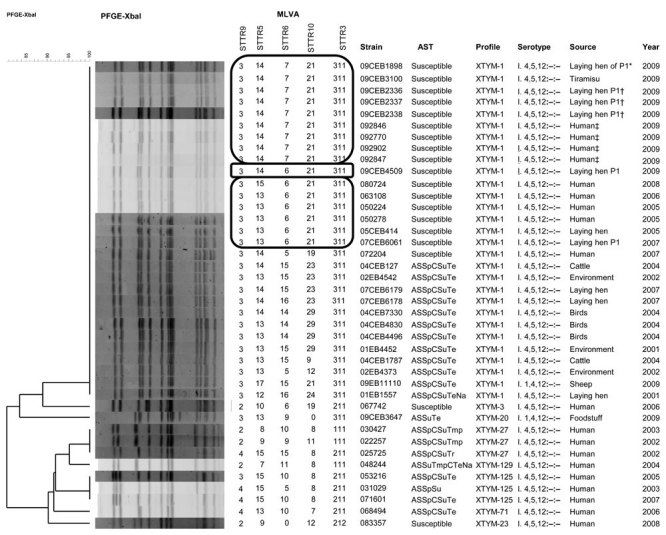
Phylogenetic analysis of XbaI–pulsed-field gel electrophoresis (PFGE) profiles obtained among a subset of 40 selected nonmotile Salmonella enterica serotype 1,4,[5],12:–:– strains isolated from humans and nonhumans during 2001–2009, France. Multilocus variable number tandem repeat analysis (MLVA), strain code, antimicrobial susceptibility testing (AST), PFGE profile found in our PFGE Typhimurium database, antigenic formula (serotype), and source and year of isolation are indicated to the right of the PFGE profiles. The MLVA groups indicated in the rectangles are the strains with the MLVA type found in our food poisoning investigation with its single- and double-locus variant with 1 repetition difference. P1 indicates the egg producer implicated in the tiramisu food poisoning. A, amoxicillin; S, streptomycin; Sp, spectinomycin; C, chloramphenicol; Su, sulfonamide; Te, tetracycline; Tmp, trimethoprim; Na, nalidixic acid. *Auto sampling; †authority sampling I; ‡tiramisu consumption; §authority sampling II.
Figure.
Minimum-spanning tree of multilocus variable number tandem repeat analysis (MLVA) of Salmonella enterica serotype 1,4,[5],12:–:– strains isolated from humans and nonhumans during 2001–2009, France. Each MLVA type is indicated by 1 node or branch tip, displayed as circles that are connected by branches of minimum-spanning tree. The length and the color of the branches represent genetic distances (changes in loci) between 2 neighboring types. The sizes of the different color circles depend on their population size. Wedges in circles indicate the proportion of isolates from respective sources with a particular MLVA type. A complex is shown in the black rectangle, based on maximum neighbor distance of changes at 2 loci and minimum size of 2 types. This specific complex linked the tiramisu food poisoning strains and other loci-derived strains for 2005–2009.
Footnotes
Suggested citation for this article: Le Hello S, Brisabois A, Accou-Demartin M, Josse A, Marault M, Francart S, et al. Foodborne outbreak and nonmotile Salmonella enterica variant, France. Emerg Infect Dis [serial on the Internet]. 2012 Jan [date cited]. http://dx.doi.org/10.3201/eid1801.110450
References
- 1.Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars, 9th ed. Paris: World Health Organization Collaborating Center for Reference and Research on Salmonella, Institut Pasteur; 2007. [Google Scholar]
- 2.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, et al. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010;161:26–9. 10.1016/j.resmic.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 3.Barbour EK, Nabbut NH, Al-Nakhli HM. A non-motile Salmonella mutant (6,7:–:–) and two serological variants encountered in Saudi Arabia. Zentralbl Bakteriol Mikrobiol Hyg [A]. 1984;256:548–52. [DOI] [PubMed] [Google Scholar]
- 4.Chadfield MS, Christensen JP, Madsen M, Sonne-Hansen J, Bisgaard M. Application of molecular methods for identification of strains classified as Salmonella enterica serovar 6,7:–:– by conventional serotyping. Avian Pathol. 2002;31:271–6. 10.1080/03079450220136585 [DOI] [PubMed] [Google Scholar]
- 5.Bruner DW, Edwards PR. A note on the monophasic non-specific Salmonella types. J Bacteriol. 1939;37:365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Switt AI, Soyer Y, Warnick LD, Wiedmann M. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:–. Foodborne Pathog Dis. 2009;6:407–15. 10.1089/fpd.2008.0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauser E, Tietze E, Helmuth R, Junker E, Blank K, Prager R, et al. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:–, an emerging health risk for humans. Appl Environ Microbiol. 2010;76:4601–10. 10.1128/AEM.02991-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins KL, Kirchner M, Guerra B, Granier SA, Lucarelli C, Porrero MC, et al. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro Surveill. 2010;15:19580. [PubMed] [Google Scholar]
- 9.International Organization for Standardization. ISO 6579:2002. Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella spp. [cited 2011 Oct 24]. http://www.iso.org/
- 10.Weill FX, Guesnier F, Guibert V, Timinouni M, Demartin M, Polomack L, et al. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J Clin Microbiol. 2006;44:700–8. 10.1128/JCM.44.3.700-708.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–50. 10.1128/JCM.43.3.1045-1050.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsson JT, Torpdahl M, Petersen RF, Sorensen G, Lindstedt BA, Nielsen EM. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill. 2009;16;14:pii:19174. [PubMed]
- 13.Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, et al. Salmonella Typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol. 2002;2:39–45. 10.1016/S1567-1348(02)00089-8 [DOI] [PubMed] [Google Scholar]
- 14.Commission Regulation (EU) N°517/2011 of 25 May 2011 implementing Regulation (EC) No.°2160/2003 of the European Parliament and the Council as regards a union target for the reduction of the prevalence of certain Salmonella serotypes in laying hens of Gallus gallus and amending Regulation (EC) No. 2160/2003 and Commission Regulation (EU) No. 200/2010. [cited 2011 Oct 24].
- 15.Note de Service DGAL/SDSSA/N2010–8026 du 27 janvier 2010. Ministère de l’Alimentation de l’Agriculture et de la Pêche, Paris, France [cited 2011 Oct 24]. http://agriculture.gouv.fr/bulletin-officiel