Abstract
SUMOylation is emerging as an important mechanism for modulating protein function in many cell types. A large variety of proteins have been proposed as SUMO targets based on the presence of a consensus SUMOylation core motif (ψ-K-x-D/E). In neurons these include multiple synaptic proteins but it has not been established whether proteins carrying this motif are SUMOylated either in vitro or in vivo. Here we use a bacterial SUMOylation assay to systematically test for SUMO-1 modification of a selection of neuronal proteins containing one or more amino acid sequences predicted as high-probability SUMOylation sites in computer-based searches. Of the 39 proteins analysed only 14 sites were posttranslationally modified by SUMO-1, including the group III metabotropic glutamate receptors and the kainate receptor subunit GluR7. These results identify new candidate proteins that may be involved in the SUMO regulation of synaptic activity and also demonstrate that the presence of the ψ-K-x-D/E motif is not sufficient to indicate that a protein can be SUMOylated in this bacterial system.
Keywords: SUMO, Posttranslational modification, SUMOylation, Metabotropic glutamate receptors, Group III mGluRs, Kainate receptor, GluR7, Synaptic protein
SUMO is an ~11 kDa protein which is conjugated to lysine residues in substrate proteins in an enzymatic cascade analogous to that of the ubiquitin pathway [15]. Yeast contain only one SUMO protein, Smt3, whereas mammals possess 4 SUMO isoforms, designated SUMO-1 to SUMO-4 [2]. SUMO-1 shares ~18% homology with ubiquitin, however, despite the low sequence homology, SUMO proteins have a very similar three-dimensional structure to ubiquitin [1,21]. The SUMO proteins can be classed into two subfamilies – SUMO-1 and SUMO-2/3. In their conjugatable forms, SUMO-2 and -3 differ only in 3 N-terminal residues and have yet to be functionally distinguished (and are therefore often collectively referred to as SUMO-2/3), however they only share ~50% homology with SUMO-1 [10,17]. The role of SUMO-4 is unclear since it does not appear to be conjugated to substrate proteins due to a proline residue which prevents maturation of the immature polypeptide [24].
SUMO is conjugated to target proteins via four sequential enzymatic steps. The four C-terminal residues are cleaved from nascent SUMO-1 by members of the SENP family of proteins, exposing a di-glycine motif which is conjugated to target proteins. This conjugatable SUMO is then activated in an ATP-dependent manner by the E1 enzyme, a dimer of SAE1/SAE2, and passed to the active-site cysteine of the E2 ‘conjugating’ enzyme Ubc9 [10,15,21]. In many cases, Ubc9 is sufficient for the transfer of activated SUMO to target proteins, however a number of E3 ‘ligase’ proteins have been described, which facilitate transfer of SUMO from Ubc9 to the substrate in vivo. However, each of the described E3 enzymes appears to act by binding Ubc9 and/or the substrate, bringing them into a position more conducive to SUMO transfer. Thus, the actual transfer of SUMO to the substrate is always dependent on Ubc9. Because of this, SUMOylation generally occurs in a consensus motif, which is directly bound by Ubc9 [10,29]. This consensus motif can be described as ψ-K-x-D/E, where ψ is a large hydrophobic residue, K is the target lysine, x can be any residue, and D/E are aspartate or glutamic acid (acidic residues).
Several computer algorithms have been designed that search protein sequences for potential SUMOylation sites. However, these are limited since the consensus sequence contains only four residues and is relatively degenerate. For example, of 5884 open reading frames (ORFs) in Saccharomyces cerevisiae, there are 2799 occurrences of the motif [IVL]-K-x-E in 1913 ORFs [15]. Thus, although it has not been established experimentally, only a small proportion of these ‘hits’ are likely to be bona fide SUMO substrates. Attempts to improve SUMO site prediction have focused on defining extended SUMOylation consensus motifs. The negatively-charged SUMOylation motif (NDSM) is based on the observation that many SUMO substrates contain an acidic patch of amino acids downstream of the ψ-K-x-D/E core motif that interacts with a corresponding basic patch on Ubc9 [36]. Similar to the NDSM, the phosphorylation-dependent SUMOylation motif (PDSM) is defined by a phosphorylated serine residue five residues downstream of the ψ-K-x-D/E motif [13]. In this case rather than being encoded in the primary protein sequence, the acidic patch is provided by phosphorylation of the downstream serine that can act as a signal for SUMOylation.
Recently, as part of a study of SUMOylation of the kainate receptor subunit GluR6 we showed that there are multiple, as yet unidentified synaptic SUMO substrates [20]. Furthermore, together with others we have shown that ischemia causes a massive upregulation of protein SUMOylation in brain that may a represent a neuroprotective mechanism [4,19,37]. A key question, therefore, is what proteins are targets for SUMOylation in neurons. To identify potential synaptic SUMO substrates we performed a bioinformatic screen of candidate synaptic proteins for high-probability SUMOylation sites using SUMOplot, and the NDSM motif. We then directly tested proteins that scored highly in these screens in biochemical SUMOylation assays. Because immunoprecipitation of SUMOylated proteins from mammalian cells is particularly challenging due to the very low levels of SUMO modification and the extensive deSUMOylation activity (for reviews see [10,15,21]), we used a recombinant bacterial SUMOylation assay [33,34]. In all we tested 39 proteins with a total of 58 high-probability SUMOylation sequences including 4 NDSMs. Our data show that the presence of consensus sequences is a relatively poor indicator of actual protein SUMOylation in this assay system.
Plasmid Constructs
pE1E2S1, a bacterial expression vector containing a fusion of SAE1/2, Ubc9 and SUMO-1 [33,34] was obtained from Dr Hisato Saitoh (Kumamoto University, Japan). GST-tagged bacterial expression vector receptor C-termini and GST-syntenin were generated by Dr Helene Hirbec while in our lab. GST-fusions of synaptotagmin II and III were from Dr Gianpietro Schiavo (Cancer Research UK, London, UK), GST-fusions of sorting nexins 1, 2, 4 and 27 were from Dr Peter Cullen (University of Bristol), GST-fusion of full-length p53 was from Dr Kevin Gaston (University of Bristol), 6×His-tagged β-2-adaptin was from Dr Tomas Kirchhausen (Harvard Medical School, MA, USA) and GST-tagged CaV2.2 I-II linker region was from Dr Annette Dolphin (University College London, UK).
Antibodies
The sources and dilutions of antibodies were; goat polyclonal anti-GST (GE Healthcare: 1:1000), rabbit polycolonal anti-β-2-adaptin (Santa Cruz, 1:200) rabbit polyclonal anti-SUMO-1 (Santa Cruz, 1:200), mouse monoclonal anti-6×His (Roche, 1:1000) mouse monoclonal anti-CamKIIα (Santa Cruz, 1:200), mouse monoclonal anti-HA-tag antibody (Santa Cruz, 1:200) and HRP-conjugated anti-mouse, anti-goat or anti-rabbit antibodies (Sigma, 1:10000).
Bacterial SUMOylation Assay
The bacterial SUMOylation assay was performed as described previously [33].
Purification of GST-tagged Proteins
One millilitre of induced bacterial culture was spun down at 16,000 × g for 1 min and the supernatant discarded. To each pellet, 0.5 ml lysis buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 20 mM NEM, 1% (v/v) Triton X-100) was added, and the cells lysed by sonication. Insoluble material was removed by spinning at 16,000 × g for 10 min and the supernatant collected. To each supernatant, 2μl 2-β-marcaptoethanol was added to neutralise unreacted NEM, which could interfere with GST binding to glutathione by alkylation of the cysteine residue of glutathione. Thirty microlitres (bed volume) of glutathione sepharose (Sigma) was added and the sample incubated on a rotating wheel at 4 °C for 30 min. Beads were then washed four times with PBS. Proteins were then eluted from the beads by boiling in SDS-PAGE sample buffer.
SDS-PAGE and Western Blotting
Proteins were separated on 6–12% acrylamide gels and transferred onto PVDF membrane (Millipore). Membranes were blocked in TBST containing 5% (w/v) powdered skimmed milk before incubation with the primary antibody diluted in TBST/5% milk for 1–16 h. After washing with TBST, membranes were incubated with secondary antibodies diluted in TBST/5% milk for 1 h and after further extensive washing in TBST, developed with enhanced chemiluminescence substrate (Roche).
The presence of E1, E2 and SUMO is the minimal requirement for target protein SUMOylation [23]. E3 ligase enzymes facilitate SUMO transfer in vivo but in many cases, are not essential [6,23,27]. In the bacterial SUMOylation assay a single vector encoding E1, E2 and SUMO-1 or SUMO-2 is cotransfromed with a vector expressing the potential SUMO target protein. This system has been validated for multiple known target proteins and it is effective for both short fragments and full-length proteins [34]. It should be noted, however, that the lack of SUMOylation in this bacterial assay does not necessarily preclude the possibility that the endogenous protein might be SUMOylated under specific conditions in the host cell where, for example, E3 proteins are present. Protein SUMOylation is detected by probing the bacterial lysates for the protein of interest by Western blots in the presence or absence of the SUMO vector. Where SUMOylation occurs an additional higher molecular weight band of the substrate protein is detected. Alternatively the potential SUMO target can by purified. This usually involves glutathione affinity chromatography of GST-tagged candidate proteins or protein fragments and allows both band shift analysis and anti-SUMO antibody Western blotting. We have previously used this approach to determine the site of SUMO modification of GluR6 [20].
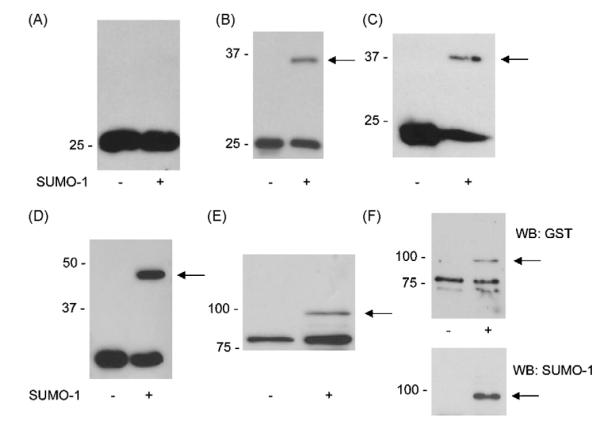
In this study we initially performed experiments to confirm the efficacy of the bacterial system in our hands by testing a selection of relatively well characterised SUMOylation targets, either as short peptide fragments or protein domains containing the known SUMOylation site, or as full-length protein (Fig. 1). The SUMO targets used were PML [31], tested as an 11 amino acid fragment fused to GST; RanGAP [22], tested as the entire 6×His-tagged C2 domain and full-length GST-tagged p53 [27]. All of these targets were consistently and robustly SUMOylated in the bacterial assay whereas free GST was not. These results confirm that proteins known to be SUMOylated in mammalian cells are efficiently SUMOylated in this bacterial system. Further, the fact that GST itself, which contains a high-probability SUMOylation site is not SUMOylated shows that the system is discriminatory and does not SUMOylate all proteins containing consensus sequences. Thus, our control data indicate that the bacterial SUMOylation system provides a useful biochemical screening tool, beyond simple prediction from sequence analysis, to identify targets that can be SUMOylated in vivo.
Fig. 1.
Efficient SUMOylation of known substrate proteins. Vectors encoding GST, or known SUMOylation substrates were transformed into bacteria with or without the SUMOylation plasmid (indicated by ‘+’ or ‘−’). Crude bacterial lysates were then subjected to Western blotting for GST or 6×His, as appropriate. SUMOylated species are indicated by an arrow. (A) GST alone, (B) GST-tagged PML (residues 485–495), (C) 6×His-tagged RanGAP (entire C2 domain), (D) GST-tagged GluR6a (intracellular C-terminus), (E) GST-p53 (full length), (F) GST-p53 was purified on glutathione beads and probed for GST (upper panel) or SUMO-1 (lower panel), confirming the band shift seen in (E) is due to SUMOylation of p53.
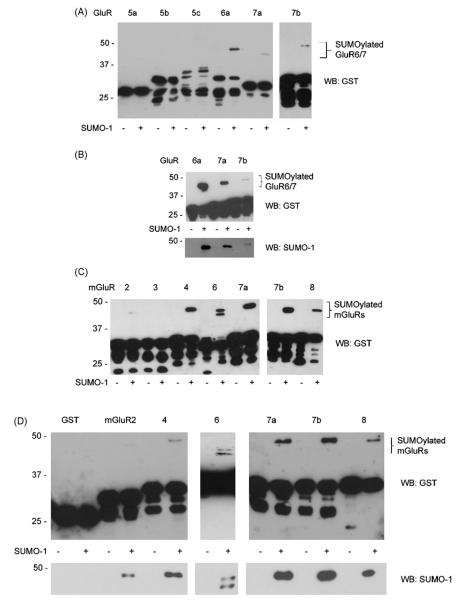
We next tested a series of candidate synaptic proteins (Table 1). The intracellular C-terminal domains of both metabotropic and ionotropic glutamate receptors are major sites of protein:protein interaction that dictate the trafficking and function of these receptors [8,25]. Database searches revealed potential SUMOylation sites in many of these C-terminal regions. As shown in Fig. 2, we reconfirmed SUMOylation of the kainate receptor subunit GluR6 and, interestingly, identified the subunit isoforms GluR7a and 7b as additional SUMOylation targets. The presynaptic Group III metabotropic glutamate receptor mGluR8 has also been previously reported as a SUMO target [32]. Here we confirmed that observation but additionally show that mGluRs 2, 4, 6, 7a and 7b are all also robustly SUMOylated. In contrast the NMDA receptor subunit NR1a and the metabotropic GABAB receptor subunits were not subject to SUMO conjugation in this assay system. The AMPAR subunits (GluR1-4) were not SUMOylated although intriguingly GluR2 and GluR3 occasionally showed a very weak and inconsistent SUMO signal.
Table 1.
List of proteins screened and their SUMOylation status in the bacterial assay
| Protein | Region | High-probability SUMOplot sites | NDSMs | Potentially modified sites | SUMO modification in bacterial assay |
|---|---|---|---|---|---|
| Ionotropic glutamate receptor subunits | |||||
| GluR1 | Intracellular CT | − | |||
| GluR2S | Intracellular CT | 1 | 1 | + | |
| GluR2L | Intracellular CT | − | |||
| GluR3 | Intracellular CT | 1 | + | ||
| GluR4 | Intracellular CT | − | |||
| GluR5a | Intracellular CT | − | |||
| GluR5b | Intracellular CT | − | |||
| GluR5c | Intracellular CT | − | |||
| GluR7a | Intracellular CT | 1 | 1 | ++ | |
| GluR7b | Intracellular CT | 1 | 1 | ++ | |
| NR1a | Intracellular CT | − | |||
| Metabotropic glutamate receptor isoforms | |||||
| mGluR1 | Intracellular CT | − | |||
| mGluR2 | Intracellular CT | 1 | + | ||
| mGluR3 | Intracellular CT | − | |||
| mGluR4 | Intracellular CT | 1 | 1 | +++ | |
| mGluR5a | Intracellular CT | 1 | − | ||
| mGluR5b | Intracellular CT | 1 | − | ||
| mGluR6 | Intracellular CT | 1 | ++ | ||
| mGluR7a | Intracellular CT | 1 | 1 | +++ | |
| mGluR7b | Intracellular CT | 1 | 1 | +++ | |
| mGluR8a | Intracellular CT | 1 | 1 | 1 | +++ |
| GABAB receptor subunits | |||||
| GABAB1(a–c) | Intracellular CT | − | |||
| GABAB1(d) | Intracellular CT | − | |||
| GABAB2 | Intracellular CT | − | |||
| Receptor interacting proteins | |||||
| β-2-adaptin | Full length | 8 | 1 | − | |
| CamKIIα | Full length | 4 | − | ||
| NSF | Full length | 3 | 1 | − | |
| PICK1 | Full length | 2 | − | ||
| PSD-95 | Full length | 2 | − | ||
| GISP | Full length | 6 | 3 | ++ | |
| Trafficking pathway proteins | |||||
| SNX1 | Full length | 3 | − | ||
| SNX2 | Full length | 4 | 1 | ++ | |
| SNX4 | Full length | 2 | − | ||
| SNX27 | Full length | 4 | − | ||
| Presynaptic proteins | |||||
| β-SNAP | Full length | − | |||
| Synaptotagmin II | Full length | 4 | 1 | − | |
| Synaptotagmin III | Full length | 1 | − | ||
| Syntenin | Full length | 3 | − | ||
| CaV2.2 | I–II Loop | 3 | − | ||
Fig. 2.
Proteins identified as SUMOylation substrates. (A) The intracellular C-termini of the kainate receptor subunits GluR5–7 were expressed as GST-fusion proteins in bacteria with (+) or without (−) the SUMOylation plasmid. Crude bacterial lysates were Western blotting for GST. (B) Proteins showing a band shift when co-expressed with the SUMOylation plasmid in (A) were purified on glutathione beads and subjected to western blotting for GST and SUMO-1, to confirm the band shift seen corresponded to SUMO-1 modification. (C) GST-fusions of mGluR intracellular C-termini. (D) GST-mGluR C-termini showing a band shift in (C) were purified and Western blotting for GST and SUMO-1.
Since SUMOylation can modulate protein:protein interactions[10,21] we also tested the SUMOylation potential of the important synaptic receptor interacting proteins PICK1 [7], β2-adaptin [18], GISP [16], PSD-95 [12] and syntenin [14] (Table 1). Of these proteins only GISP showed evidence of SUMOylation in the bacterial assay despite each containing high-probability sites and an NDSM in β-2-adaptin.
Several candidate presynaptic proteins were also tested. The vesicle release proteins synaptotagmin II and III [26] and β-SNAP [30] showed no indication of SUMOylation, again despite high-probability SUMOylation sites in synaptotagmin II and III, and an NDSM in synaptotagmin II (Table 1). Finally in this series we tested some proteins involved in generalised protein trafficking pathways in mammalian cells. We chose the sorting nexin family of proteins as models [3]. Sorting nexins 1, 2, 4 and 27 each contain high-probability SUMOylation sites, however of these, only SNX2 showed any evidence of SUMO modification.
It is becoming increasingly apparent that SUMOylation has multiple and diverse roles in cell function both inside and outside the nucleus [10,21]. Several previous studies have identified novel SUMO targets in yeast and mammalian cells using proteomics-based approaches [5,9,11,28,35] and the number of validated cytosolic and plasma membrane SUMO targets is growing rapidly but there remain many hundreds of potential candidate proteins that have not yet been tested.
In this study we used a targeted approach to test a selection of predicted candidate synaptic proteins for SUMO modification in an efficient and discriminating bacterial assay. Our results suggest that the majority of proteins containing consensus high-probability SUMOylation sites are not in fact SUMOylated (Table 1).
One possible reason for our data is that the bacterial assay we used can generate falsely negative results, i.e. proteins that are SUMOylated at synapses are not efficiently SUMOylated in this reconstituted system. However, in the initial characterisation of the bacterial system a range of full-length mammalian proteins including thymine DNA glycosylase (TDG), p53 and Tonalli-related SP-ring finger domain (TONAS) [33,34] were shown to be efficiently SUMOylated. Furthermore, all of the proteins we tested that are already known to be SUMOylated yielded positive results. We demonstrate that this bacterial system efficiently SUMOylates short peptides (PML and GluR6), a defined protein domain (RanGAP) and a full-length protein (p53) of known SUMO targets. In each case these targets were robustly modified to the extent that 30–60% of the total expressed target protein was SUMOylated. Therefore, although this system is unlikely to be a fully accurate representation of the SUMOylation state of proteins in vivo in every case the positive control known SUMO target protein tested was efficiently SUMOylated. These data suggest that the bacterial assay provides a useful and, as far as we can ascertain, reliable method for testing whether candidate proteins identified using sequence analysis information are indeed bona fide SUMO targets.
Of the 58 high-probability SUMOplot sites (including 4 NDSMs) tested, only 14 are genuinely SUMOylated. Of these 14, 1 is an NDSM, 11 are high-probability SUMOplot sites, and 3 were not predicted by either method (Table 1). Based on these data, SUMOplot predictions yield a ~19% success rate at predicting SUMOylated proteins, and the NDSM has an improved ~25% success rate. However, while the NDSM is present in a large number of genuinely SUMOylated proteins and has contributed to understanding the molecular mechanisms of SUMO modification, the presence of this motif is not sufficient to definitively predict SUMO modification.
PIAS1 binding to group III mGluRs and SUMOylation the C-terminus of mGluR8 has been reported previously [32].Wehave extended those initial observations by demonstrating that all group III mGluRs can be SUMOylated. We also show that, in addition to the kainate receptor subunit GluR6, the subunit isoforms GluR7a and 7b can also be SUMOylated. Our results also suggest that the sorting nexin SNX2, which is a component of the mammalian retromer complex, can be SUMOylated.
Interestingly, three of the proteins SUMOylated in bacteria, GluR3, mGluR2 and mGluR6, were not predicted as SUMO targets by either SUMOplot or NDSM. While SUMOylation of these proteins will require further confirmation in mammalian cells, it has been reported that mGluR6 binds the SUMO E3 enzyme PIAS1 and that mGluR8 is SUMOylated [32]. Despite being highly reproducible, we are somewhat cautious of the mGluR2 SUMOylation result. It was modified to a much lower level than either previously validated SUMO substrates or group III mGluRs, it has significant sequence identity with mGluR3 which was not SUMOylated and, unlike all of the group III mGluRs, the group II mGluR2 has not been reported to bind PIAS1. In addition, SUMOylation of GluR3 was particularly weak and inconsistent, indicating that GluR3, along with mGluR2, may be a false positive result in this assay, suggesting that rather than underestimating the potential for target protein SUMOylation, the bacterial system may overestimate it.
Overall it appears that ψ-K-x-D/E sites are not necessarily required for Ubc9 binding and SUMOylation. Conversely, many proteins with very high-probability consensus sequences are not SUMOylated. Taken together these data suggest that secondary protein structure is an important consideration. Clearly, if a protein possesses high-probability SUMOylation sites but they are hidden deep inside the core of the protein structure it is unlikely to be SUMOylated without conformational alteration. If SUMOylation site prediction is to improve beyond its current boundaries, it will likely have to take account of secondary and possibly tertiary protein structure. At its simplest, Ubc9 may predominantly modify relatively unstructured regions of proteins which allow its binding to lysine residues whereas a more complicated scenario is that Ubc9 requires complex binding faces to utilise non adjacent residues in the primary structure, for efficient modification to occur.
Acknowledgements
We are grateful to the MRC, the BBSRC and the EU (GRIPPANT) for financial support. We wish to thank again the many labs that provided cDNA constructs for this study. We also thank Philip Rubin and Patrick Tidball for technical assistance and Dan Rocca, Stéphane Martin, Sriharsha Kantamneni and David Holman for advice.
References
- [1].Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- [2].Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- [3].Carlton JG, Cullen PJ. Sorting nexins. Curr. Biol. 2005;15:R819–R820. doi: 10.1016/j.cub.2005.10.012. [DOI] [PubMed] [Google Scholar]
- [4].Cimarosti H, Lindberg C, Bomholt SF, Rønn LCB, Henley JM. Increased protein SUMOylation following focal cerebral ischemia. Neuropharmacology. 2008;54:280–289. doi: 10.1016/j.neuropharm.2007.09.010. [DOI] [PubMed] [Google Scholar]
- [5].Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- [6].Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- [7].K K. Dev. PDZ domain protein–protein interactions: a case study with PICK1. Curr. Top. Med. Chem. 2007;7:3–20. doi: 10.2174/156802607779318343. [DOI] [PubMed] [Google Scholar]
- [8].Enz R. The trick of the tail: protein–protein interactions of metabotropic glutamate receptors. Bioessays. 2007;29:60–73. doi: 10.1002/bies.20518. [DOI] [PubMed] [Google Scholar]
- [9].Ganesan AK, Kho Y, Kim SC, Chen Y, Zhao Y, White MA. Broad spectrum identification of SUMO substrates in melanoma cells. Proteomics. 2007;7:2216–2221. doi: 10.1002/pmic.200600971. [DOI] [PubMed] [Google Scholar]
- [10].Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- [11].Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 2005;280:5004–5012. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- [12].Gomperts SN. Clustering membrane proteins: it’s all coming together with the PSD-95/SAP90 protein family. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- [13].Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hirbec H, Martin S, Henley JM. Syntenin is involved in the developmental regulation of neuronal membrane architecture. Mol. Cell Neurosci. 2005;28:737–746. doi: 10.1016/j.mcn.2004.12.005. [DOI] [PubMed] [Google Scholar]
- [15].Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- [16].Kantamneni S, Correa SA, Hodgkinson GK, Meyer G, Vinh NN, Henley JM, Nishimune A. GISP: a novel brain-specific protein that promotes surface expression and function of GABA(B) receptors. J. Neurochem. 2007;100:1003–1017. doi: 10.1111/j.1471-4159.2006.04271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lapenta V, Chiurazzi P, van der Spek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- [18].Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36:661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- [19].Lee YJ, Miyake S, Wakita H, McMullen DC, Azuma Y, Auh S, Hallenbeck JM. Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J. Cereb Blood Flow Metab. 2007;27:950–962. doi: 10.1038/sj.jcbfm.9600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2007;8:948–959. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2, Biochem. Biophys. Res. Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- [24].Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation, Biochem. Biophys. Res. Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- [25].Palmer CL, Cotton L, Henley JM. The molecular pharmacology and cell biology of {alpha}-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol. Rev. 2005;57:253–277. doi: 10.1124/pr.57.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- [27].Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rosas-Acosta G, Langereis MA, Deyrieux A, Wilson VG. Proteins of the PIAS family enhance the sumoylation of the papillomavirus E1 protein. Virology. 2005;331:190–203. doi: 10.1016/j.virol.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- [30].Stenbeck G. Soluble NSF-attachment proteins. Int. J. Biochem. Cell Biol. 1998;30:573–577. doi: 10.1016/s1357-2725(97)00064-2. [DOI] [PubMed] [Google Scholar]
- [31].Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tang Z, El Far O, Betz H, Scheschonka A. Pias1 interaction and sumoylation of metabotropic glutamate receptor 8. J. Biol. Chem. 2005;280:38153–38159. doi: 10.1074/jbc.M508168200. [DOI] [PubMed] [Google Scholar]
- [33].Uchimura Y, Nakamura M, Sugasawa K, Nakao M, Saitoh H. Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Anal. Biochem. 2004;331:204–206. doi: 10.1016/j.ab.2004.04.034. [DOI] [PubMed] [Google Scholar]
- [34].Uchimura Y, Nakao M, Saitoh H. Generation of SUMO-1 modified proteins in E. coli: towards understanding the biochemistry/structural biology of the SUMO-1 pathway. FEBS Lett. 2004;564:85–90. doi: 10.1016/S0014-5793(04)00321-7. [DOI] [PubMed] [Google Scholar]
- [35].Wykoff DD, O’Shea EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- [36].Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J. Cereb Blood Flow Metab. 2007:1–11. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]