Abstract
WTX encodes a tumor suppressor, frequently inactivated in Wilms tumor, with both plasma membrane and nuclear localization. WTX has been implicated in β-catenin turnover, but its effect on nuclear proteins is unknown. We report an interaction between WTX and p53, derived from the unexpected observation of WTX, p53 and E1B 55K colocalization within the characteristic cytoplasmic body of adenovirus transformed kidney cells. In other cells without adenovirus expression, the C terminal domain of WTX binds to the DNA binding domain of p53, enhances its binding to CBP, and increases CBP/p300-mediated acetylation of p53 at Lys 382. WTX knockdown accelerates CBP/p300 protein turnover and attenuates this modification of p53. In p53-reconstitution experiments, cell cycle arrest, apoptosis, and p53-target gene expression are suppressed by depletion of WTX. Together, these results suggest that WTX modulates p53 function, in part through regulation of its activator CBP/p300.
INTRODUCTION
Inactivation of WTX/AMER1/FAM123B is the most frequent genetic event in sporadic Wilms tumor, reported in up to 30% of cases (Rivera et al., 2007). The location of WTX on the X chromosome is noteworthy in that “one hit” somatic inactivation, targeting the single allele in males or the active X allele in females, is sufficient for gene inactivation. Germline mutations of WTX have recently been shown to cause Osteopathia Striata with Cranial Sclerosis (OSCS), an X-linked syndrome associated with bone malformations and mental retardation (Jenkins et al., 2009). Inactivation of Wtx in the mouse leads to abnormalities in mesenchymal differentiation, most prominent in bone, adipose tissue, kidney and heart (Moisan et al., 2011). WTX therefore plays a major role in both tumor suppression and normal differentiation. WTX does not contain recognizable domains and its functional properties are yet to be defined. Protein interaction studies have suggested that WTX associates with the APC complex and may negatively regulate β-catenin stability (Grohmann et al., 2007; Major et al., 2007), while recent studies have also suggested a positive effect on WNT signaling through tethering CK1γ, GSK3β, Axin and LRP6 (Tanneberger et al., 2011). Cellular localization studies indicate that most WTX resides at the plasma membrane (Grohmann et al., 2007), although a shorter splice form appears primarily in the nucleus, and inhibition of nuclear export leads to accumulation of full length WTX within specific nuclear structures known as paraspeckles (Rivera et al., 2009). While the nuclear function of WTX is unclear, it modulates the transcriptional activity of WT1, another Wilms tumor suppressor that encodes a master transcriptional regulator of kidney and genitourinary development (Rivera and Haber, 2005; Schedl and Hastie, 1998). Thus, WTX may exhibit distinct functions in nuclear, cytoplasmic, and plasma membrane compartments.
DNA tumor viruses have provided crucial tools to understand essential cellular regulatory mechanisms, including transcription, cell cycle control, and DNA replication (Levine, 2009). Viral proteins target the cellular machinery, characteristically inactivating pRB family members to trigger aberrant cellular proliferation, while suppressing the compensatory p53-mediated apoptotic stimuli. The adenovirus protein E1B 55K is remarkable in that it suppresses p53-mediated transactivation (Yew et al., 1994), while also preventing the activating acetylation of p53 by PCAF (Liu et al., 2000), mediating nuclear export of p53 through sumoylation of p53 (Muller and Dobner, 2008; Wimmer et al., 2010), and functioning as an E3 ubiquitin ligase to mediate the degradation of p53 (Harada et al., 2002; Querido et al., 2001). Ultimately, E1B 55K sequesters p53 within a single perinuclear cytoplasmic body, recently identified as an aggresome (Liu et al., 2005; Zantema et al., 1985). In addition to p53, the cytoplasmic E1B body also targets other important cellular regulatory proteins such as components of the Mre11-Rad50-Nbs1 (MRN) complex, HDAC1/mSin3A, and WT1 (Liu et al., 2005; Maheswaran et al., 1998; Punga and Akusjarvi, 2000). The striking localization of proteins within the E1B body in adenovirus-infected cells may provide a clue to their interaction partners.
The TP53 tumor suppressor gene is frequently inactivated in most tumor types, although such mutations are very rare in Wilms tumor (Bardeesy et al., 1995; Bardeesy et al., 1994). Upon activation by genotoxic stress, p53 transactivates a set of well characterized target genes involved in cell cycle arrest and apoptosis (Laptenko and Prives, 2006; Vogelstein et al., 2000; Vousden and Prives, 2009). The activation of p53 itself is tightly regulated at the post-translational level, being modulated by phosphorylation, acetylation, and ubiquitylation. Among these, the acetylation of p53, resulting in enhanced stability and sequence specific DNA binding, is mediated by the histone acetyltransferases CBP/p300, PCAF, and hMOF/TIP60 (Gu and Roeder, 1997; Liu et al., 1999; Sakaguchi et al., 1998; Sykes et al., 2006; Tang et al., 2008), and negatively regulated by complexes containing the histone deacetylases HDAC1/2 or the NAD-dependent histone deacetylase SIRT1 (Juan et al., 2000; Luo et al., 2001; Luo et al., 2000; Vaziri et al., 2001). The key p53 regulator MDM2 has been shown to inhibit p53 acetylation by forming a ternary complex with CBP/p300 and p53, and by ubiquitylation of the same lysine residues that are targeted by acetylation (Ito et al., 2001; Kobet et al., 2000). While the precise consequences of p53 acetylation at different lysine residues by diverse acetyl transferases/deacetylases are uncertain, they may modulate differential activation of p53 target genes.
Here we report a physical and functional interaction between 2 WTX and p53 tumor suppressors, initially identified by the unexpected observation of WTX localization within the cytoplasmic E1B body of adenovirus transformed human embryonic kidney (HEK) 293 cells. WTX enhances the acetylation of p53 by CBP/p300, in part by regulating the turnover of these acetyltransferases and enhancing the p53-CBP interaction. Together, our observations suggest a role for WTX in the regulation of p53 activity.
RESULTS
Ectopic expression of WTX in HEK293 cells dissociates the E1B 55K-p53 containing cytoplasmic body and activates p53
In an effort to delineate WTX-dependent pathways, we generated doxycycline-inducible expression in the human embryonic kidney cell line HEK 293, and studied two clones (F-5 and F-18) with tightly regulated expression of full length WTX (Figure S1 A). Microarray expression profiles 12 hrs following WTX induction revealed enrichment of 13 up-regulated and 8 down-regulated pathways by Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005), with less than 5% False Discovery Rate (Tables S1 and S2). Notably, among these 13 up-regulated pathways, 9 pathways were functionally related to p53, indicating that endogenous p53 might be activated following ectopic expression of WTX (Table 1, Figure S1B). The p53-dependence of these WTX-mediated changes was confirmed by shRNA-mediated p53 knockdown, which diminished the induction of these transcripts following WTX expression (Figure S1C). Moreover, two activating post-translational modifications of p53 characteristically induced by ionizing radiation (IR), namely phosphorylation at Ser 15 and acetylation at Lys 382, were increased upon induction of WTX (Figure S1D). While activation of p53 is observed following a number of oncogenic or DNA damage-inducing stimuli, the effect of WTX appeared to be direct, based on cellular localization and protein association studies (see below).
Table 1.
Up-regulated gene sets in WTX inducible HEK293 cells (FDR < 5%). See also Table S1, S2 and Figure S1.
| NAME | SIZE | SIG | NES | FDR q- val |
RANK AT MAX |
|---|---|---|---|---|---|
| CELL CYCLE REGULATOR * | 24 | 7 | 2.131 | 0.001 | 969 |
| P53 PATHWAY * | 16 | 7 | 2.079 | 0.003 | 2384 |
| P53 GENES ALL * | 17 | 7 | 2.078 | 0.003 | 1202 |
| DNA DAMAGE SIGNALING * | 88 | 37 | 2.077 | 0.003 | 4473 |
| WNT TARGETS | 25 | 16 | 2.054 | 0.003 | 4058 |
| RADIATION SENSITIVITY * | 24 | 7 | 1.883 | 0.009 | 1482 |
| APOPTOSIS * | 67 | 25 | 1.877 | 0.009 | 3294 |
| HEMATOPOESIS RELATED TRANSCRIPTION FACTORS |
84 | 32 | 1.874 | 0.009 | 3319 |
| ETS PATHWAY | 18 | 9 | 1.841 | 0.012 | 5278 |
| P53 SIGNALING * | 93 | 35 | 1.812 | 0.017 | 4618 |
| P53 HYPOXIA PATHWAY * | 19 | 6 | 1.784 | 0.021 | 1285 |
| NGF PATHWAY | 19 | 6 | 1.718 | 0.036 | 2592 |
| DEATH PATHWAY * | 33 | 14 | 1.715 | 0.037 | 3245 |
NES: Normalized enrichment score
FDR: False discovery rate
p53 related pathways
HEK293 cells are remarkable in that endogenous p53 is inactivated by expression of the adenovirus type 5 E1B 55K oncoprotein, in part through its recruitment to the cytoplasmic E1B body. Surprisingly, we observed that WTX expression triggered disruption of the characteristic adenovirus E1B body in these cells and release of p53 into the nucleus (Figure 1). Indeed, p53 was clearly localized to the E1B body in uninduced F-5 cells, but doxyclicline induction of WTX triggered the disappearance of these cytoplasmic p53 foci and increased nuclear localization of p53 (Figure 1A). Similarly, induction of WTX led to dissociation of E1B 55K with relocalization from the focal cytoplasmic body to a larger cytoplasmic aggregation along with plasma membrane distribution (Figure 1B).
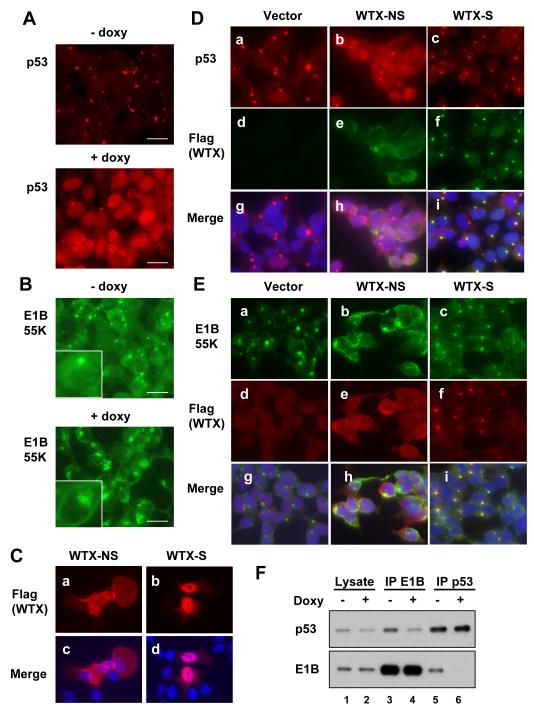
Figure 1. Cellular localization of p53, E1B 55K and WTX in the adenovirus-transformed HEK293 cells.
(A and B) Immunostaining of p53 (A) and E1B 55K (B) in HEK 293-derived F-5 cells with doxycycline-regulated WTX, encoding both full length and internally spliced transcripts. Flag-WTX was induced for 18 hr (+doxy) leading to relocalization of both p53 and E1B 55K from the characteristic cytoplasmic body. Insets in Figure 1B show magnified images of individual cells (Size bar: 20 μm).
(C) Normal cellular distribution of Flag-WTX-NS (non-spliceable cDNA) and Flag-WTX-S (spliced cDNA), demonstrated following transfection of constructs into U2OS cells (DAPI-stained nuclei in merged images).
(D) Immunostaining of p53, Flag-WTX-NS or Flag-WTX-S in lentiviral WTX-inducible HEK293 cells (pool). The normally nuclear WTX-S is recruited to the cytoplasmic body in HEK 293 cells, along with p53. WTX-NS maintains its plasma membrane and cytoplasmic localization, but disrupts the cytoplasmic body. p53 is redistributed to the nucleus from its characteristic localization in the cytoplasmic body.
(E) Immunostaining of E1B 55K, Flag-WTX-NS and Flag-WTX-S in lentiviral WTX-inducible HEK293 cells shows redistribution of E1B 55K to the plasma membrane in WTX-NS-expressing cells.
(F) Immunoprecipitation-Western blotting analysis, demonstrating that the coimmunoprecipitation of E1B 55K and p53 is reduced, following WTX induction in HEK 293 cells.
WTX cDNA encodes two distinct transcripts, resulting from an alternative splice junction that is internal to the cDNA (Rivera et al., 2009). In-frame deletion of 831 nucleotides produces a shortened spliced transcript (WTX-S), whose product is mainly nuclear in localization. Although full-length WTX is the predominant endogenous transcript, WTX-S expression becomes significant under experimental conditions due to internal splicing of the ectopic cDNA (examples: Figures 2C and 5B). To identify the localization of the unspliced full length WTX, we made use of a non-spliceable cDNA (WTX-NS) in which a nucleotide substitution at the splice donor site prevents generation of WTX-S (Rivera et al., 2009). Since antibodies that recognize WTX are inadequate for immunofluorescence studies, we generated Flag-tagged constructs to monitor their cellular localization. Consistent with previous reports using GFP-fusion constructs (Rivera et al., 2009), Flag-WTX-NS and Flag-WTX-S in U2OS cells were localized mainly to the plasma membrane and nucleus, respectively (Figure 1C). However, in E1B 55K-expressing HEK 293 cells, WTX-S was instead colocalized with both p53 and E1B 55K within the cytoplasmic body (Figure 1D and 1E). WTX-NS in HEK293 remained localized primarily in the plasma membrane, but its expression led to the disruption of the E1B body, the appearance of large cytoplasmic aggregates of E1B 55K, and remarkably, to plasma membrane localization of E1B 55K. p53 itself was released from the E1B body by ectopic expression of WTX-NS and accumulated in the nucleus (Figure 1D and 1E), contributing to the observed increased expression of p53 target genes. Thus, while WTX-S is recruited to the E1B 55K cytoplasmic body, the strong membrane targeting signal of WTX-NS may serve to redirect the localization of E1B 55K itself. Consistent with these cellular imaging changes, immunoprecipitation-immunoblotting analysis demonstrated that the physical interaction between E1B 55K and p53 was reduced by the ectopic expression of full length WTX (Figure 1F).
Figure 2. Physical associations between WTX, p53, and E1B 55K.
(A) Co-immunoprecipitation of Flag-WTX, E1B 55K and p53 in WTX inducible HEK 293-derived F-5 cells. Immunoprecipitation of cellular lysates using anti-Flag, E1B 55K antibodies or control IgG, followed by immunoblotting with antibodies against E1B 55K, p53 or Flag. Arrowheads indicate Flag-WTX-NS (NS) and Flag-WTX-S (S).
(B) shRNA knockdown of p53 does not reduce co-immunoprecipitation of Flag-WTX and E1B 55K in F-5 cells. Arrowheads indicate Flag-WTX-NS (NS) and Flag-WTX-S (S).
(C) WTX and p53 are coimmunoprecipated in U2OS cells, which lack adenovirus E1B 55K. Flag-WTX and TP53 were co-transfected, followed by immunoprecipitation using either anti-Flag or anti-p53 antibodies and immunoblotting. Arrowheads indicate Flag-WTX-NS (NS) and Flag-WTX-S (S).
(D) Mapping WTX-binding domain within p53 by immunoprecipitation-Western blotting in H1299 cells. Flag-WTX and various HA-p53 fragments were transfected into H1299 cells, immunoprecipitated with anti-Flag antibody and analyzed by immunoblotting. The DNA binding domain of p53 binds to WTX (Renilla: non-specific (NS) protein control). The WTX domain involved in binding p53 (aa 561-1135) is shown in Supplementary (Figure S2B).
(E) Mapping WTX-binding domain within E1B 55K. Flag-WTX and various HA-E1B 55K fragments were transfected into H1299 cells, immunoprecipitated using anti-Flag antibody and analyzed by immunoblotting. The central p53 binding domain of E1B 55K binds to WTX (V: empty vector).The WTX domain binding to E1B 55K (aa 561-850) is shown in Supplementary (Figure S2C).
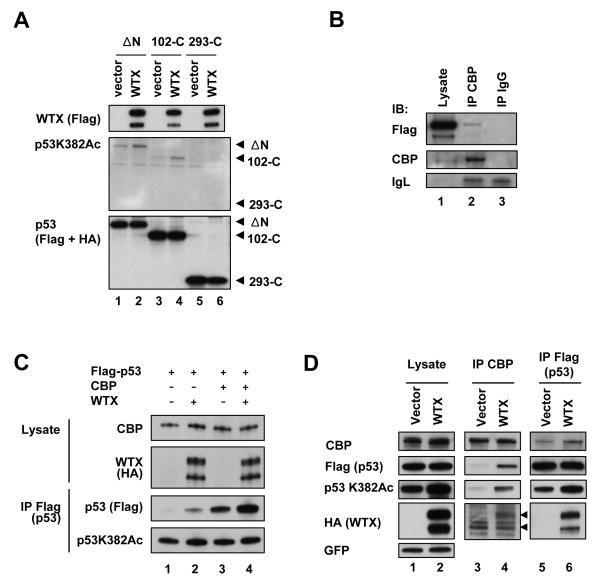
Figure 5. Enhancement of CBP/p300-mediated acetylation of p53 by WTX.
(A) Enhanced acetylation of truncated p53 constructs by WTX. Western analysis of tagged (Flag or HA) p53 constructs, following cotransfection with WTX into H1299 cells. p53 constructs are deleted for the N-terminal 60 amino acids encoding the transactivation domain (ΔN, Flag-p53), the N-terminal 102 amino acids (102-C, HA-p53: also shown as p53 construct C in Figure 2D), or the N-terminal 293 amino acids (293-C, HA-p53: construct E in Figure 2D). The CBP/p300 binding domain of p53 has been mapped to amino acids 1-83.
(B) Co-immunoprecipitation of WTX and endogenous CBP by immunoprecipitation-immunoblotting analysis in H1299 cells. (IgG: negative control).
(C) WTX enhancement of CBP-mediated p53 acetylation. H1299 cells were transfected with indicated combinations of plasmids and acetylation of p53 was analyzed by immunoprecipitation-Western blot analysis using antibodies against Lys382 acetylated p53, Flag (total p53), CBP and HA (WTX). Protease inhibitor MG132 was added to cells to prevent degradation of transfected p53 and produce comparable baseline expression.
(D) Enhancement of p53-CBP interaction by WTX. Flag-p53 and CBP were co-transfected into H1299 cells along with HA-WTX or empty vector, followed by immunoprecipitation with anti-Flag (p53) or anti-CBP antibody, and co-immunoprecipitation of p53 and CBP were analyzed. Arrowheads indicate the positions for WTX-NS and WTX-S. GFP is a transfection control. See also Figure S5.
Of note, the interaction between WTX and the p53-EIB 55K complex identified by cellular imaging studies was further supported by an independent line of investigation. Identification of WTX-interacting proteins in HEK293 cells, using immunoprecipitation-mass spectrometry analysis, revealed several proteins known to be associated with the E1B body, including E1B 55K itself, p53, Mre11 and Rad50, consistent with a physical interaction between WTX and known components of the E1B body (Table S3).
WTX interacts with both p53 and E1B 55K
To further define the protein interaction between WTX, p53 and/or EIB 55K, we undertook immunoprecipitation-immunoblotting studies using HEK293 (F-5 clone) cell lysates. Both E1B 55K and p53 were coprecipitated with Flag-tagged WTX, following extraction using high salt (500 mM NaCl) conditions (Figure 2A, lane 2). Reciprocally, both WTX and p53 were found in E1B 55K immunoprecipitates (Figure 2A lane 3). These observations may be explained either by a ternary complex including WTX, p53 and E1B 55K, or by distinct pairwise interactions. Since the E1B 55K-p53 interaction was reduced by overexpression of WTX (Figure 1F), we reasoned that WTX might compete for binding with these proteins, leading to their dissociation. We first tested whether WTX can interact with p53 and E1B 55K independent of each other. The WTX-E1B 55K interaction was not affected by shRNA knockdown of p53 in HEK293 cells (Figure 2B). WTX was also coimmunoprecipitated with transfected E1B 55K in p53(-/-) H1299 cells (Figure 2E lane 6). Similarly, the WTX-p53 interaction was independent of E1B 55K, as demonstrated by their coimmunoprecipitation from U2OS and H1299 cells that do not express E1B 55K (Figures 2C and S2B). Together, these observations suggest distinct protein interactions involving WTX-p53 and WTX-E1B 55K.
We mapped the interaction domains for these interactions using a panel of deletion constructs. E1B 55K is known to bind the N-terminal transactivation domain of p53 (Kao et al., 1990). WTX interacted with the central DNA binding domain of p53 (amino acids 102-292, Figure 2D lane 8), whereas the N-terminal domain of p53 was dispensable to its interaction with WTX (Figure 2D lane 7). In contrast, the central domain of E1B 55K which is required for binding to p53 (Grand et al., 1999) was also indispensable for its binding to WTX (amino acids 225-355, Figure 2E lanes 7 and 8). Thus, WTX and p53 may compete with each other for binding to E1B 55K. The WTX domains required for binding to p53 or E1B 55K were also mapped using coimmunoprecipiation studies in H1299 cells. The C-terminal half of WTX (aa 561-1135) was sufficient for both p53 and E1B binding. Binding of WTX to E1B 55K mapped to amino acids 561-850 of WTX, but binding to p53 required additional C-terminal sequences (lanes 15 of Figure S2B and C), suggesting that these interacting regions within WTX are overlapping but not identical (Figure S2). Notably, both E1B 55K and p53 interact with the two splice isoforms of WTX (lanes 12 of Figure S2B and C). However, the physical disruption of the E1B body appears to require the membrane association domain of WTX-NS, which is lacking in WTX-S (Figures 1D and E). While HEK293 cells express low levels of endogenous WTX, ectopic WTX expression levels are required to perturb the p53-E1B 55K interaction. Taken together, domain mapping experiments suggest that the dissociation of the E1B 55K-p53 interaction may result primarily from competition by WTX for binding to E1B 55K.
WTX modulates acetylation of p53 at Lys 373/382
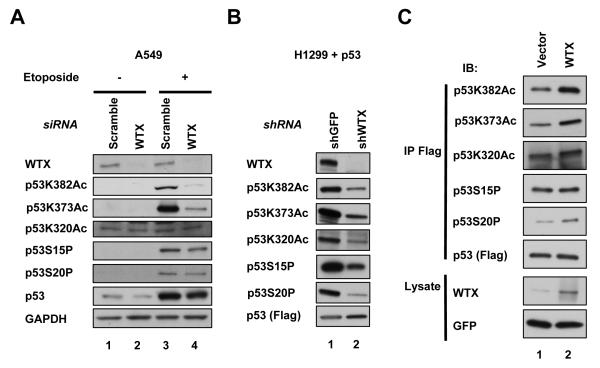
To test for functional consequences of WTX expression on p53 activation, we measured phosphorylation of Ser 15 and 20 and acetylation of Lys 373, 382 and 320, the most extensively studied post-translational modifications of p53, following either ectopic expression or knockdown of WTX. Acetylation of Lys 373/382 and Lys 320 represent target sites for the histone acetyl transferases CBP/p300 and PCAF, respectively. In WTX overexpressing HEK 293 cells, we observed enhanced phosphorylation of p53 Ser 15 and acetylation of Lys 382, but not acetylation of Lys 320 or phosphorylation of Ser 20 (Figure S1D and data not shown). Consistent changes in Lys 373/382 acetylation were evident in non-adenovirus-transformed cells: coexpression of p53 and WTX in H1299 cells enhanced acetylation of Lys 382 (Figure 3C). In these experiments, the proteasome inhibitor MG132 was used to equalize ectopic p53 protein levels which are suppressed by co-transfection of WTX. Conversely, a reduction in p53 Lys 373/382 acetylation was observed in WTX-depleted p53-reconstituted H1299 cells (Figure 3B). Variable but consistent reduction in baseline and DNA damage (etoposide or UV)-induced acetylation of endogenous p53 at Lys 373/382 was also observed following depletion of WTX in A549 and U2OS cells lines, using either of two different small interfering (si) RNA or two short hairpin (sh) RNA constructs (Figures 3A, S3 A-D). Thus, expression of WTX was positively correlated with Lys 373/382 acetylation of p53. We also observed that acetylation of p53 Lys320 and phosphorylation of Ser 15 and 20 were positively associated with WTX but only in the context of ectopic expression of p53 (Figures 3B and C), and hence there were not as consistent as the WTX-enhanced Lys 373/382 acetylation.
Figure 3. Modulation of p53 Lys 382 acetylation by WTX.
(A) Analysis of etoposide-induced (20 μM for 2 hours) modifications of endogenous p53, including Lys 373, 382 and 320 acetylation, and Ser 15 and 20 phosphorylation, in A549 cells following WTX knockdown.
(B) Immunoblotting analysis of baseline p53 modifications following expression of Flag-p53 in H1299 cells with shRNA knockdown of WTX versus control (GFP).
(C) Analysis of p53 modifications, following ectopic expression of Flag-p53 and WTX in H1299 cells. Cells were treated for 6 hours with the proteasome inhibitor MG132 before harvesting cells to prevent degradation of p53. GFP was used for transfection control. See also Figure S3.
Since transfection of wild type p53 triggers cell death, we made use of inactive p53 mutants, to ensure that the p53 acetylation observed was not secondary to p53 activation itself. Transfection of the inactive P278A mutant (p53*) also showed increased acetylation at Lys 382 in the presence of ectopic WTX (Figure S3E, lane 2). Finally, we tested whether this effect could be an indirect result of WTX-induced genotoxic stress. Apoptosis caused by overexpression of full length WTX is attributable to WTX-NS, whereas ectopic expression of the spliced form WTX-S has no effect on cellular proliferation (data not shown). However, all forms of WTX constructs mediated comparable levels of p53 Lys 382 acetylation, suggesting that this effect is not a secondary consequence of cellular stress caused by WTX overexpression (Figure S3E). Together, these results suggest that WTX can affect p53 activation, and that this effect is independent of E1B 55K.
WTX regulates protein stability of CBP/p300 and enhances CBP-p53 interaction
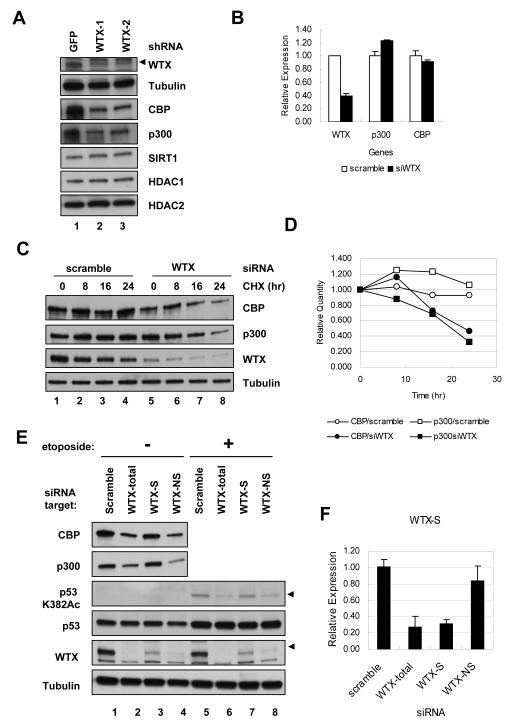
Acetylation of p53 Lys 373/382 is mediated by the acetyl transferases CBP/p300 and it is negatively regulated by the deacetylases HDAC1, HDAC2, and SIRT1 (Gu and Roeder, 1997; Juan et al., 2000; Luo et al., 2001; Vaziri et al., 2001). Since WTX itself does not encode recognizable acetyltransferase or deacetylase domains, we tested whether WTX might modulate the activity of these bona fide p53 regulators. Knockdown of WTX did not alter the gross nuclear localization of these p53 modifier proteins. In addition, siRNA knockdown of HDAC1, HDAC2 or SIRT1 did not alter the enhanced p53 acetylation mediated by WTX overexpression (data not shown). However, suppression of WTX resulted in significantly reduced expression of CBP, and less consistently p300. No change was evident in steady state levels of HDAC1, HDAC2, or SIRT1 (Figure 4A and S4A). The downregulation of CBP and p300 proteins was not due to a reduction in their respective mRNA levels (Figure 4B), but it was associated with increased protein turnover (Figures 4C and D, S4B and C), which was rescued by the proteasome inhibitor MG132 (Figure S4D), indicating involvement of ubiquitin-mediated degradation. Among the WTX splice variants, ectopic WTX-S was capable of mediating the same effects on p53 acetylation as full length WTX-NS (Figure S3E), but its knockdown had little impact on endogenous CBP/p300 and p53 acetylation, given its low baseline expression in the cells tested (Figures 4E, F and S4E). Since available antibodies recognize only full-length WTX, effective knockdown of WTX-S was measured by quantitative real-time PCR.
Figure 4. Reduced protein stability of CBP/p300 following depletion of WTX.
(A) Immunoblotting analysis of the known p53 acetyl transferases and deacetylases CBP, p300, SIRT1, HDAC1 and HDAC2 in U2OS cells following knockdown of WTX, using either of two independent shRNAs, demonstrating reduced expression of CBP/p300 following WTX depletion. Arrowhead indicates the WTX migration (endogenous full length WTX), as opposed to background bands.
(B) Quantitative real-time PCR analysis of CBP and p300 mRNA level in WTX depleted U2OS cells, demonstrating unaltered mRNA levels.
(C) Increased CBP and p300 protein turnover following WTX knockdown in U2OS cells measured by immunoblotting analysis. Cycloheximide (CHX, 100 μg/ml) was added for indicated time at 40 hr after transfection, a time at which WTX knockdown is achieved but before the downregulation of CBP/p300 occurs.
(D) Quantitation of immunoblotting results in (C), using QuantityOne software (Bio-Rad), with CBP/p300 levels normalized to tubulin.
(E) Downregulation of CBP/p300 and reduction of etoposide induced p53 acetylation following siRNA targeting of each WTX isotype in U2OS cells. WTX-1 targets both WTX-NS and WTX-S (total), whereas WTX-2 only targets WTX-NS. A specific siRNA was designed to target WTX-S. Arrowheads indicate the correct bands (Note: There is no WTX-S band in Western blot).
(F) Quantitative real time PCR analysis for WTX-S transcript with duplicated samples with (E), demonstrating specificity of each siRNA. Error bars indicate standard error of mean. See also Figure S4.
The downregulation of CBP/p300 by WTX knockdown was dependent upon the presence of E3 ubiquitin ligases. Knockdown of the F-box protein Skp2, a component of the SCF Skp2 E3 ubiquitin ligase complex which has been reported to regulate protein stability of p300 (Kitagawa et al., 2008), as well as knockdown of the F-box protein β-TrCP, a component of SCF β-TrCP E3 ubiquitin ligase complex and a known WTX interacting partner (Major et al., 2007), abrogated CBP/p300 downregulation following WTX depletion. However, knockdown of Trim28, a RING domain protein that serves as E3 ubiquitin ligase for p53 (Doyle et al., 2010), did not have any effect (Figure S4F and G). These results point to specific protein degradation pathways through which WTX may regulate CBP and p300 at the post-transcriptional level.
CBP/p300 interacts with the N-terminal transactivation domain of p53 (Scolnick et al., 1997), mediating acetylation of lysine residues and functioning as a transcriptional coactivator. In this context, it is interesting that WTX enhances acetylation of both wild type and, to a lesser degree, N-terminal truncated p53 proteins (ΔNp53 and 102-C). The truncated p53 constructs lack the CBP/p300 binding domain, but retain the WTX binding domain (Figure 5A). Thus, WTX itself may be capable of bridging the interaction between CBP/p300 and p53, enabling CBP/p300-mediated acetylation of the truncated p53 proteins. In fact, in p53-null H1299 cells, low levels of flag-tagged WTX were co-immunoprecipitated with endogenous CBP, suggesting WTX-CBP binding independent of p53 (Figure 5B). In cotransfection experiments, expression of WTX enhanced the CBP mediated Lys 382 acetylation of wild type p53 (Figure 5C). A similar effect was observed with an inactive missense p53 mutant (Figure S5A). Notably, ectopic expression of WTX did not increase CBP protein levels, suggesting that other factors may be limiting when WTX is abundant (Figure S5B). However, even in the absence of CBP/p300 stabilization, the effect of ectopic WTX on p53 acetylation was remarkable (Figure 5C). Finally, co-immunoprecipitation experiments showed that the physical interaction between CBP and p53, along with p53 acetylation, was enhanced in the presence of ectopic WTX (Figure 5D). Taken together, these results suggest that WTX modulates p53 acetylation through its stabilization of CBP/p300, as well as its enhancement of the CBP/p300-p53 complex.
Alteration of p53 response by WTX
Recent studies using systematic point mutations of lysine residues in p53 have shown that specific acetylation of a subset of p53 residues by CBP/p300 is indispensable for induction of some p53 target genes important for cell cycle arrest and apoptosis, but that complete loss of all known acetylation residues is required to abolish the p53 response (Feng et al., 2005; Krummel et al., 2005; Tang et al., 2008). Given the effect of WTX expression on the modification of p53 Lys 373/382 and on downregulation of CBP/p300, we tested for an effect of WTX on p53 functional properties. Transfection of wild type TP53 into p53-null H1299 cells triggered apoptosis, which was most evident at 48 hrs. shRNA-mediated knockdown of WTX strongly suppressed this effect, as measured both by subG1 cell populations at 48 hr, and by cleaved caspase 3 and PARP cleavage as early as 24 hr (Figures 6A and S6B-E). The fraction of cells arrested in G1 phase at 24 hr, prior to the initiation of apoptosis was also reduced by WTX knockdown (Figure S6A). In addition, etoposide induced apoptosis in A549 cells which harbor endogenous wild type TP53 was also significantly reduced by WTX knockdown (Figure S6F and S6G).
Figure 6. Altered p53 response in WTX-depleted cells.
(A) Sub-G1 (apoptotic) cell population, 48 hours following transfection of TP53 or empty vector into H1299 cells expressing shRNA against WTX or GFP. Suppression of p53-mediated apoptosis by WTX knockdown is shown in two independent experiments.
(B) Quantitative real-time PCR analysis of selected p53 target genes, following treatment with etoposide (20 μM for 48 hours) of p53-reconstituted H1299 cells harboring shRNA against WTX or GFP. Error bars indicate standard error of mean. See also Figure S6.
We analyzed the expression of characteristic p53 target genes following treatment with the DNA damaging agent etoposide in stably p53-reconstituted H1299 cells, with or without WTX knockdown. The baseline level of reconstituted p53 expression in these cells normalized to GAPDH was comparable to the physiological expression in U2OS cells as measured by real time PCR (data not shown). Etoposide-mediated induction of p53 target genes such as p21, Apaf1, and Bax were significantly repressed by WTX-mediated knockdown (Figures 6B and S6H). Similarly, in other cell lines with endogenous wild type TP53, WTX knockdown suppressed the etoposide-induced expression of Gadd45A, MDM2, and RRM2B (U2OS cells), and of MDM2 and RRM2B (A549 cells) (Figures S6I and J). Taken together, these results suggest that depletion of WTX attenuates the p53 response, an effect that is correlated with its modulation of CBP/p300.
DISCUSSION
We have demonstrated that the Wilms tumor suppressor WTX enhances p53 function. While this unexpected observation was triggered by the striking colocalization of WTX with E1B 55K and p53 proteins within the E1B cytoplasmic body of adenovirus-transformed kidney cells, the effect of WTX on p53 postranslational modification is independent of E1B 55K. Instead, it is associated with WTX-modulated stability of the p53 acetyl transferases CBP/p300 and enhanced association between p53 and CBP. In parallel to the reported WTX modulation of β-catenin degradation in the cytoplasm, these observations suggest an effect of WTX on turnover of nuclear proteins.
The highly evolved transforming proteins of adenovirus have proven critical in identifying key cellular proteins whose inactivation is important to allow unregulated host cell proliferation. Thus adenovirus E1A plays a major role in scavenging Rb family members from E2F transcription factors and triggering cell cycle progression, while E1B 55K targets p53 to suppress the physiological checkpoint response to oncogene-induced proliferation (Reviewed in (Berk, 2005). It is in this context that the targeting of WTX by E1B 55K suggests a significant role for WTX in the regulation of important cellular proliferation pathways. In addition to the striking cellular colocalization of these proteins, overexpression of WTX may compete with p53 for binding to E1B 55K, leading to release of p53 from the cytoplasmic E1B body, its accumulation in the nucleus, Ser 15 phosphorylation, Lys 382 acetylation, and activation of the p53-dependent transcriptional program. While further experiments are required to determine the significance of WTX inactivation for adenoviral replication and transformation, these observations implicate this tumor suppressor in the well studied p53 pathway.
The binding of WTX to p53 was not identified in previous immunoprecipitation-mass spectrometric analyses (Major et al., 2007). However, these studies relied on a database-annotated WTX C-terminus sequence that proved incorrect, lacking the WTX amino acids 786 to 1135, which encompass the domain required for p53 binding. The functional association of WTX with p53 involves the stabilization of CBP/p300 as well as enhancement of p53-CBP binding, which leads to the activating acetylation of p53 on Lys 373/382. As such, WTX may function as a scaffolding molecule for p53-CBP binding and part of a complex regulating protein turnover. This is analogous to its proposed cytoplasmic function in regulating β-catenin degradation, where it has been reported to negatively regulate β-catenin stability by recruiting it to the APC-Axin complex and SCF β-TrCP E3 ubiquitin ligase (Major et al., 2007). Our data suggests that downregulation of CBP/p300 by WTX depletion is dependent on the presence of F-box proteins β-TrCP and Skp2. Thus, further mechanistic insight into the role of WTX within the protein turnover machinery will be required to understand its apparently distinct effects on β-TrCP mediated protein degradation. In addition, it is of interest that the F-box protein Skp2 counteracts p53 activation, both through ubiquitin-mediated degradation of p300 and by inhibition of the p300-p53 interaction (Kitagawa et al., 2008). In these contexts, WTX may antagonize Skp2 in modulating CBP/p300 stability and p53 function, since WTX appears to enhance the physical interaction between CBP and p53 proteins in addition to regulating CBP/p300 turnover.
Cellular protein levels of CBP/p300 are tightly regulated, as demonstrated by the severe developmental defects and mental retardation associated with CBP haploinsufficiency in Rubinstein-Taybi Syndrome, as well as in CBP hemizygous mouse models (Goodman and Smolik, 2000; Janknecht, 2002; Petrij et al., 1995). As such, the impact of WTX mutation or deletion on CBP/p300-dependent processes may have significant consequences in Wilms tumors. We also note that p53 acetylation is a standard measure for a number of modifying enzymes, including CBP/p300, that also target chromatin and other cellular components, raising the possibility that the functional properties of WTX during normal development and tumorigenesis may not be limited to its effect on p53 activity. While our studies point to a strong association between WTX modulation of CBP/p300 and resulting alterations in p53 function, we note that other WTX-mediated effects may also contribute to p53 function. For instance, WTX may also affect p53 function directly or indirectly through phosphorylation of Ser 15 and 20, although p53 phosphorylation changes were not as consistent when analyzing endogenous proteins.
The precise mechanism by which somatic WTX inactivation 2 enhances embryonic kidney tumorigenesis remains unknown. Neither the human germline WTX mutation syndrome OSCS, nor the recently generated Wtx-mouse knockout are tumor prone, although both cases have reported expansion of a population of renal precursor cells, which may be susceptible for malignant transformation (Fukuzawa et al., 2010; Jenkins et al., 2009; Moisan et al., 2011). It is possible that WTX-mediated regulation of p53 activity contributes to tumorigenesis. Since Wilms tumors rarely display mutations of p53 itself (Bardeesy et al., 1995; Bardeesy et al., 1994), abrogation of p53 function through accessory pathways may indeed be relevant. Alternatively, the tumorigenic consequences of WTX inactivation may involve additional targets of CBP/p300 or other proteins whose turnover is affected by WTX. Similarly, p53 regulation by WTX is unlikely to underlie the developmental consequences of WTX inactivation on mesenchymal differentiation as p53 knockout mice develop normally (Donehower et al., 1992; Jacks et al., 1994).
Finally, we note the parallels between the functional properties of the first Wilms tumor suppressor identified, WT1, and the recently discovered WTX. WT1 encodes a zinc finger transcription factor, subject to complex alternative splicing, whose major isoforms mediate transactivation of a cellular differentiation program (Lee and Haber, 2001; Reddy and Licht, 1996). WT1-mediated transactivation is dependent in part on its physical interaction with CBP/p300, and it has been shown to bind in vivo to p53, modulating its functional properties (Maheswaran et al., 1993; Wang et al., 2001). Remarkably, WT1 expression in adenovirus-transformed cells is also localized to the E1B 55K cytoplasmic body (Maheswaran et al., 1998). We have recently demonstrated the coimmunoprecipitation of WT1 and WTX and the modulation of WT1 transactivational activity by expression of WTX (Rivera et al., 2009). Taken together, these observations suggest that WTX has a distinct function relating to the regulation of critical developmental and tumor suppressor pathways. Further definition of these mechanisms will contribute to understanding the role of WTX in development and cancer.
MATERIALS AND METHODS
Microarray
F-5 and F-18 cells were treated with 1 μg/ml doxycycline for 12 hr and RNA was extracted using RNeasy kit (Qiagen). Purified RNA was processed and hybridized with U133 Plus 2.0 chip (Affymetrix) at the Molecular Profiling Laboratory at MGH Cancer Center for microarray analysis. Microarray results were subjected to the Gene Set Enrichment Analysis (GSEA) using the Molecular Signatures Database (MSigDB) ver. 2 (http://www.broadinstitute.org/gsea/msigdb/collections.jsp#C2) to analyze enriched biological pathways.
Immunoflurescence microscopy
Cells grown on coverslips were fixed with 4% formaldehyde, permeabilized with 0.5 % Triton-X 100, and stained with antibodies against indicated proteins. Alexa-fluor 488 or 568 conjugated secondary antibodies (Invitrogen) were used for visualization. Images were obtained from Nikon Eclipse 90i fluorescence microscope (Nikon, Melville, NY) with NIS-Element AR2.30 software (Nikon).
Quantitative real-time PCR
RNA was extracted using RNeasy kit (Qiagen) and cDNA was synthesized using SuperScript III (Invitrogen). Quantitative real-time PCR was performed using Power SYBR Green and ABI 7500 Real-Time PCR system (Applied Biosystems). The ddCt method was used to calculate relative quantity of expression using GAPDH as an internal control. Primers used were listed in Supplemental information.
Flow cytometry
For measuring sub G1 population, cells were fixed with ice-cold 70% ethanol and resuspended in PBS containing 25 μg/ml Propidium Iodide and 100 μg/ml RNase A. Cleaved Caspase-3 was stained as described in manufacturer’s protocol (Cell Signaling Technology). FACS Calibur and CellQuest Pro software (BD Bioscience) was used for collecting data. FlowJo software (Tree Star, Inc.) was used for data analysis.
Immunoprecipitation
Total or nuclear lysates were incubated with antibodies for hours at 4°C followed by addition of protein G sepharose beads (GE Healthcare) and subsequent incubation for 1 hr. Beads were washed with binding buffer (50 mM Tris pH7.4, 150 mM NaCl, 1% Nonidet P-40) or radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS). The bound proteins were eluted by boiling in 1x SDS loading buffer and subjected to Western blotting.
Supplementary Material
WTX releases p53 from suppression by adenovirus E1B 55K.
WTX activates p53 through Lys 382 acetylation.
WTX modulates protein stability of the acetyl transferases CBP/p300.
WTX enhances the interaction between p53 and CBP.
ACKNOWLEDGMENTS
This work was supported by NIH 5R37CA058596, P50CA101942, HHMI (DAH); NIH T32CA009216 (WJK); NIDDK, the Burroughs Wellcome Fund, HHMI (MNR). The authors would like to thank Drs. Nick Dyson, Shyamala Maheswaran and Ho-June Lee for helpful discussions and critical comments. We are grateful to Drs. Anders Näär and Joshua Black for providing reagents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The full microarray data set has been deposited in Gene Expression Omnibus (GEO) under the accession number GSE34715.
REFERENCES
- Bardeesy N, Beckwith JB, Pelletier J. Clonal expansion and attenuated apoptosis in Wilms’ tumors are associated with p53 gene mutations. Cancer Res. 1995;55:215–219. [PubMed] [Google Scholar]
- Bardeesy N, Falkoff D, Petruzzi MJ, Nowak N, Zabel B, Adam M, Aguiar MC, Grundy P, Shows T, Pelletier J. Anaplastic Wilms’ tumour, a subtype displaying poor prognosis, harbours p53 gene mutations. Nat Genet. 1994;7:91–97. doi: 10.1038/ng0594-91. [DOI] [PubMed] [Google Scholar]
- Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa R, Holman SK, Chow CW, Savarirayan R, Reeve AE, Robertson SP. WTX mutations can occur both early and late in the pathogenesis of Wilms tumour. J Med Genet. 2010;47:791–794. doi: 10.1136/jmg.2010.080663. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Grand RJ, Parkhill J, Szestak T, Rookes SM, Roberts S, Gallimore PH. Definition of a major p53 binding site on Ad2E1B58K protein and a possible nuclear localization signal on the Ad12E1B54K protein. Oncogene. 1999;18:955–965. doi: 10.1038/sj.onc.1202358. [DOI] [PubMed] [Google Scholar]
- Grohmann A, Tanneberger K, Alzner A, Schneikert J, Behrens J. AMER1 regulates the distribution of the tumor suppressor APC between microtubules and the plasma membrane. J Cell Sci. 2007;120:3738–3747. doi: 10.1242/jcs.011320. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Harada JN, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination 1 machinery. J Virol. 2002;76:9194–9206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Janknecht R. The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol Histopathol. 2002;17:657–668. doi: 10.14670/HH-17.657. [DOI] [PubMed] [Google Scholar]
- Jenkins ZA, van Kogelenberg M, Morgan T, Jeffs A, Fukuzawa R, Pearl E, Thaller C, Hing AV, Porteous ME, Garcia-Minaur S, et al. Germline mutations in WTX cause a sclerosing skeletal dysplasia but do not predispose to tumorigenesis. Nat Genet. 2009;41:95–100. doi: 10.1038/ng.270. [DOI] [PubMed] [Google Scholar]
- Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- Kao CC, Yew PR, Berk AJ. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Lee SH, McCormick F. Skp2 suppresses p53-dependent apoptosis by inhibiting p300. Mol Cell. 2008;29:217–231. doi: 10.1016/j.molcel.2007.11.036. [DOI] [PubMed] [Google Scholar]
- Kobet E, Zeng X, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci U S A. 2000;97:12547–12552. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Lee SB, Haber DA. Wilms tumor and the WT1 gene. Exp Cell Res. 2001;264:74–99. doi: 10.1006/excr.2000.5131. [DOI] [PubMed] [Google Scholar]
- Levine AJ. The common mechanisms of transformation by the small DNA tumor viruses: The inactivation of tumor suppressor gene products: 1 p53. Virology. 2009;384:285–293. doi: 10.1016/j.virol.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Colosimo AL, Yang XJ, Liao D. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol. 2000;20:5540–5553. doi: 10.1128/mcb.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol. 2005;79:14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Englert C, Lee SB, Ezzel RM, Settleman J, Haber DA. E1B 55K sequesters WT1 along with p53 within a cytoplasmic body in adenovirus-transformed kidney cells. Oncogene. 1998;16:2041–2050. doi: 10.1038/sj.onc.1201741. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Park S, Bernard A, Morris JF, Rauscher FJ, 3rd, Hill DE, Haber DA. Physical and functional interaction between WT1 and p53 proteins. Proc Natl Acad Sci U S A. 1993;90:5100–5104. doi: 10.1073/pnas.90.11.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- Moisan A, Rivera MN, Lotinun S, Akhavanfard S, Coffman EJ, Cook EB, Stoykova S, Mukherjee S, Schoonmaker JA, Burger A, et al. The WTX Tumor Suppressor Regulates Mesenchymal Progenitor Cell Fate Specification. Dev Cell. 2011;20:583–596. doi: 10.1016/j.devcel.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Dobner T. The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle. 2008;7:754–758. doi: 10.4161/cc.7.6.5495. [DOI] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Punga T, Akusjarvi G. The adenovirus-2 E1B-55K protein interacts with a mSin3A/histone deacetylase 1 complex. FEBS Lett. 2000;476:248–252. doi: 10.1016/s0014-5793(00)01739-7. [DOI] [PubMed] [Google Scholar]
- Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JC, Licht JD. The WT1 Wilms’ tumor suppressor gene: how much do we really know? Biochim Biophys Acta. 1996;1287:1–28. doi: 10.1016/0304-419x(95)00014-7. [DOI] [PubMed] [Google Scholar]
- Rivera MN, Haber DA. Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer. 2005;5:699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- Rivera MN, Kim WJ, Wells J, Driscoll DR, Brannigan BW, Han M, Kim JC, Feinberg AP, Gerald WL, Vargas SO, et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science. 2007;315:642–645. doi: 10.1126/science.1137509. [DOI] [PubMed] [Google Scholar]
- Rivera MN, Kim WJ, Wells J, Stone A, Burger A, Coffman EJ, Zhang J, Haber DA. The tumor suppressor WTX shuttles to the nucleus and modulates WT1 activity. Proc Natl Acad Sci U S A. 2009;106:8338–8343. doi: 10.1073/pnas.0811349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl A, Hastie N. Multiple roles for the Wilms’ tumour suppressor gene, WT1 in genitourinary development. Mol Cell Endocrinol. 1998;140:65–69. doi: 10.1016/s0303-7207(98)00031-8. [DOI] [PubMed] [Google Scholar]
- Scolnick DM, Chehab NH, Stavridi ES, Lien MC, Caruso L, Moran E, Berger SL, Halazonetis TD. CREB-binding protein and p300/CBP27 associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanneberger K, Pfister AS, Brauburger K, Schneikert J, Hadjihannas MV, Kriz V, Schulte G, Bryja V, Behrens J. Amer1/WTX couples Wnt-induced formation of PtdIns(4,5)P(2) to LRP6 phosphorylation. EMBO J. 2011 doi: 10.1038/emboj.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang W, Lee SB, Palmer R, Ellisen LW, Haber DA. A functional interaction with CBP contributes to transcriptional activation by the Wilms tumor suppressor WT1. J Biol Chem. 2001;276:16810–16816. doi: 10.1074/jbc.M009687200. [DOI] [PubMed] [Google Scholar]
- Wimmer P, Schreiner S, Everett RD, Sirma H, Groitl P, Dobner T. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene. 2010;29:5511–5522. doi: 10.1038/onc.2010.284. [DOI] [PubMed] [Google Scholar]
- Yew PR, Liu X, Berk AJ. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- Zantema A, Fransen JA, Davis-Olivier A, Ramaekers FC, Vooijs GP, DeLeys B, Van der Eb AJ. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.