Abstract
Methicillin resistant Staphylococcus aureus (MRSA) have emerged among livestock in several countries. In this study, we describe the results of a screening performed in pigs and raw pork samples in Thailand. Ten pork samples and 15 nasal swabs from pigs were collected from 2 markets and 1 pig farm in the Samuth Songkhram province in Thailand. MRSA were isolated using selective isolation procedures and confirmed by mecA PCR. The MRSA were characterized by antimicrobial susceptibility testing, pulsed field gel electrophoresis (PFGE), spa typing, SCCmec typing, and MLST. Resistance and virulence markers were screened using a microarray. Five of the pork samples and six pig nasal swabs were positive for MRSA. All 11 isolates belonged to spa type t337 but showed diversity in antimicrobial resistance patterns and PFGE profiles. Additionally, the isolates were sequence-typed; ST9, ST2136, ST2278 belonging to the clonal complex; CC9. All isolates harbored SCCmec IX and were resistant to 7 out of 14 tested antimicrobials; additional resistances to all antimicrobials tested were found in some of the pork and pig isolates and 1 pork isolate was resistant to 13 antimicrobials tested. Microarray analysis identified blaZ, aac-aphD, vga(A), tetM, and a tet efflux marker, in all strains and additionally ermB and aadD, cat and fex(A) in the pork isolates. None of the isolates were found PVL-positive, but enterotoxins were identified in all isolates. To our knowledge, only a few descriptions of MRSA in livestock and food products in Thailand have been observed but this is the first observation of MRSA CC9 associated with SCCmec IX in pork. This study indicates a likely widespread distribution of MRSA in pig and pork in Thailand and further investigation on the prevalence and importance of livestock associated MRSA in Thailand is needed.
Keywords: MRSA, SCCmec type IX, spa type t337, Thailand, pig, pork
Introduction
Methicillin resistant Staphylococcus aureus (MRSA) have recently emerged in livestock, mainly pigs, but also in other animal species (Catry et al., 2010). Most isolates have belonged to CC398, which is also present in MSSA isolates from pigs (Voss et al., 2005; van Belkum et al., 2008; Hasman et al., 2010), but other MRSA clones have also been found in pigs in Europe and North America, including CC1, CC5, CC8, CC9, CC30, and CC971. In China and Malaysia, isolates belonging to CC9 have been more frequently observed (Cui et al., 2009; Neela et al., 2009). However in South East Asia, there is still limited data on the occurrences of MRSA in livestock. In this study, we present the first description of MRSA isolated from pork in Thailand; S. aureus of spa type t337 belonging to CC9, harboring the SCCmec element type IX.
Materials and Methods
Bacterial isolates
The S. aureus included in this study were isolated from samples collected the 10th of January 2011 from two open-air market shops in the city of Samuth Songkhram and in one nearby pig farm with breeding facilities, within the Samuth Songkhram province in Thailand. By convenience sampling 10 pork samples of 25 g were obtained from the market, 5 different cuts from each shop, and 15 nasal swabs from live pigs from the farm. Meat samples and nasal swabs were inoculated in Mueller Hinton broth (Becton Dickinson, Franklin Lakes, NJ, USA) supplemented with 6.5% NaCl (225 and 10 mL, respectively) which was incubated at 37 ± 1°C for 18–24 h. One milliliter of MH culture was transferred to Tryptone Soya Broth (TSB; Oxoid, Cambridge, UK) supplemented with 4 mg/L cefoxitin and 75 mg/L aztreonam, and incubated at 37 ± 1°C in 18–24 h. Ten microliters was streaked on Brilliance MRSA agar (Oxoid, Cambridge, UK) and incubated at 37 ± 1°C for 24–48 h. Presumptive MRSA colonies were identified as denim blue colonies and these colonies were isolated on blood agar plates and thereafter kept frozen at −70°C until further examination.
Antimicrobial susceptibility testing
MIC testing was performed using a commercially prepared, dehydrated panel, Sensititre, from TREK Diagnostic Systems, Ltd. (East Grinstead, England). The following antimicrobials and epidemiological cut-off values or clinical breakpoints were used in the study: cefoxitin, FOX (R > 4 mg/L); chloramphenicol, CHL (R > 16 mg/L); ciprofloxacin, CIP (R > 1 mg/L); erythromycin, ERY (R > 1 mg/L); florfenicol, FFN (R > 8 mg/L); gentamicin, GEN (R > 2 mg/L); penicillin, PEN (R > 0.125 mg/L); spectinomycin, SPE (R > 64 mg/L); streptomycin, STR (R > 16 mg/L); sulfamethoxazole, SMX (R > 128 mg/L); tetracycline, TET (R > 1 mg/L); tiamulin, TIA (R > 16 mg/L); trimethoprim, TMP (R > 8 mg/L); sulfamethoxazole + trimethoprim, SXT (R > 2 mg/L). The quinolone-resistant determining region (QRDR) regions of staphylococcal topoisomerase genes were amplified and sequenced in one isolate; 3-P7 (N) using the primers described by Schmitz et al. (1998).
Detection of the mecA gene, spa-, and SCCmec types
The presence of the mecA gene was determined using a multiplex PCR directed against the 16S ribosomal RNA (presence of DNA), the mecA gene, and the nucA gene (presence of S. aureus) as described previously by Maes et al. (2002). MRSA isolates were spa – typed using a conventional PCR assay described by Hasman et al. (2010), and a multiplex PCR was performed to identify the SCCmec cassette (Kondo et al., 2007).
MLST typing
MLST was performed by the method previously described by Enright et al. (2000), on all of isolates. The individual MLST types were assigned to the specific CC types utilizing the eBURST2.
Pulsed field gel electrophoresis
Pulsed field gel electrophoresis (PFGE) was performed according to Murchan et al. (2003) using SmaI restriction. The comparative analysis of the PFGE profiles was performed by using the Bionumerics software version 4.6 (Applied Maths, Sint-Martens-Latem, Belgium) and the Dice correlation coefficient for band matching with a 1.0% position tolerance and an optimization at 1.0%.
Genotyping
All isolates were characterized using the Identibac Staphylococcus aureus genotyping microarray performed at the Alere Laboratories (Alere Labs; Jena, Germany), investigating the presence of resistance genes and virulence markers. The array results were interpreted according to manufacturer recommendations.
Results
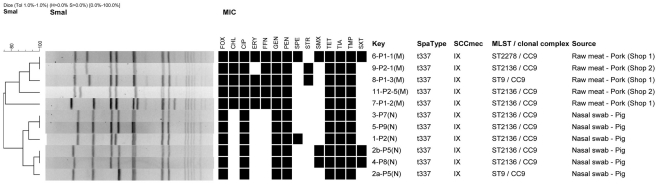
Among the 25 samples, 11 isolates; 5 from raw pork and 6 from pigs were isolated, identified as MRSA and confirmed to harbor the mecA gene. The mecA gene was carried on the SCCmec element type IX. Among the isolates only a single spa type was identified; t337. MLST typing revealed the pork isolate as belonging to three MLST types; ST2136 (n = 3), ST9 (n = 1), and ST2278 (n = 1) where the pig isolates belonged to ST2136 (n = 5) and ST9 (n = 1) but both within CC9 (Figure 1). ST2278 was assigned as a new MLST types not previously observed. ST9 and ST2278 both differed with one allelic difference compared to the predominating MLST; ST2136 in this study.
Figure 1.
Dendrographic analysis of PFGE (SmaI) of methicillin resistant Staphylococcusaureus isolates originating from pork and pigs in Thailand. Black squares in the dendrogramme represent the isolates as being resistant. The codes of the antimicrobials are listed in the methods section. All isolates were collected the 10th of January 2011.
A high level of antimicrobial resistance was observed among all of the MRSA isolates showing resistance to 7 out of the 14 antimicrobials tested (Figure 1). This included FOX, CIP, GEN, PEN, TET, TIA, and TMP, respectively. All five pork isolates conferred resistance to both CHL and FFN. Additionally, four (80%) of the five pork isolates also conferred resistance to ERY (Figure 1).
The genotypic characterization of the strains showed the presence of blaZ, aac-aphD, vga(A), tetM, and a tet efflux marker, besides the mecA gene. Some of the pork samples were found additionally positive to ermB and aadD, cat and fex(A).
Two single base-pair substitutions were identified in the QRDRs of one of the pig isolates resistant to ciprofloxacin. The isolate had a mutation at codon 84 (TCA [Ser] → TTA [Leu]) in gyrA and codon 80 (TCC [Ser] → TTC [Phe]) in grlA. All pig strains and some pork meat strains harbored several enterotoxin genes (entG, entI, entM, entN, and entO). PVL, TSST-1, and exfoliative toxins were not found in any of the isolates (Table 1).
Table 1.
Distribution of antimicrobial and virulence factors Staphylococcusaureus isolates originating from pork and pigs in Thailand.
| Country | Source | No. of isolates | No. and (%) of isolates with |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial resistance genes |
Virulence markers |
|||||||||||||||
| blaZ1 | ermB2 | aac-aphD3 | aadD3 | vga(A)4 | tetM5 | Tetefflux5 | catgenes6 | fex(A)7 | entG8 | entI8 | entM8 | entN8 | entO8 | |||
| Thailand | Pig nasal swab | 6 | 6 (100) | 0 (0) | 6 (100) | 0 (0) | 6 (100) | 6 (100) | 6 (100) | 0 (0) | 0 (0) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) |
| Thailand | Raw pork | 5 | 5 (100) | 4 (80) | 5 (100) | 4 (80) | 5 (100) | 5 (100) | 5 (100) | 1 (20) | 5 (100) | 4 (80) | 3 (60) | 3 (60) | 4 (80) | 5 (100) |
| Total | 11 | 11 (100) | 4 (36) | 11 (100) | 4 (36) | 11 (100) | 11 (100) | 11 (100) | 1 (9) | 5 (45) | 10 (91) | 9 (82) | 9 (82) | 10 (91) | 11 (100) | |
The following genes confer resistance to: 1penicillin; 2erythromycin; 3gentamicin; 4tiamulin; 5tetracycline; 6chloramphenicol; 7florfenicol; 8enterotoxins. The isolates did not harbor the following antimicrobial resistance genes: ermA, ermC, dfrA, and tetK. The isolates did not harbor the following virulence genes; Panton Valentine Leucocidin PVL- LukS-PV and Luk F-PV, Toxic Shock syndrome Toxin – TSST-1 markers and Exfoliative toxins markers etA, etB, etD.
Pulsed field gel electrophoresis revealed a high diversity among the 11 isolates with eight unique SmaI patterns divided into two groups (pork and pigs) defined by 75% similarity. Three PFGE clusters with indistinguishable types were observed, of which one cluster consisted of isolates from two raw pork isolates displaying a different antimicrobial resistance profile. The two remaining clusters consisted of pairs of pork isolates with identical antimicrobial resistance patterns, by pairs (Figure 1).
Discussion
Methicillin resistance in livestock associated MRSA (LA-MRSA) has mostly been related to ST398 carrying the SCCmec IV and V elements (Catry et al., 2010). However, CC9 MRSA has been found as the dominant subtype in isolates originating from pigs in Asia; China, and Malaysia where the isolates contained the SCCmec III and SCCmec V, respectively (Cui et al., 2009; Neela et al., 2009). We confirmed this observation as all isolates in this study were spa type t337 and MLST of the isolates confirmed its association with CC9.
Recently, a novel SCCmec element; IX was described related to CC398 in a healthy human originating from Thailand (Li et al., 2011). We found that pig and pork isolates belonging to CC9-related MLST type and of spa type t337 have acquired the same SCCmec element IX.
This specific genotype has recently only been described in another study where four pigs from a farm situated in Lampoon province, Thailand belonged to ST9/t337 carrying the ccrAB type 1 and mec class C2 equivalent to SCCmec element IX (Anukool et al., 2011). This finding strongly supports that LA-MRSA belonging to CC9-related MLST type and of spa type t337 has acquired the SCCmec element IX in Thailand.
The finding of pork isolates clustering separately from isolates from live animals is likely a consequence of the small sample sizes. Thus, there is no evidence if the pig farm were supplier to the shops at the market. However, this hypothesis needs to be further elucidated, as does the potential importance for human health.
In addition to the acquired methicillin resistance, the isolates conferred resistance to several antimicrobial agents including critical important antimicrobials for human health. In comparison to the antimicrobial resistance profile of the four pig isolates described by Anukool et al., we found similar high level of resistance to macrolides and chloramphenicol. However, this high frequency was only observed in pig isolates in contrast to the pork isolates where no resistances to these compounds were observed. In the study by Anukool et al. (2011) the authors suggest based on a negative result testing for the presence of the cfr gene that cat genes should be responsible for resistance to chloramphenicol. We however, found that the fexA gene was present in 80% of the tested pig isolates and only one isolate harbored cat genes. The high antimicrobial resistant profile among the isolates might be a result of the high selective pressure to antimicrobials used in the agriculture sector (Carlet et al., 2011). This hypothesis is supported by the genetic diversity indicating that the isolates have been in the reservoir for some time. The resistant nature of these isolates and the harbored enterotoxins pose a great concern as they may cause problems in the treatment of humans with S. aureus infections. Likewise, MRSA might also pose a threat by possible spread into the community either by direct occupational contact to animals or by a more remote hypothesis of foodborne transmission.
In this study, we described the spread of the mecA gene to the S. aureus spa type t337 CC9, harbored on the novel SCCmec IX element. We advocate the Thai authorities to embark on a broader investigation to measure the prevalence of LA-MRSA and to assess what actions to initiate in respect of control and prevention of the MRSA among livestock.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Samut Songkhram provincial livestock office for granting permission, give advice, and provide the staff for guidance to the farm. Additionally, to Mr. Jacob Dyring Jensen from DTU-Food for outstanding technical support. FUNDING: This work was supported by the World Health Organization Global Foodborne Infections Network (GFN) and by a grant from the European Commission (CONCORD, agreement number 222718). Additionally, the travel to Thailand was funded by Jens Hansens Mindelegat.
Footnotes
References
- Anukool U., O’Neill C. E., Butr-Indr B., Hawkey P. M., Gaze W. H., Wellington E. M. (2011). Meticillin-resistant Staphylococcus aureus in pigs from Thailand. Int. J. Antimicrob. Agents 38, 86–87 10.1016/j.ijantimicag.2011.02.018 [DOI] [PubMed] [Google Scholar]
- Carlet J., Collignon P., Goldmann D., Goossens H., Gyssens I. C., Harbarth S., Jarlier V., Levy S. B., N’Doye B., Pittet D., Richtmann R., Seto W. H., van der Meer J. W. M., Voss A. (2011). Society’s failure to protect a precious resource: antibiotics. Lancet 23, 369–371 10.1016/S0140-6736(11)60401-7 [DOI] [PubMed] [Google Scholar]
- Catry B., Van Duijkeren E., Pomba M. C., Greko C., Moreno M. A., Pyörälä S., Ruzauskas M., Sanders P., Threlfall E. J., Ungemach F., Törneke K., Munoz-Madero C., Torren-Edo J. (2010). Scientific advisory group on antimicrobials (SAGAM). Reflection paper on MRSA in food-producing and companion animals: epidemiology and control options for human and animal health. Epidemiol. Infect. 138, 626–644 10.1017/S0950268810000014 [DOI] [PubMed] [Google Scholar]
- Cui S., Li J., Hu C., Jin S., Li F., Guo L., Ran L., Ma Y. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683 10.1093/jac/dkp275 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H., Moodley A., Guardabassi L., Stegger M., Skov R. L., Aarestrup F. M. (2010). Spa type distribution in Staphylococcus originating from pigs, cattle and poultry. Vet. Microbiol. 141, 326–331 10.1016/j.vetmic.2009.09.025 [DOI] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Ma X. X., Watanabe S., Kreiswirth B. N., Etienne J., Hiramatsu K. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51, 264–274 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Skov R. L., Han X., Larsen A. R., Larsen J., Sørum M., Wulf M., Voss A., Hiramatsu K., Ito T. (2011). Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-esistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55, 3046–3050 10.1128/AAC.00085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes N., Magdalena J., Rottiers S., De Gheldre Y., Struelens M. J. (2002). Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative Staphylococci and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 40, 1514–1517 10.1128/JCM.40.4.1514-1517.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchan S., Kaufmann M. E., Deplano A., de Ryck R., Struelens M., Zinn C. E., Fussing V., Salmenlinna S., Vuopio-Varkila J., Solh N. E., Cuny C., Witte W., Tassios P. T., Legakis N., van Leeuwen W., van Belkum A., Vindel A., Laconcha I., Garaizar J., Haeggman S., Olsson-Liljequist B., Ransjo U., Coombes G., Cookson B. (2003). Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41, 1574–1585 10.1128/JCM.41.4.1574-1585.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neela V., Arif M. Z., Shamsudin M. N., van Belkum A., Khoon L. Y., Ghaznavi E. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47, 4138–4140 10.1128/JCM.01363-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz F. J., Hofmann B., Hansen B., Scheuring S., Lückefahr M., Klootwijk M., Verhoef J., Fluit A., Heinz H. P., Köhrer K., Jones M. E. (1998). Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 41, 481–484 10.1093/jac/41.6.662 [DOI] [PubMed] [Google Scholar]
- van Belkum A., Melles D. C., Peeters J. K., van Leeuwen W. B., van Duijkeren E., Huijsdens X. W., Spalburg E., de Neeling A. J., Verbrugh H. A. (2008). Dutch Working Party on Surveillance and Research of MRSA-SOM. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerging Infect. Dis. 14, 479–483 10.3201/eid1403.0760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerging Infect. Dis. 11, 1965–1966 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]