Abstract

A series of eight peptides corresponding to the amino acid sequence of the hinge region of IgG and 17 newly synthesized peptide analogues containing a piperidine moiety as a replacement of a glycine residue were tested as potential inhibitors of the bacterial IgG degrading enzyme of Streptococcus pyogenes, IdeS. None of the peptides showed any inhibitory activity of IdeS, but several piperidine-based analogues were identified as inhibitors. Two different analysis methods were used: an SDS-PAGE based assay to detect IgG cleavage products and a surface plasmon resonance spectroscopy based assay to quantify the degree of inhibition. To investigate the selectivity of the inhibitors for IdeS, all compounds were screened against two other related cysteine proteases (SpeB and papain). The selectivity results show that larger analogues that are active inhibitors of IdeS are even more potent as inhibitors of papain, whereas smaller analogues that are active inhibitors of IdeS inhibit neither SpeB nor papain. Two compounds were identified that exhibit high selectivity against IdeS and will be used for further studies.
Introduction
Proteolytic activities play a decisive role in all aspects of cellular life, including cell division processes, metabolism and catabolism, protein translocation, immune defense mechanisms, and more.1 The human bacterial pathogen Streptococcus pyogenes (or group A streptococcus) is the causative agent of a great variety of infections, ranging from mucocutaneous infections of the throat and skin to life threatening conditions including necrotizing fasciitis and streptococcal toxic shock syndrome.2 More than 600 million people are estimated to suffer from streptococcal pharyngitis alone, and mucocutaneous infections cause substantial morbidity and considerable economic loss to society.3 Postinfectious sequelae include serious inflammatory diseases such as acute rheumatic fever (ARF), rheumatic heart disease (RHD), and poststreptococcal glomerulonephritis (PSGN). Inflammatory autoimmune diseases such as guttate psoriasis have also been associated with streptococcal infections4 although the underlying molecular mechanisms still remain to be solved.
S. pyogenes employs two papain-like cysteine proteases to adapt to the dynamic environment in its human host and to evade the human immune response: the classical streptococcal cysteine protease SpeB and the immunoglobulin G (IgG) degrading protease, IdeS.5,6 Both enzymes adopt a canonical papain-like structural fold and show, despite the lack of sequence similarity, large structural similarities.7−10 Besides IdeS, also SpeB and papain have the ability to cleave the IgG heavy chain. The SpeB cleavage site is identical to IdeS cleavage at a defined site between glycine residues 236 and 237, creating one F(ab′)2 fragment and two identical 1/2Fc fragments.6,11,12 Papain cleavage occurs at the peptide bond between histidine in position 224 and threonine in position 225 of the hinge region of IgG, thereby generating two Fab fragments and one Fc fragment.13 However, the proteases have distinguished substrate recognition properties: SpeB and papain exhibit a broad proteolytic activity and degrade or activate a wide variety of substrates.1,14 IdeS, on the other side, is highly specific and recognizes only IgG as substrate.6,12,15 Furthermore, IdeS, in contrast to papain and other prokaryotic cysteine proteases, including SpeB and the staphylococcal cysteine protease StpA,16 is not inhibited by the classical cysteine protease inhibitor E64.6,12 This interesting property is explained by an unusually narrow active site cleft that does not offer enough space to accommodate the P3 residue of E64 and thus points to distinct substrate recognition properties.7
Given the essential role of IdeS in the evasion of IgG mediated immune responses, there is a high medical interest to identify specific inhibitors for prokaryotic cysteine proteases. Furthermore, IdeS is currently evaluated as a therapeutic agent to treat conditions in which antibodies reacting against human antigens misdirect the human immune response toward the body’s own cells. The efficient removal of pathogenic IgG is an important clinical challenge, and several animal models have provided the proof of principle for the use of IdeS as a therapeutic agent.17−19 However, an IdeS specific inhibitor would also allow the external control of proteolytic activity in these applications, which might prove to be a valuable tool in treatment.
However, because of the structural similarity of papain-like proteases, it is not a simple task to identify inhibitors that efficiently block prokaryotic proteases without affecting several essential protease functions in the human host. Compounds reported to inhibit IdeS, including alkylating agents,6 Z-LVG CHN26 and TPCK/TLCK,15 are also efficient inhibitors of other cysteine proteases and do not exhibit any selectivity toward IdeS. Recently, we showed that TPCK/TLCK analogues containing aldehyde-based warheads act as reversible inhibitors of IdeS, however their selectivity was not studied.20 The rationale for the approach in the present study was to identify specific inhibitors for IdeS based on the fact that a noncovalent inhibitor lacking an electrophilic warhead would have to depend on other specific interactions with the enzyme, which therefore should increase the selectivity and thus harbor the potential to be specific.
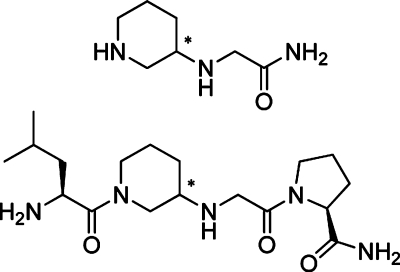
IdeS does only hydrolyze IgG and neither synthetic or natural peptides containing the P4–P1 subsites of the IgG hinge region, nor peptides with sequences covering the IdeS cleavage site are cleaved by the protease.12 Because such peptide-based substrates are not hydrolyzed by IdeS, they have in the present study instead been investigated for their putative inhibitory capacity on the streptococcal cysteine proteases IdeS and SpeB and also on papain. The tested peptides were of different length, from four up to eight amino acids, covering the P4–P′4 residues of IgG. In addition, a series of di-, tri-, and tetrapeptide analogues based on the amino acid sequence of IgG surrounding the IdeS cleavage site have been synthesized and were tested for potential inhibitory activity. In the analogues, one of the two glycine residues at the cleavage site, Gly236 or Gly237, was replaced by a piperidine moiety, thus forming either pip236G- or Gpip237-fragments (Figure 1).
Figure 1.

In the synthesized analogues, a piperidine moiety replaces one of the two glycine residues at the IdeS cleavage site. Thereby, a new stereogenic center is introduced at different positions in the two fragments (marked with an asterisk).
The piperidine moiety can be inserted through a short and efficient synthetic route, and the strategy used allows further extension both N- and C-terminally to the desired lengths of the peptide analogues. The piperidine ring rigidifies the structure of the analogues compared to the original peptide backbone, yet the structure is flexible as the six-membered ring can adopt different chair conformations. A new stereogenic center is introduced at different positions in the two fragments (Figure 1), and the environment around the scissile amide bond is changed by the different polarity profiles of the analogues. The hydrogen bond acceptor/donor ability is also altered.
Peptides and synthesized peptide analogues were screened for their ability to inhibit IdeS, SpeB, and papain. Interestingly, several of the synthesized peptide analogues affected the activity of the enzymes, however, different inhibitory profiles were observed for the three cysteine proteases.
Results and Discussion
Synthesis of Peptide Analogues
Short and efficient synthetic routes to the peptide analogues discussed in this study have been developed. The synthesis starts by formation of the new stereogenic center via a reductive amination with N-protected 3-piperidone and either glycine or enantiopure l-proline. The stereoisomers were separated and further modified or used in peptide coupling reactions with enantiopure l-amino acids to form the desired peptide analogues. Separation of the stereoisomers made it possible to keep track of the absolute configuration of the created stereogenic center throughout the synthesis. Further, no racemization or epimerization were observed in any of the amino acid coupling reactions.
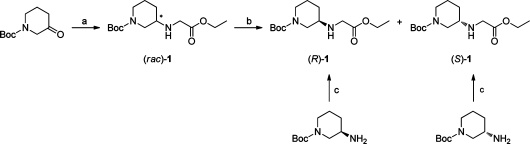
Reductive amination of Boc-protected 3-piperidone with ethyl glycinate and NaBH(OAc)3 gave the racemate 1 in moderate yield (62%) (Scheme 1). The racemate was separated into the pure enantiomers using preparative HPLC.
Scheme 1.
Reagents and conditions: (a) ethyl glycinate, NaBH(OAc)3, AcOH, molecular sieves, dichloromethane, room temp, 20 h (62%); (b) preparative HPLC; (c) ethyl glyoxylate, NaBH(OAc)3, AcOH, dichloromethane, room temp, 4 h (56% and 62%).
To determine the absolute configuration of the two enantiomers, reference compounds were synthesized via reductive amination of ethyl glyoxylate with (R)- and (S)-3-amino-1-Boc-piperidine to afford (R)- and (S)-1, respectively, with known configuration at the stereogenic center (Scheme 1). Comparison of the retention times from analytical HPLC allowed an unequivocal assignment of the configuration of the enantiomers isolated via preparative HPLC. Unfortunately, the synthesized reference compounds showed too low ee (90% and 86%, respectively) to be used as starting material for the synthesis of the analogues. However, 2 g of (rac)-1 was separated into (R)- and (S)-1 in 11 injections in sufficient amount for the synthetic route to follow.
As shown in Scheme 2, aminolysis of the isolated (R)- and (S)-1 with ammonia in methanol afforded the amides (R)- and (S)-2, respectively, in high yields (92% and 90%) without the need for purification. The 13C NMR spectra showed a considerable broadening of the signals assigned to the carbon atoms adjacent to the nitrogen in the piperidine ring, indicating that the Boc-group gives rise to rotamers. Removal of the Boc protecting group by TFA gave the GlyGly-NH2 analogues (R)- and (S)-pipG (3) in quantitative yields. The 1H NMR spectra showed high purity for both compounds. Furthermore, the coupling pattern in the 1H NMR spectra implies that the piperidine ring adopts one single chair conformation with the glycine amide residue in an equatorial position. The suggested ring conformation is shown in the Supporting Information.
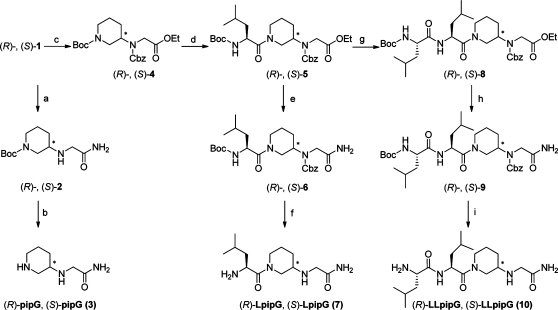
Scheme 2.
Reagents and conditions: (a) NH3 in methanol, room temp, 72 h (92 and 90%); (b) TFA, room temp, 12 h (quant yields); (c) Cbz-OSu, Et3N, dichloromethane, 0 °C → room temp, 17 h (82% and 84%); (d) (i) TFA, room temp, 16 h, (ii) Boc-l-Leu × H2O, EDC, HOBt, dichloromethane, molecular sieves, Et3N, room temp, 15 h (91% and 79% over two steps); (e) NH3 in methanol, room temp, 72 h (65% and 73%); (f) (i) H2, Pd/C, ethanol, room temp, 72 h, (ii) TFA, room temp, 4 h (84% and 75% over two steps); (g) (i) TFA, room temp, 16 h, (ii) Boc-l-Leu × H2O, EDC, HOBt, dichloromethane, molecular sieves, Et3N, room temp, 15 h (88 and 91%, over two steps); (h) NH3 in methanol, room temp, 168 h (63% and 86%); (i) (i) H2, Pd/C, ethanol, 3 h, (ii) TFA, room temp, 14 h (90% and 92% over two steps).
Routes to the analogues of the tripeptide LeuGlyGly-NH2, (R)- and (S)-LpipG (7), and the tetrapeptide LeuLeuGlyGly-NH2, (R)- and (S)-LLpipG (10), are shown in Scheme 2. The isolated enantiomers (R)- and (S)-1 were protected using Cbz-succinimide (Cbz-OSu) to afford (R)- and (S)-4, respectively, in high yields (82% and 84%). The 13C NMR spectra recorded at ambient temperature showed rotamers from both the Boc- and Cbz-groups, however the signals coalesced in spectra recorded at 50 °C. The enantiomers (R)- and (S)-4 were Boc-deprotected followed by coupling with Boc-l-leucine to afford high yields of (R)- and (S)-5, respectively (91% and 79%). Aminolysis afforded amides (R)- and (S)-6, which were deprotected to the end products (R)- and (S)-LpipG (7), respectively (yields: 84% and 75% over two steps). Boc-Deprotection of (R)- and (S)-5 and coupling to Boc-l-leucine gave (R)- and (S)-8, respectively, in high yields (88% and 91%). A subsequent aminolysis afforded (R)- and (S)-9. The reaction times were rather long and purification was needed which resulted in lower yields (63% and 86%). Finally, the Cbz- and Boc-protecting groups were removed by catalytic hydrogenation (Pd/C) and TFA hydrolysis, respectively, to afford the end products (R)- and (S)-LLpipG (10) in high yields (90% and 92%). The total yield for the synthesis of (R)- and (S)-LLpipG (10) over eight steps from (R)- and (S)-1, respectively, was 37% and 48%.
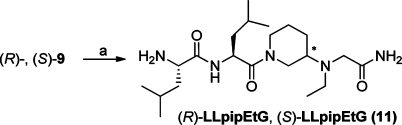
N-Ethylated byproducts were formed during the Cbz-deprotection to (R)- and (S)-LpipG (7) as well as (R)- and (S)-LLpipG (10) using catalytic hydrogenation (Pd/C 10%). Isolation of the byproducts after prolonged reaction time and subsequent TFA hydrolysis gave (R)- and (S)-LLpipEtG (11) (Scheme 3). Already after three hours, byproduct formation was observed. To verify the structure of the byproduct, Cbz-deprotected (R)-9 was converted to the corresponding ethylated product in 76% yield via reductive amination using acetaldehyde and NaBH(OAc)3 (reaction not shown). The NMR spectra of the synthesized compound and that of (R)-LLpipEtG (11) were identical. It has been proposed that a Pd-catalyzed oxidation of ethanol to acetaldehyde can occur during the hydrogenation reaction.21,22 The acetaldehyde forms an imine with the Cbz-deprotected secondary amine, and a subsequent reduction provides the ethylated byproduct.
Scheme 3.
Reagents and conditions: (a) (i) H2, Pd/C, ethanol, room temp, 144 h (23 and 13%), (ii) TFA in dichloromethane, room temp, 16 h (99% and 98%).
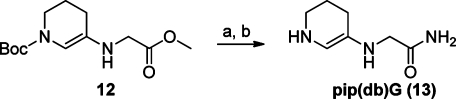
The unsaturated enamine byproduct 12 (Scheme 4) was collected from a reductive amination reaction of 1-Boc-3-piperidone with methyl glycinate performed in a highly concentrated reaction mixture. The opportunity to use the unsaturated piperidine as a scaffold for the synthesis of analogues was taken. Attempts to Cbz-protect 12 using the conditions described for (S)-4 gave only unreacted starting material back. Boc-Deprotection by TFA in dichloromethane was performed, but subsequent coupling with Boc-l-leucine did not provide satisfying results. However, 12 was amidated by treatment with NH3 in methanol and subsequent Boc-deprotection gave pip(db)G (13) in 90% (Scheme 4).
Scheme 4.
Reagents and conditions: (a) NH3 in methanol, room temp, 72 h (93%); (b) TFA/dichloromethane, room temp, 12 h (97%).
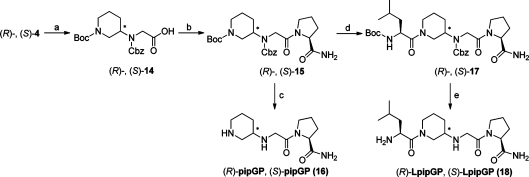
The synthetic route to the LeuGlyGlyPro-NH2 analogues (R)- and (S)-LpipGP (18), in which Gly236 in IgG is mimicked by the piperidine moiety, is shown in Scheme 5. Starting from the Cbz-protected core compounds (R)-and (S)-4, hydrolysis of the esters in excellent yields (95% and 99% of (R)- and (S)-14, respectively) followed by coupling to l-proline amide afforded (R)- and (S)-15, respectively, in high yields (82% and 78%). A small portion of (R)- and (S)-15 were deprotected to provide the GlyGlyPro-NH2 analogues (R)- and (S)-pipGP (16). Interestingly, the NMR spectra of the two diastereomers were highly similar. Furthermore, the coupling pattern in the 1H NMR spectra resembled the pattern seen for (R)- and (S)-pipG (3), implying that also here the monosubstituted piperidine ring adopts a defined chair conformation. To continue, removal of the Boc-protecting groups in (R)- and (S)-15 and subsequent couplings with Boc-l-leucine mediated by HOBt and EDC gave the epimers (R)- and (S)-17 in moderate to high yields (64% and 90%, respectively). The lower yield is due to the need of two purification steps. Also here, the NMR spectra recorded at different temperatures identified rotamers. The 1H NMR spectrum of (R)-17 showed five doublets assigned to the two methyl groups in the leucine side chain, implying that the molecule adopts several different conformations at room temperature. Finally, the two protecting groups were removed as described above to give the end products (R)- and (S)-LpipGP (18) in good yields (72% and 73%, respectively) both with excellent purities (100%) when analyzed by reversed phase HPLC. After the deprotection, the 1H NMR spectrum of both (R)- and (S)-LpipGP (18) showed two doublets assigned to the two nonequivalent methyl groups, as expected. The total yield for the synthesis of (R)- and (S)-LpipGP (18) over seven steps was 29% and 43%, respectively.
Scheme 5.
Reagents and conditions: (a) LiOH 1 M aq, THF/MeOH/H2O 3:1:1, 0 °C 30 min, room temp, 3 h (95 and 99%); (b) l-Pro-NH2, EDC, HOBt, Et3N, dichloromethane, room temp, 16 h (82% and 78%); (c) (i) H2, Pd/C, ethanol, room temp, 24 h, (ii) TFA, room temp, 4 h (quant yields over two steps); (d) (i) TFA, room temp, 16 h, (ii) Boc-l-Leu × H2O, EDC, HOBt, dichloromethane, molecular sieves, Et3N, room temp, 15 h (64% and 90% over two steps); (e) (i) H2, Pd/C, ethanol, room temp, 48 and 24 h, (ii) TFA, room temp, 4 h (72% and 73% over two steps).
The corresponding tetrapeptide analogues in which the piperidine moiety replaces Gly237 in IgG ((R)- and (S)-LGpipP (23)) were also synthesized (Scheme 6). Reductive amination of Boc-protected 3-piperidone with l-proline amide and NaBH(OAc)3 afforded 19 in 68% yield and a diastereomeric ratio of 60:40. The diastereomers were separated by preparative HPLC to give (R)- and (S)-19, and the absolute configurations were determined by chemical correlation.23
Scheme 6.
Reagents and conditions: (a) (i) l-Pro-NH2, NaBH(OAc)3, HOAc, dichloromethane, room temp, 20 h (68%), (ii) preparative HPLC; (b) (i) TFA/dichloromethane, room temp, (ii) Boc-Gly, EDC, HOBt, dichloromethane, 0 °C → room temp, 15 h, 45 and 47% over two steps); (c) TFA/dichloromethane, room temp, 20 h (quant yields); (d) (i) TFA/dichloromethane, room temp, 20 h (quant yields), (ii) Boc-l-Leu, EDC, HOBt, Et3N, dichloromethane, 0 °C → room temp, 15 h (93% and 78%); (e) TFA/dichloromethane, room temp, 12 h (quant yield and 98%).
Boc-protected glycine was coupled to each deprotected diastereomer using EDC and HOBt to afford (R)- and (S)-20 in 45% and 47% yield, respectively. Deprotection with TFA in dichloromethane gave quantitative yields of (R)- and (S)-GpipP (21) as TFA salts. A subsequent coupling to Boc-protected l-leucine gave (R)- and (S)-22, respectively, in high yields (93% and 78%). Deprotection with TFA in dichloromethane overnight afforded (R)- and (S)-LGpipP (23) in excellent yields. The total yield for the synthesis of (R)- and (S)-LGpipP (23) in five steps from (R)- and (S)-19 was 42% and 36%, respectively.
IgG-Based Peptides As Potential Inhibitors of IdeS
Eight different peptides based on the amino acid sequence of the IgG hinge region (Figure 2) were tested for putative inhibitory activity toward IdeS.24
Figure 2.
Peptides tested for inhibitory activity.
The inhibitory activity was screened in a gel-electrophoresis (SDS-PAGE) based assay to detect IgG cleavage products.25 None of the peptides was found to be active as inhibitors of IdeS (data not shown). Thus, although the sequences of the tested peptides are identical to the IgG sequence around the IdeS cleavage site, these short peptides did apparently not bind strongly enough to the enzyme to affect enzymatic activity.
Inhibitory Capacity of Peptide Analogues toward IdeS
In total 17 piperidine-containing analogues of the peptide sequences GG, LLGG, and LGGP, all based on the IdeS cleavage site in human IgG1, were tested for their inhibitory activity toward IdeS. Inhibitory effects were screened by SDS-PAGE and quantified by surface plasmon resonance spectroscopy (Figure 3).15
Figure 3.
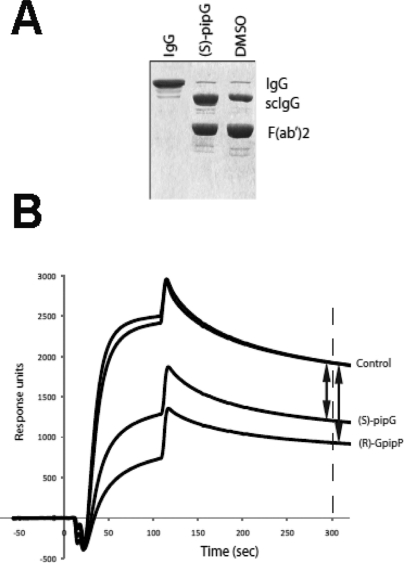
(A) Representative nonreducing SDS-PAGE analysis of IgG and IgG hydrolyzed by IdeS in the presence ((S)-pipG ((S)-3)) or absence (DMSO) of peptide analogues. scIgG indicates single-chain cleaved IgG; F(ab′)2 indicates double-chain cleaved IgG. (B) Representative surface plasmon resonance spectroscopy curves of uncleaved IgG (control) and cleaved IgG in presence of peptide analogues (S)-pipG ((S)-3) and (R)-GpipP ((R)-21). Differences in response units are indicated by arrows. (S)-pipG ((S)-3) has greater inhibitory activity than (R)-GpipP ((R)-21).
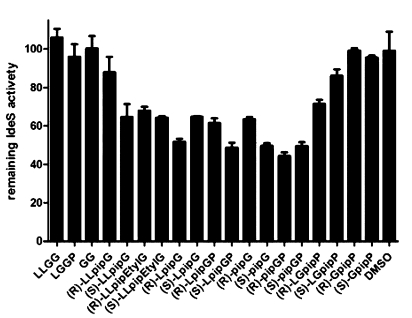
Whereas the corresponding peptides were inactive, the peptide analogues showed a significant inhibitory activity (Figure 4). The most potent inhibitors of IdeS activity are (R)-pipGP ((R)-16), (S)-LpipGP ((S)-18), (S)-pipGP ((S)-16), (S)-pipG ((S)-3), and (R)-LpipG ((R)-7). In general, peptide analogues in which the piperidine moiety replaces the glycine corresponding to G236 in IgG (pip236G) appear to be stronger inhibitors than analogues with the piperidine moiety replacing G237 of IgG (Gpip237), independently of C-terminal or N-terminal amino acid extensions. For example, (R)-pipGP ((R)-16) is more active than (R)-GpipP ((R)-21) and (S)-LpipGP ((S)-18) is more active than (S)-LGpipP ((S)-23). The two (R)- and (S)-epimers of each analogue pair were tested, but no clear trend how stereochemistry of the piperidine moiety affected potency could be seen (Figure 4).
Figure 4.
IdeS activity in presence of peptides and peptide analogues using IgG1 as substrate. IdeS activity is expressed relative to a standard curve obtained in absence of inhibitors. (R) and (S) indicate the different stereoisomers of each analogue; “pip” represents the piperidine moiety replacing glycine residues. The enzyme activity was determined for IdeS at a final concentration of 0.8 μM in the presence of 4.8 × 103 molar excess of peptides/peptide analogues.
The ethylated analogues (R)- and (S)-LLpipEtG (11) have also been tested, both showing comparable inhibitory activity against IdeS as (S)-LLpipG ((S)-10). This implies that the secondary amine is not involved in hydrogen bond donation and that there is space enough for an ethyl group. Interestingly, the structural difference between LpipGP (18) and pipG (3) is two amino acids, but they show similar inhibitory activity. This indicates that the extra amino acids are not contributing to more favorable interactions to accomplish more efficient binding or are too flexible to gain favorable binding energies. Thus, when evaluating ligand efficiency (LE), where binding energy per atom is considered, (S)-pipG ((S)-3) is the most favorable peptide analogue in the series.26
The pKa values for (R)- and (S)-pipG (3) and (R)- and (S)-LpipGP (18) have been determined (Table 1). Interestingly, the two pKa values differ considerably. For (R)- and (S)-pipG (3), the acyclic amine bears the electron withdrawing acetamide group, thus the piperidine nitrogen would be expected to be protonated first. For (R)- and (S)-LpipGP (18), the primary amine would be expected to have the pKa of an amino acid amide, i.e., between 8 and 9, which is what is found. The acyclic secondary amines will not be protonated at physiological pH, and the pip moiety is therefore able to act as a glycine isostere.
Table 1. pKa Values for the pipG and LpipGP Analoguesa.
| measured |
predicted |
|||
|---|---|---|---|---|
| compd | pKa1 | pKa2 | pKa1 | pKa2 |
| (R)-pipG | 4.41 | 10.11 | 5.40 | 9.84 |
| (S)-pipG | 4.35 | 10.06 | ||
| (R)-LpipGP | 5.13 | 8.46 | 7.35 | 9.13 |
| (S)-LpipGP | 5.21 | 8.53 | ||
Specificity of Peptide Analogues
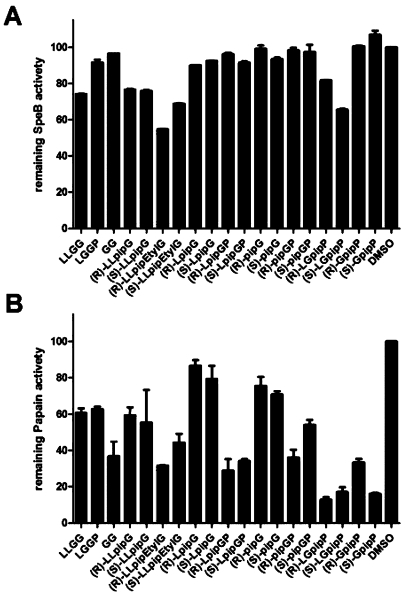
IdeS exhibits pronounced substrate specificity in comparison to other IgG cleaving cysteine proteases of the papain superfamily.12 Still, the extended structural similarity of papain-like proteases raised the question whether piperidine containing peptide analogues could have inhibitory capacity toward other IgG cleaving proteases or in fact harbor a potential specificity toward IdeS. Therefore, peptides and peptide analogues were also screened for activity toward the streptococcal cysteine protease SpeB and the family protease papain (Figure 5).
Figure 5.
Inhibitory profile of peptides and peptide analogues on the activity of streptococcal cysteine protease SpeB (A) and family class protease papain (B). Activity is expressed relative to enzyme activity in absence of putative inhibitors. (R) and (S) indicate the different stereoisomers of each analogue; “pip” represents the piperidine moiety replacing glycine residues. The enzyme activity was determined for papain at a final concentration of 0.9 μM and SpeB at final concentration of 0.28 μM in the presence of 4.8 × 103 molar excess of peptides/peptide analogues.
Interestingly, very different inhibitory profiles were obtained that apparently distinguish the proteases from the profile obtained for IdeS and also from each other (Figures 4 and 5). In general, the inhibitory effect of all the tested compounds was much less pronounced on SpeB compared to inhibition of IdeS. Hardly any of the pip236G-fragment containing analogues had any inhibitory effect on SpeB activity (Figure 5A). Notable is however that an inhibitory capacity was observed for the LLGG peptide. In contrast to the effect observed on IdeS activity (Figure 4), the degree of inhibition was not improved when testing LLpipG (10) peptide analogues in which Gly236 was replaced with a piperidine moiety. However, a clearly improved degree of inhibition could be observed when testing the ethylated analogues (R)- and (S)-LLpipEtG (11). In addition, (R)- and (S)-LGpipP (23) containing a similar tertiary amine fragment as the ethylated analogues also inhibited SpeB activity, which is in contrast to the effect observed for IdeS. Thus, the absence of the possibility of hydrogen bond donation and the rigid structure of neighboring piperidine and proline rings appear to increase the interaction between the LGpipP (23) analogues and SpeB. Consequently, LpipGP (18) analogues do not affect SpeB activity.
The specificity study was extended further to include the cysteine protease papain. Like SpeB also, papain exhibits a broad proteolytic activity and degrades a wide variety of substrates, including IgG. The analysis of putative inhibitory effects of peptides and piperidine-containing analogues on papain revealed that the enzyme was clearly very sensitive toward most compounds tested (Figure 5B). In analogy to SpeB, the positioning of the piperidine in the peptide sequence favors a replacement of the glycine corresponding to G237, resulting in the most efficient papain inhibitors in the series, with (R)-LGpipP ((R)-23) being the most active analogue.
In summary, the most active inhibitors of IdeS, (R)-pipGP ((R)-16) and (S)-LpipGP ((S)-18), are even more active toward papain but show no inhibitory activity toward SpeB. However, both (S)-pipG ((S)-3) and (R)-LpipG ((R)-7) show promising selectivity profiles as they are both potent inhibitors of IdeS but are hardly affecting papain and do not inhibit SpeB. These compounds were investigated further to evaluate whether they might be a suitable starting point for the development of specific IdeS protease inhibitors.
Various amounts of inhibitor and substrate were used to determine the inhibition constant (Ki) for these two compounds. Because a mechanism of competitive inhibition was postulated, initial velocities were determined in the presence or absence of inhibitor. The binding affinity of IdeS for IgG has been previously determined, and a high affinity was observed with a Kd of 2.5 μM and a Km for complete enzymatic cleavage ranging from 6.8 to 18.9 μM depending on the IgG subtype.12
Because of the high affinity substrate binding, a considerable excess of inhibitor was expected to be necessary to affect initial velocities and to successfully compete for IgG. The Ki values for (S)-pipG ((S)-3) and (R)-LpipG ((R)-7) were determined to approximately 6.7 and 5.7 mM, respectively. Although these concentrations are too high for in vivo evaluation, they were surprisingly low considering the high affinity of IdeS for IgG. Importantly, initial velocities of papain activity remained unchanged by the presence of the inhibitors and therefore Ki values could not be determined reliably, but the values exceed those observed for IdeS at least 16–267-fold, respectively.
Thus, the aminopiperidine containing analogues (S)-pipG ((S)-3) and (R)-LpipG ((R)-7) exhibit effective and specific IdeS inhibitor properties and will serve as a starting point for further development of more potent inhibitors. Initially, an attempt to increase the rigidity of the compound will be undertaken which hopefully will increase the potency of the inhibitor. Synthesis of such compounds is ongoing.
Conclusion
A series of 17 piperidine-containing peptide analogues have been efficiently synthesized. The peptide analogues were tested in inhibition assays addressing cysteine protease activities with focus on the streptococcal IgG degrading enzyme IdeS. Several of the synthesized piperidine-containing peptide analogues are active IdeS inhibitors, but the piperidine moiety does not necessarily improve the inhibitory capacity of the analogues toward other cysteine proteases. To the best of our knowledge, these are the first noncovalently bound inhibitors of IdeS, also showing selectivity toward SpeB and papain.
Experimental Section
Synthesis. General
Starting materials and reagents were purchased from Sigma-Aldrich or Bachem and were used as such. The peptides were purchased from CASLO Laboratory ApS, Lyngby, Denmark. The purity of the peptides was >98%. Analytical TLC was performed on silica plated aluminum sheets (grade 60 F254, Merck). The spots were visualized by treatment with a dip solution [KMnO4 (1.5 g), K2CO3 (10 g), NaOH (10%) (1.25 mL) in water (200 mL)], followed by heating. Flash chromatography was performed on Merck silica gel 60 (0.040–0.063 mm). 1H and 13C NMR spectra were recorded on a JEOL Eclipse 400 spectrometer at 400 and 100 MHz, respectively, at ambient temperature and in CDCl3 if not otherwise stated. 13C NMR spectra were also recorded on a Varian 500 spectrometer at 125 MHz. Chemical shifts are reported in ppm, with the solvent residual peak as internal standard (CHCl3 δH 7.26, δC 77.0, CH3OH δH 3.30, δC 49.0). DEPT, COSY, TOCSY, and HSQC spectra were recorded to validate structural assignments of the NMR signals. Separation of the stereoisomers of 1 and 19 were performed on a preparative HPLC Shimadzu system equipped with a Chiralpak AD 250 mm × 20 mm column (Daicel Chemical CO., Tokyo, Japan) with a mobile phase consisting of hexane/ethanol 95:5 or hexane/2-propanol 90:10 at a flow rate of 15 or 10 mL/min, respectively. Detection was carried out at 240 nm. Chiral HPLC analysis was performed on a Varian system equipped with a Chiralpak AD 4.6 mm × 250 mm column (Daicel Chemical CO., Tokyo, Japan) at a flow rate of 1 mL/min. Detection was carried out at 240 nm. Analytical reversed phase HPLC was performed on a Waters 2690 system (photodiode array detector at 220 nm and flow rate 1.0 mL/min) equipped with a 250 mm × 4.6 mm Reprosil PUR C18aq column. Optical rotation was measured with a Perkin-Elmer 341 LC polarimeter at 20 °C. Infrared spectra were recorded on a Perkin-Elmer 16 PC FTIR spectrometer, and only the major peaks are listed. HRMS analyses were run at BioAnSer, Gothenburg, Sweden. The purity of all tested compounds was determined using analytical reversed phase HPLC on a Waters 2690 system (photodiode array detection at 220 nm and a flow rate of 1.0 mL/min) equipped with a 250 mm × 4.6 mm Reprosil PUR C18aq column. (S)- and (R)-pipG (3) were further analyzed using a reversed phase Waters Acquity UPLC system equipped with a BEC C18 column, connected to a MS instrument with a Waters SQD single quadrupole detector. All compounds showed a purity >95% except (S)-pipG ((S)-3) which was 90% pure. Measurement of the aqueous dissociation constant, pKa values, of (R)- and (S)-LpipGP (18) and (R)- and (S)-pipG (3) were performed at AstraZeneca, R&D Mölndal, Sweden.27
Ethyl 2-[(1-tert-Butoxycarbonylpiperidin-3-yl)amino]acetate ((rac)-1)
NaBH(OAc)3 (5.3 g, 25.0 mmol) was added to a solution of 1-Boc-piperidin-3-one (4.98 g, 25.0 mmol), acetic acid (2.25 g, 37.5 mmol), glycine ethyl ester hydrochloride (3.5 g, 25.0 mmol), and molecular sieves (4.5 g) in dichloromethane (100 mL). The reaction mixture was stirred at room temperature for 20 h, diluted with dichloromethane (100 mL), and quenched by the addition of aqueous NaOH (3 M) (20 mL). The aqueous phase was extracted with dichloromethane (2 × 100 mL). The combined organic phases were dried (Na2SO4), filtered, and concentrated in vacuo. Purification by flash chromatography (heptane/ethyl acetate 1:2, Rf = 0.18) gave (rac)-1 as a transparent oil (4.4 g, 62%). Resolution of a portion of the racemate (2.0 g) by chiral preparative HPLC gave the enantiomers of 1.
(−)-Ethyl 2-[(1-tert-Butoxycarbonyl-(R)-piperidin-3-yl)amino]acetate ((R)-1)
Resolution of (rac)-1 (2.0 g) gave enantiomerically pure (R)-1 (779 mg). Analytical chiral HPLC (hexane/2-propanol 95:5) tR 9.4 min; 95% purity, ee 100%. 1H NMR δ 4.18 (q, J = 7.1 Hz, 2H), 3.80–3.73 (m, 2H), 3.55–3.41 (AB system, 2H), 2.86–2.51 (m, 3H), 2.20–1.80 (m, 2H), 1.73–1.64 (m, 1H), 1.47–1.21 (m, 2H), 1.44 (s, 9H), 1.26 (t, J = 7.1 Hz, 3H). 13C NMR δ 172.2, 154.6, 79.2, 60.6, 53.3, 48.6, 48.1, 43.9, 31.1, 28.2, 23.2, 14.0; [α]D20 −8.5 (c 1.0, CHCl3). IR (neat) v 3515, 3333, 2976, 2933, 2860, 1738, 1691, 1423, 1157 cm–1. HRMS (FT-ICR-MS) calcd for C14H26N2O4 [M + H]+ 287.1965; found 287.1962. For NMR spectral assignments, see the Supporting Information.
(+)-Ethyl 2-[(1-tert-Butoxycarbonyl-(S)-piperidin-3-yl)amino]acetate ((S)-1)
Resolution of (rac)-1 (2.0 g) gave enantiomerically pure (S)-1 (802 mg). Analytical chiral HPLC (hexane/2-propanol 95:5) tR 8.7 min, 94% purity, ee 100%. 1H NMR δ 4.18 (q, J = 7.1 Hz, 2H), 3.81–3.73 (m, 2H), 3.51–3.37 (AB system, 2H), 2.93–2.48 (m, 3H), 2.03–1.80 (m, 3H), 1.73–1.64 (m, 1H), 1.49–1.21 (m, 10H), 1.26 (t, J = 7.1 Hz, 3H). 13C NMR δ 172.2, 154.6, 79.1, 60.5, 53.3, 48.9, 48.1, 43.9, 31.0, 28.2, 23.2, 13.9; [α]D20 +8.5 (c 1.0, CHCl3). IR (neat) v 3515, 3333, 2976, 2933, 2860, 1738, 1691, 1423, 1157 cm–1. HRMS (FT-ICR-MS) calcd for C14H26N2O4 [M + H]+ 287.1965; found 287.1962.
Both enantiomers of 1 were also obtained separately by reductive alkylation of 3-amino-1-tert-butyloxycarbonylpiperidine with defined stereochemistry. These products were used as reference material.
(−)-Ethyl 2-[(1-tert-Butoxycarbonyl-(R)-piperidin-3-yl)amino]acetate ((R)-1)
Ethyl glyoxylate in toluene (50%) (0.5 mL, 2.5 mmol) was added dropwise to a mixture of (R)-3-amino-1-tert-butyloxycarbonylpiperidine (500 mg, 2.5 mmol) and Na2SO4 (900 mg) in dichloromethane (9 mL). The reaction mixture was stirred at room temperature for 2 h. NaBH(OAc)3 (529 mg, 2.5 mmol) and AcOH (225 mg, 3.74 mmol) were added, and the stirring continued for 90 min. The mixture was diluted with dichloromethane (20 mL) and quenched by addition of aqueous NaOH (3 M) (8 mL). The aqueous phase was extracted with dichloromethane (1 × 20 mL). The combined organic phases were dried (Na2SO4) and filtered and the solvent was removed in vacuo. Purification by flash chromatography (heptane/ethyl acetate 1:3, Rf = 0.17) gave (R)-1 as a transparent oil (410 mg, 60%). Analytical chiral HPLC (hexane/2-propanol 95:5) tR 9.3 min; ee 90%. NMR data were in agreement with those reported for the pure enantiomer (R)-1.
(+)-Ethyl 2-[(1-tert-Butoxycarbonyl-(S)-piperidin-3-yl)amino]acetate ((S)-1)
Compound (S)-1 was synthesized from (S)-3-amino-1-tert-butyloxycarbonylpiperidine (500 mg, 2.5 mmol) using the same procedure as described for (R)-1 to afford (S)-1 as a transparent oil (380 mg, 56%). Analytical chiral HPLC (hexane/2-propanol 95:5) tR 8.5 min; ee 86%. NMR data were in agreement with those reported for the pure enantiomer (S)-1.
(−)-2-[(1-tert-Butoxycarbonyl-(R)-piperidin-3-yl)amino]acetamide ((R)-2)
Compound (R)-2 was synthesized from (R)-1 (210 mg, 0.73 mmol) and NH3 in methanol (7 N) (4.2 mL) as described for (S)-2 to give the product as a white solid (174 mg, 92%). Analytical HPLC (hexane/ethanol 97:3) tR 84.4 min, purity 100%. 1H NMR δ 7.05 (br s, 2H), 5.53 (br s, 1H), 3.97–3.54 (m, 2H), 3.38–3.25 (AB system, 2H), 3.16–2.67 (m, 2H), 2.56 (app tt, J = 8.1, 8.1, 3.7, 3.7 Hz, 1H), 1.97–1.84 (m, 1H), 1.73–1.63 (m, 1H), 1.51–1.23 (m, 2H), 1.45 (s, 9H). 13C NMR δ 175.1, 154.6, 79.2, 53.7, 49.5, 48.7, 43.8, 30.9, 28.1, 22.9; [α]D20 −4.6 (c 1.0, CHCl3). IR (KBr) v 3398, 2977, 2933, 1683, 1638, 1423, 1262, 1180, 1155 cm–1. HRMS (FT-ICR-MS) calcd for C12H23N3O3 [M + H]+ 258.1812; found 258.1809. For NMR spectral assignment, see the Supporting Information.
(+)-2-[(1-tert-Butoxycarbonyl-(S)-piperidin-3-yl)amino]acetamide ((S)-2)
NH3 in methanol (7 N) (4.3 mL) was added to (S)-1 (214 mg, 0.75 mmol), and the reaction mixture was stirred at room temperature for 72 h. The solvent and excess of reagent were removed in vacuo. The remaining was trituated with hexane/diethyl ether (1:1) (1 mL) to give (S)-2 as a white solid (173 mg, 90%). Analytical chiral HPLC (hexane/ethanol 97:3) tR 76.0 min, purity 100%. 1H NMR δ 7.06 (br s, 2H), 5.62 (br s, 1H), 3.94–3.56 (m, 2H), 3.39–3.24 (AB system, 2H), 3.32–2.72 (m, 2H), 2.56 (app tt, J = 8.1, 8.1, 3.7, 3.7 Hz, 1H), 1.95–1.86 (m, 1H), 1.73–1.57 (m, 2H), 1.45 (s, 9H), 1.39–1.23 (m, 2H). 13C NMR δ 175.0, 154.5, 79.1, 53.6, 49.4, 48.6, 43.7, 30.8, 28.0, 22.8; [α]D20 +5.1 (c 1.0, CHCl3). IR (KBr) v 3398, 2977, 2933, 1683, 1638, 1423, 1262, 1180, 1155 cm–1. HRMS (FT-ICR-MS) calcd for C12H23N3O3 [M + H]+ 258.1812; found 258.1810.
(−)-2-((R)-Piperidin-3-ylamino)acetamide × 2TFA ((R)-pipG) ((R)-3)
Deprotection of (R)-2 (137 mg, 0.53 mmol) using TFA (364 mg, 3.2 mmol) was performed as described for (S)-pipG ((S)-3) to give the TFA salt of (R)-pipG ((R)-3) as a white solid (210 mg, quant). 1H NMR (CD3OD) δ 4.00–3.88 (AB system, 2H), 3.73 (dd, J = 12.1, 4.0 Hz, 1H), 3.63–3.51 (app tt, J = 11.4, 11.4, 4.1, 4.1 Hz, 1H), 3.40 (app dd, J = 12.9, 3.6 Hz, 1H), 3.08 (app t, J = 11.7 Hz, 1H), 2.96 (dt, J = 12.7, 12.7, 3.2 Hz, 1H), 2.32–2.24 (m, 1H), 2.14–2.04 (m, 1H), 1.87–1.78 (m, 1H), 1.71 (app ddt, J = 12.5, 12.5, 3.6 Hz, 1H). 13C NMR (CD3OD) δ 168.5, 53.0, 46.6, 45.0, 44.6, 26.2, 21.6; [α]D20 −9.3 (c 0.41, CH3OH). IR (neat) v 3399, 3012, 2822, 1683, 1436, 1205, 1130 cm–1. HRMS (FT-ICR-MS) Calcd for C7H15N3O [M + H]+ 158.1288; found 158.1287; pKa1 = 5.13, pKa2 = 8.46. For NMR spectral assignment, see the Supporting Information.
(+)-2-((S)-Piperidin-3-ylamino)acetamide × 2TFA ((S)-pipG) ((S)-3)
TFA (375 mg, 3.3 mmol) was added to (S)-2 (141 mg, 0.55 mmol) in dichloromethane (3 mL), and the reaction mixture was stirred at room temperature for 12 h. The solvent and excess of reagent were removed in vacuo to give the TFA-salt of (S)-pipG ((S)-3) as a white solid (210 mg, quant). 1H NMR (CD3OD) δ 4.00–3.85 (AB system, 2H), 3.76–3.65 (m, 1H), 3.56 (app tt, J = 11.4, 11.4, 3.7, 3.7, 1H), 3.44–3.34 (m, 1H), 3.08 (app t, J = 11.7 Hz, 1H), 2.96 (app dt, J = 12.8, 12.8, 3.3 Hz, 1H,), 2.33–2.21 (m, 1H), 2.15–2.01 (m, 1H), 1.88–1.60 (m, 2H). 13C NMR (CD3OD) δ 168.4, 53.0, 46.6, 45.0, 44.6, 26.2, 21.6; [α]D20 +6.6 (c 0.41, CH3OH). IR (neat) v 3383, 3014, 2824, 1676, 1432, 1202, 1130 cm–1. HRMS (FT-ICR-MS) calcd for C7H15N3O [M + H]+ 158.1288; found 158.1287. The purity was 90% according to UPLC analysis. pKa1 = 5.21, pKa2 = 8.53.
(−)-Ethyl 2-[(Benzyloxycarbonyl)(1-tert-butoxycarbonyl-(R)-piperidin-3-yl)amino]acetate ((R)-4)
Compound (R)-4 was synthesized from (R)-1 (802 mg, 2.8 mmol) and N-(benzyloxycarbonyloxy)succinimide (698 mg, 2.8 mmol) with Et3N (312 mg, 3.1 mmol) in dichloromethane (11 mL) as described for (S)-4 to give the product as a transparent oil (0.98 g, 82%). Analytical chiral HPLC (hexane/2-propanol 95:5) tR 19.8 min, 98% purity, ee 100%. 1H NMR (rotamers) δ 7.59–7.20 (m, 5H), 5.19 and 5.10 (rot, s, 2H), 4.34–3.72 (m, 6H), 2.80–2.43 (m, 3H), 2.03–1.87 (m, 1H), 1.80–0.98 (m, 1H), 1.43–1.17 (m, 2H), 1.43 and 1.37 (rot, s, 9H), 1.26 and 1.17 (rot, t, J = 7.1 Hz, 3H). 13C NMR δ 169.7, 155.8, 155.1, 154.5, and 154.3 (rot), 136.1, 128.3, and 128.2 (rot), 127.8, 127.5, 79.4, 67.4, and 67.1 (rot), 60.9, 53.1, and 52.9 (rot), 47.0 and 46.5, 45.6 and 45.3, 43.2, 28.7, 28.1, 24.4, 13.9; [α]D20 −14.9 (c 1.0, CHCl3). IR (neat) v 3500, 2976, 2864, 1749, 1693, 1416, 1152 cm–1. HRMS (FT-ICR-MS) calcd for C22H32N2O6 [M + H]+ 421.2333; found 421.2333. For NMR spectral assignment, see the Supporting Information.
(+)-Ethyl 2-[(Benzyloxycarbonyl)(1-tert-butoxycarbonyl-(S)-piperidin-3-yl)amino]acetate ((S)-4)
Et3N (290 mg, 2.9 mmol) in dichloromethane (2 mL) was added to a solution of (S)-1 (745 mg, 2.6 mmol) in dichloromethane (8 mL) at 0 °C, and N-(benzyloxycarbonyloxy)succinimide (648 mg, 2.6 mmol) was added in portions. The reaction mixture was stirred at room temperature for 17 h, diluted with dichloromethane (30 mL), and washed with brine (2 × 15 mL). The organic phase was dried (Na2SO4), filtered, and concentrated in vacuo. Purification by flash chromatography (heptane/ethyl acetate 2:1, Rf = 0.23) gave (S)-4 as a transparent oil (0.94 g, 84%). Analytical chiral HPLC (hexane/2-propanol 95:5) tR 18.7 min, 98% purity, ee 100%. 1H NMR (rotamers) δ 7.48–7.19 (m, 5H), 5.18 and 5.11 (rot, s, 2H), 4.31–3.79 (m, 6H), 2.80–2.41 (m, 3H), 2.04–1.89 (m, 1H), 1.80–1.32 (m, 12H), 1.27 and 1.17 (rot, t, J = 7.1 Hz, 3H). 13C NMR δ 169.8, 155.9, 155.1, 154.5, and 154.4 (rot), 136.2, 128.3, and 128.2 (rot), 127.8, 127.6, 79.5, 67.5, and 67.1 (rot), 61.0, 53.2, and 53.0 (rot), 47.1 and 46.5 (rot), 45.6 and 45.4 (rot), 43.3, 28.7, 28.2, 24.4, 13.9; [α]D20 +14.0 (c 1.0, CHCl3). IR (neat) v 3499, 2976, 2863, 1747, 1693, 1415, 1152 cm–1; HRMS (FT-ICR-MS) calcd for C22H32N2O6 [M + H]+ 421.2333; found 421.2334.
(−)-Ethyl 2-[(Benzyloxycarbonyl)(1-(tert-butoxycarbonyl-l-leucinyl)-(R)-piperidin-3-yl)amino]acetate ((R)-5)
Deprotection of (R)-4 (870 mg, 2.07 mmol) with TFA (2.0 mL, 27.0 mmol) in dichloromethane (8 mL) was performed as described for (S)-5 to give the Boc-deprotected intermediate. The crude intermediate and Boc-l-Leu × H2O (551 mg, 2.21 mmol) were coupled as described for (S)-5 using EDC (424 mg, 2.21 mmol), HOBt (299 mg, 2.21 mmol), and Et3N (813 mg, 8.04 mmol) in dichloromethane (11 mL) with molecular sieves (600 mg). Purification by flash chromatography (heptane/ethyl acetate 1:1, Rf = 0.2) gave (R)-5 as a transparent oil (1.0 g, 91% yield over two steps). Analytical chiral HPLC (hexane/ethanol 70:30) tR 7.5 min, 99% purity, de 97%. 1H NMR (CD3OD) δ 7.41–7.25 (m, 5H), 5.19–5.10 (m, 1H), 5.06 (s, 2H), 4.64–4.38 (m, 2H), 4.23–3.84 (m, 6H), 3.22–3.04 (m, 1H), 3.03–2.91 (m, 1H), 2.76–2.61 (m, 1H), 2.55–2.41 (m, 1H), 1.99–1.56 (m, 2H), 1.55–1.07 (m, 14H), 1.00–0.72 (m, 6H). 13C NMR (CD3OD) δ 173.7 and 173.4 (rot), 171.8 and 171.7 (rot), 157.6, 157.2, 137.8, 129.5, 129.1, 128.8, 80.5, 68.6, 62.3, 55.0, 50.2, 46.7, 46.0, 43.1, 42.4, and 42.1 (rot), 29.0, 28.7, 26.0, 25.4, 23.6, 22.2, 14.4; [α]D20 −31.5 (c 1.0, CH3OH). IR (KBr) v 3423, 2959, 1753, 1708, 1642, 1445, 1205 cm–1. HRMS (FT-ICR-MS) calcd for C28H43N3O7 [M + H]+ 534.3174; found 534.3176.
(+)-Ethyl 2-[(Benzyloxycarbonyl)(1-(tert-butoxycarbonyl-l-leucinyl)-(S)-piperidin-3-yl)amino]acetate ((S)-5)
TFA (2.0 mL, 27.0 mmol) was added to (S)-4 (950 mg, 2.26 mmol) in dichloromethane (8 mL) and stirred at room temperature for 16 h. The solvent and excess of reagents were removed in vacuo to give the Boc-deprotected intermediate. Boc-l-Leu × H2O (620 mg, 2.49 mmol), EDC (477 mg, 2.49 mmol), and HOBt (336 mg, 2.49 mmol) were stirred in dichloromethane (6 mL) with molecular sieves (600 mg) at 0 °C for 15 min. Into this solution, the Boc-deprotected intermediate and Et3N (9.14 mg, 9.04 mmol) in dichloromethane (5 mL) were added and the reaction mixture was stirred at room temperature for 15 h. The mixture was diluted with dichloromethane (20 mL) and washed with aqueous citric acid (10%) (2 × 10 mL), saturated aqueous NaHCO3 (2 × 10 mL), and brine (1 × 10 mL). The organic phase was dried (Na2SO4) and filtered. The solvent was removed in vacuo. Purification by flash chromatography (heptane/ethyl acetate 1:1, Rf = 0.18) gave (S)-5 as a transparent oil (950 mg, 79% yield over two steps). Analytical chiral HPLC (hexane/ethanol 70:30) tR 6.4 min, purity 100%. 1H NMR (CD3OD) δ 7.49–7.22 (m, 5H), 5.44–5.15 (m, 1H), 5.07 (s, 2H), 4.72–4.47 (m, 2H), 4.24–3.65 (m, 6H), 3.58–3.30 (m, 1H), 3.12–2.81 (m, 1H), 2.69–2.48 (m, 1H), 2.47–2.23 (m, 1H), 2.12–1.90 (m, 2H), 1.89–1.10 (m, 15H), 1.06–0.72 (m, 6H). 13C NMR (CD3OD) δ (174.0, 173.8 and 173.4, rot), 171.8, 171.6, (157.7, 157.0 and 156.8, rot), 137.8 and 137.6 (rot), 129.5 and 129.4 (rot), 129.2 and 129.1 (rot), 128.8 and 128.7 (rot), 80.4 and 80.1 (rot), 68.5 and 68.3 (rot), 62.2, 54.5, and 54.3 (rot), 50.4, 48.0, 46.4, and 46.2 (rot), 45.8, 30.1, 29.7, and 29.5 (rot), 28.8, 26.2, 25.9, and 25.7 (rot), 23.7, 22.0, 14.5; [α]D20 +25.7 (c 1.0, CH3OH). IR (KBr) v 3425, 2958, 1754, 1720, 1640, 1442, 1204 cm–1. HRMS (FT-ICR-MS) calcd for C28H43N3O7 [M + H]+ 534.3174; found 534.3174.
(−)-2-[(Benzyloxycarbonyl)(1-(tert-butoxycarbonyl-l-leucinyl)-(R)-piperidin-3-yl)amino]acetamide ((R)-6)
Compound (R)-6 was synthesized from (R)-5 (200 mg, 0.38 mmol) and NH3 in methanol (7 N) (4.0 mL) as described for (S)-2. Purification by flash chromatography (dichloromethane/methanol 95:5, Rf = 0.09) gave (R)-6 as a white solid (122 mg, 65%). Analytical chiral HPLC (hexane/ethanol 60:40) tR 12.3 min, 95% purity, de 90%. 1H NMR (CD3OD, 50 °C) δ 7.39–7.23 (m, 5H), 5.12 (br s, 2H), 4.49–4.44 (m, 1H), 4.11–3.80 (m, 3H), 3.18–3.04 (m, 1H), 3.03–2.89 (m, 1H), 2.77–2.61 (m, 1H), 2.56–2.40 (m, 1H), 2.03–1.91 (m, 1H), 1.85–1.27 (m, 15H), 1.02–0.73 (m, 6H). 13C NMR (CD3OD, 50 °C) δ 174.4, 173.7, and 173.4 (rot), 157.6, 137.8, 129.5, 129.1, 128.8, 80.5, 68.7, 54.9, 50.2, 47.6, 46.7, and 46.4 (rot), 43.1, 42.4, 28.8, 26.6, 25.9, 25.5, 23.5, 22.2; [α]D20 −33.9 (c 1.0, CH3OH). IR (KBr) v 3414, 2956, 1707, 1640, 1446 cm–1. HRMS (FT-ICR-MS) Calcd for C26H40N4O6 [M + H]+ 505.3021; found 505.3021.
(+)-2-[(Benzyloxycarbonyl)(1-(tert-butoxycarbonyl-l-leucinyl)-(S)-piperidin-3-yl)amino]acetamide ((S)-6)
Compound (S)-6 was synthesized from (S)-5 (156 mg, 0.29 mmol) and NH3 in methanol (7 N) (4.0 mL) as described for (S)-2. Purification by flash chromatography (dichloromethane/methanol 95:5, Rf = 0.07) gave the product as a white solid (108 mg, 73%). Analytical chiral HPLC (hexane/ethanol 60:40) tR 8.5 min, 100% purity. 1H NMR (CD3OD, 50 °C) δ 7.46–7.23 (m, 5H), 5.12 (br s, 2H), 4.49–4.38 (m, 1H), 4.20–3.79 (m, 3H), 3.26–3.10 (m, 1H), 3.09–2.93 (m, 1H), 2.77–2.62 (m, 1H), 2.56–2.38 (m, 1H), 2.05–1.61 (m, 4H), 1.60–1.24 (12H), 1.00–0.75 (m, 6H). 13C NMR (CD3OD, 50 °C) δ 174.4, 173.8, and 173.5 (rot), 157.7, 137.8, 129.5, 129.0, 128.8, 80.4, 68.7, 54.7, 50.6, 47.4, 46.4, 43.6, 42.8, and 42.2 (rot), 28.8, 26.3, 26.0, 25.6, 23.6, 22.1; [α]D20 +13.2 (c 1.0, CH3OH). IR (KBr) v 3421, 2957, 1703, 1638, 1456 cm–1. HRMS (FT-ICR-MS) calcd for C26H40N4O6 [M + H]+ 505.3021; found 505.3022.
(−)-2-[(1-l-Leucinyl-(R)-piperidin-3-yl)amino]acetamide × 2TFA ((R)-LpipG) ((R)-7)
Compound (R)-6 (115 mg, 0.23 mmol) was deprotected as described for (R)-LLpipG ((R)-10) to give the white TFA salt of (R)-LpipG ((R)-7) (95 mg, 84%). Analytical reversed phase HPLC (the mobile phase was 6.4–32.8% CH3CN in water during 30 min, 0.085% TFA throughout) tR 6.2 min, 100% purity. 1H (CD3OD) NMR δ 4.75 (app d, 1H), 4.48–4.38 (m, 1H), 3.98–3.86 (m, 2H), 3.74 (app d, 1H), 3.23–3.08 (m, 2H), 2.85 (app t, 1H), 2.32–2.18 (m, 1H), 1.98–1.51 (m, 6H), 1.05–0.95 (m, 6H). 13C NMR (CD3OD) δ 169.9, 168.5, 54.7, 50.5, 46.6, 46.4, 44.5, 41.0, 28.0, 25.3, 25.0, 23.5, 21.6; [α]D20 −11.2 (c 1.0, CH3OH). IR (KBr) v 3420, 2967, 1676, 1202, 1134 cm–1. HRMS (FT-ICR-MS) calcd for C13H26N4O2 [M + H]+ 271.2129; found 271.2128. For NMR spectral assignment, see the Supporting Information.
(+)-2-[(1-l-Leucinyl-(S)-piperidin-3-yl)amino]acetamide × 2TFA ((S)-LpipG) ((S)-7)
Compound (S)-6 (117 mg, 0.23 mmol) was deprotected as described for (R)-LLpipG ((R)-10) but with stirring for 14 h for the first deprotection. The second deprotection gave the white TFA salt of (S)-LpipG ((S)-7) (87 mg, 75%). Analytical reversed phase HPLC (the mobile phase was 6.4–32.8% CH3CN in H2O for 30 min, 0.085% TFA throughout) tR 6.6 min, 100% purity. 1H (CD3OD) NMR δ 4.55–4.40 (m, 1H), 4.25 (app d, 1H), 4.00–3.85 (m, 2H), 3.66–3.57 (m, 1H), 3.50–3.42 (m, 1H), 2.63 (app t, 1H), 2.32–2.16 (m, 1H), 2.03–1.93 (m, 1H), 1.90–1.48 (m, 6H), 1.07–0.94 (m, 6H). 13C NMR (CD3OD) δ 170.3, 168.5, 54.9, 50.5, 46.7, 46,6, 43.9, 41.1, 27.3, 25.3, 23.5, 23.1, 21,7; [α]D20 +14.8 (c 1.0, CH3OH). IR (KBr) v 3418, 2965, 1676, 1201, 1134 cm–1. HRMS (FT-ICR-MS) calcd for C13H26N4O2 [M + H]+ 271.2129; found 271.2128.
(−)-Ethyl 2-[(Benzyloxycarbonyl)(1-((tert-butoxycarbonyl-l-leucinyl)-l-leucinyl)-(R)-piperidin-3-yl)amino]acetate ((R)-8)
Deprotection of (R)-5 (649 mg, 1.22 mmol) by TFA (1.0 mL, 13 mmol) in dichloromethane (6 mL) were performed as described for (S)-5 to give the Boc-deprotected intermediate. The intermediate and the second Boc-l-Leu × H2O (334 mg, 1.34 mmol) were coupled as described for (S)-5 with EDC (257 mg, 1.34 mmol), HOBt (181 mg, 1.34 mmol), Et3N (493 mg, 4.87 mmol), and molecular sieves (200 mg) in dichloromethane (11 mL). Purification by flash chromatography (heptane/ethyl acetate 1:1, Rf = 0.14) gave (R)-8 as a transparent oil (690 g, 88% yield over two steps). Analytical chiral HPLC (hexane/ethanol 70:30) tR 9.2 min, 100% purity. 1H NMR (CD3OD) δ 7.43–7.25 (m, 5H), 5.20–5.02 (m, 2H), 5.00–4.85 (m, 1H), 4.65–4.51 (m, 1H), 4.50–4.38 (m, 1H), 4.21–3.80 (m, 8H), 3.21–3.03 (m, 1H), 3.02–2.89 (m, 1H), 2.79–2.61 (m, 1H), 2.52–2.38 (m, 1H), 1.98–1.35 (m, 13H), 1.34–1.18 (m, 2H), 1.16–1.09 (m, 3H) 1.04–0.81 (m, 12H). 13C NMR (CD3OD) δ 174.9, 172.3, and 172.1 (rot), 171.6 and 171.4 (rot), 157.5, 157.2, 157.0, and 156.7 (rot), 137.6, 129.4, 129.0, 128.8, 128.7, 80.2, 68.4, and 68.3 (rot), 62.2 and 62.1 (rot), 54.2, 54.5, 49.6, 46.5, 45.9, and 45.6 (rot), 42.8, 42.2, and 41.9 (rot), 28.7, 26.4, and 25.7 (rot), 25.6, 25.4, 23.7, 23.6, 23.5, 22.2, 22.0, 21.4, 14.5; [α]D20 −43.5 (c 1.0, CH3OH). IR (KBr) v 3318, 2957, 1754, 1709, 1640, 1445, 1206 cm–1. HRMS (FT-ICR-MS) calcd for C34H54N4O8 [M + H]+ 647.4014; found 647.4015.
(−)-Ethyl 2-[(Benzyloxycarbonyl)(1-((tert-butoxycarbonyl-l-leucinyl)-l-leucinyl)-(S)-piperidin-3-yl)amino]acetate ((S)-8)
Deprotection of (S)-5 (535 mg, 1.0 mmol) with TFA (1.0 mL, 13 mmol) and dichloromethane (6 mL) were performed as described for the synthesis of (S)-5 to give the Boc-deprotected intermediate. The intermediate and the second Boc-l-Leu × H2O (275 mg, 1.10 mmol) were coupled as described for the synthesis of (S)-5 with EDC (212 mg, 1.10 mmol), HOBt (149 mg, 1.10 mmol), Et3N (406 mg, 4.02 mmol), and molecular sieves (240 mg) in dichloromethane (11 mL). Purification by flash chromatography (heptane/ethyl acetate 1:1, Rf = 0.24) gave (S)-8 as a transparent oil (590 g, 91% yield over two steps). Analytical chiral HPLC (hexane/ethanol 70:30) tR 5.6 min, 100% purity. 1H NMR (CD3OD) δ 7.47–7.23 (m, 5H), 5.28–5.02 (m, 2H), 5.00–4.86 (m, 1H), 4.67–4.50 (m, 1H), 4.48–4.36 (m, 1H), 4.23–3.97 (m, 6H), 3.96–3.81 (m, 1H), 3.74–3.62 (m, 1H), 3.49–3.36 (m, 1H), 3.27–3.17 (m, 1H), 3.05–2.90 (m, 1H), 2.73–2.60 (m, 1H), 2.54–2.35 (m, 1H), 2.14–1.08 (m, 18H), 1.00–0.74 (m, 12H). 13C NMR (CD3OD) δ 174.8, 172.6, and 172.5 (rot), 171.7 and 171.5 (rot), 157.5, 156.8, and 156.5 (rot), 137.6 and 137.5 (rot), 129.3, 129.1, and 128.9 (rot), 128.7, 128.6, 80.2, 68.5, 68.3, and 68.1 (rot), 62.1, 54.3, 50.2, 46.3, 46.2, 45.7, 43.4, 42.7, 41.9, 30.0, 28.8, 26.2, 25.8, 23.8, 23.6, 22.4, 22.2, 22.0, 14.5; [α]D20 −9.2 (c 0.5, CH3OH). IR (KBr) v 3319, 2957, 1753, 1709, 1641, 1447, 1205 cm–1. HRMS (FT-ICR-MS) calcd for C34H54N4O8 [M + H]+ 647.4014; found 647.4014.
(−)-2-[(Benzyloxycarbonyl)(1-((tert-butoxycarbonyl-l-leucinyl)-l-leucinyl)-(R)-piperidin-3-yl)amino]acetamide ((R)-9)
Compound (R)-9 was synthesized from (R)-8 (213 mg, 0.33 mmol) and NH3 in methanol (7 N) (2.0 mL) as described for (S)-2, but the reaction mixture was stirred for 168 h. Purification by flash chromatography (dichloromethane/methanol 95:5, Rf = 0.05) gave the product as a white solid (119 mg, 63%). Analytical chiral HPLC (hexane/ethanol 70:30) tR 13.7 min, 100% purity. 1H NMR (CD3OD) δ 7.42–7.24 (m, 5H), 5.20–5.05 (m, 2H), 4.96–4.87 (m, 1H), 4.66–4.52 (m, 1H), 4.48–4.37 (m, 1H), 4.14–3.84 (m, 6H), 3.17–2.92 (m, 2H), 2.73–2.59 (m, 1H), 2.54–2.40 (m, 1H), 2.03–1.91 (m, 1H), 1.87–1.34 (m, 14H), 1.00–0.84 (m, 12H). 13C NMR (CD3OD, 50 °C) δ 175.0, 174.4, 172.6, and 172.4 (rot), 157.7, 157.5, 137.8, 129.5, 129.0, 128.7, 80.6, 68.7, 55.2, 54.8, 54.6, 47.6, 46.7, 46.1, 43.0, 42.3, 42.1, 29.1, 28.7, 26.6, 26.0, 25.7, 25.3, 23.6, 23.3, 22.2, 21.9; [α]D20 −50.0 (c 0.67, CH3OH). IR (KBr) v 3408, 2956, 1704, 1638, 1449 cm–1. HRMS (FT-ICR-MS) calcd for C32H51N5O7 [M + H]+ 618.3861; found 618.3863.
(−)-2-[(Benzyloxycarbonyl)(1-((tert-butoxycarbonyl-l-leucinyl)-l-leucinyl)-(S)-piperidin-3-yl)amino]acetamide ((S)-9)
Compound (S)-9 was synthesized from (S)-8 (394 mg, 0.61 mmol) and NH3 in methanol (7 N) (4.0 mL) as described for (R)-7. Purification by flash chromatography (dichloromethane/methanol 95:5, Rf = 0.13) gave (S)-7 as a white solid (325 mg, 86%). Analytical chiral HPLC (hexane/ethanol 70:30) tR 8.4 min, 100% purity. 1H NMR (CD3OD) δ 7.51–7.22 (m, 5H), 5.27–5.06 (m, 2H), 4.98–4.86 (m, 1H), 4.66–4.50 (m, 1H), 4.47–4.37 (m, 1H), 4.23–3.79 (m, 6H), 3.28–3.11 (m, 1H), 3.06–2.91 (m, 1H), 2.72–2.58 (m, 1H), 2.54–2.38 (m, 1H), 2.04–1.81 (m, 2H), 1.80–1.25 (m, 13H), 0.99–0.75 (m, 12H). 13C NMR (CD3OD, 50 °C) δ 174.9, 174.3, and 174.2 (rot), 172.7 and 172.3 (rot), 157.5, 137.8, and 137.7 (rot), 129.4, 128.9, 128.7, 80.5, 68.6, 56.4, 54.7, 47.3, 46.4, 46.1, 43.5, 42.8, 42.1, 42.0, 29.1, 28.8, 26.2, 25.8, 25.7, 23.6, 23.4, 22.2, 22.0; [α]D20 −9.5 (c 0.67, CH3OH). IR (KBr) v 3407, 2957, 1707, 1638, 1450 cm–1. HRMS (FT-ICR-MS) calcd for C32H51N5O7 [M + H]+ 618.3861; found 618.3861.
(−)-2-[(l-Leucinyl-(1-l-leucinyl-(R)-piperidin-3-yl))amino]acetamide × 2TFA ((R)-LLpipG) ((R)-10)
Compound (R)-9 (282 mg, 0.46 mmol) was deprotected as described for (S)-LpipGP ((S)-18) but with stirring for 3 h during the first deprotection step. Purification by flash chromatography (ethyl acetate/methanol 20:3) gave the Cbz-deprotected intermediate (189 mg). Further deprotection (180 mg) was performed as described for (S)-LpipGP ((S)-18) with TFA/dichloromethane (1:10) (3 mL) and with stirring for 14 h to give the white TFA salt of (R)-LLpipG ((R)-10) (251 mg, 90%). Analytical reversed phase HPLC (the mobile phase was 10.8–37.2% CH3CN in H2O for 30 min with 0.1% TFA throughout) tR 8.3 min, 100% purity. 1H NMR (CD3OD) δ 4.93–4.82 (m, 1H), 4.67 (app d, 1H), 4.09 (app d, 1H), 4.04–3.83 (m, 3H), 3.82–3.64 (m, 2H), 3.47–3.38 (m, 1H), 3.26–3.09 (m, 2H), 2.84 (app t, 1H), 2.32–2.11 (m, 1H), 1.99–1.41 (m, 6H), 1.04–0.76 (m, 12H). 13C NMR (CD3OD) δ 172.8, 170.7, 168.6, 55.6, 54.8, 52.7, 46.7, 46.4, 44.5, 41.8, 41.3, 28.0, 25.7, 25.1, 25.0, 23.5, 23.1, 21.9, 21.7; [α]D20 −13.6 (c 1.0, CH3OH). IR (KBr) v 3422, 2964, 1676, 1202, 1135 cm–1. HRMS (FT-ICR-MS) calcd for C19H37N5O3 [M + H]+ 384.2969; found 384.2967. For NMR spectral assignment, see the Supporting Information.
(+)-2-[(1-(l-Leucinyl-l-leucinyl)-(S)-piperidin-3-yl)amino]acetamide × 2TFA ((S)-LLpipG) ((S)-10)
Compound (S)-9 (230 mg, 0.37 mmol) was deprotected as described for (R)-LLpipG ((R)-10) to give the white TFA salt of (S)-LLpipG ((S)-10) (209 mg, 92%). Analytical reversed phase HPLC (the mobile phase was 10.8–37.2% CH3CN in H2O for 30 min) tR 8.60 min, 100% purity. 1H NMR (CD3OD) δ 4.83–4.77 (m, 1H), 4.53–4.39 (m, 1H), 4.13–3.86 (m, 3H), 3.75–3.65 (m, 1H), 3.63–3.45 (m, 2H), 3.39–3.32 (m, 1H) 2.60 (app t, 1H), 2.31–2.14 (m, 1H), 2.09–1.97 (m, 1H), 1.94–1.81 (m, 1H), 1.80–1.56 (m, 5H), 1.55–1.39 (m, 1H), 1.04–0.86 (m, 12H). 13C NMR (CD3OD) δ 173.0, 171.3, 168.7, 56.1, 55.2, 52.7, 47.1, 46.9, 44.1, 41.6, 41.3, 27.5, 25.8, 25.2, 24.4, 23.5, 23.2, 22.1, 21.8; [α]D20 +14.3 (c 1.0, CH3OH). IR (KBr) v 3419, 2966, 1674, 1202, 1137 cm–1. HRMS (FT-ICR-MS) calcd for C19H37N5O3 [M + H]+ 384.2969; found 384.2966.
Biological Studies
Purification of IdeS
For recombinant IdeS expression an ideS allele was PCR-amplified from genomic streptococcal DNA using primers 5′-TGTTACCATGGATAGTTTTTCTGCTAAT-3′ introducing a NcoI restriction site and 5′-GCTAGGTACCTTAATTGGTCTGATTCCAACTAT-3′ introducing an Acc65I restriction site for subsequent cloning into plasmid pETtrx_1a.29 IdeS was expressed in Escherichia coli strain BL21 to obtain a His-tagged Trx-IdeS protein. The fusion protein was purified on Ni2+-resin using standard protocols. The His-Trx tag was removed by enzymatic cleavage with Tev-protease in His-trap binding buffer (20 mM Tris-HCl pH8.0, 150 mM NaCl, 10% glycerol) at 30 °C for up to 20 h followed by a second round of purification on Ni2+-resin to remove uncleaved fusion protein.29 The collected flow through, containing IdeS without tag, was purified by size exclusion chromatography on a HiPrep 16/60 Sephacryl S-300 HR column (GE Healthcare). Purified protein was kept in PBS at −20 °C until use.
IdeS Activity Assay and Inhibitor Screening
IdeS (10 μL) (1.83 μM in PBS) was mixed with peptide or peptide analogue (3 μL) (30 mM in 50% DMSO, 50 mM phosphate buffer pH 7.4) and incubated for 15 min at room temperature prior to the addition of IgG1 (10 μL) (Sigma I5154, 8.07 μM in 20 mM TRIS-buffered saline pH 8.0) as substrate. The reactions were incubated for 20 min at 37 °C before termination by the addition of iodoacetamide (10 μL, 16.5 mM) (Sigma I1149). Samples were run in triplicates. Controls where IdeS had been replaced with PBS were included in duplicates for each peptide/peptide analogue. Samples were analyzed by SDS-PAGE and surface plasmon resonance spectroscopy.
Determination of Inhibition Constants
Ki values were determined by accessing velocity curves at different substrate concentrations in the presence or absence of inhibitor. Inhibitor concentrations were 5, 2, and 0.5 mM for (R)-LpipG ((R)-7) and 5, 2.5, and 1 mM for (S)-pipG ((S)-3). For IdeS, repurified polyclonal IgG at concentrations of 150, 100, 50, 20, or 10 μM, was used in reactions with 2 μM of IdeS. Reactions were analyzed by non- reducing SDS page gel analysis to allow measurements of the initial substrate cleavage. The enzyme velocity v was determined according to the following equation:
 |
1 |
For papain, Nα-benzoyl-dl-arginine 4-nitroanilide hydrochloride (Sigma) at concentrations of 6, 3, 1, 0.5, or 0.25 mM was used with 5 μM of papain and inhibitor. Ki nonlinear regression (curve fit) for competitive inhibition and enzymatic kinetics were calculated using GraphPad Prism version 5.04 for Windows, GraphPad Software, San Diego California, www.graphpad.com.
Surface Plasmon Resonance Spectroscopy Assays
Surface plasmon resonance spectroscopy assays were performed with 1042.5 RU of Protein A (Abcam) covalently bound to a CM5 chip using NHS/EDC coupling according to the manufacturer’s instructions. One part of the IgG1/IdeS reaction was mixed with two parts water before applying the sample on the Protein A-CM5 chip. Run conditions on the Biacore X100 machine were 5 μL/min flow rate, a 90 s sample injection time, HBS-EP+ (Biacore) as running buffer, and 20 mM glycine pH 2.0 as regeneration solution of the chip. Report point for amount of protein bound to the chip surface was retrieved at 190 s after sample injection stop. Amount of IgG1 cleavage was first plotted as percentage decrease between PBS-control (uncleaved IgG1) and IdeS/IgG1 samples. A standard curve for IdeS concentration was made without the presence of peptide or peptide analogue. The experimental data obtained from assays with IdeS together with peptide/peptide analogue experiments were plotted to the standard curve and presented as approximate fraction of active IdeS at any given time point.
Papain Activity Assay
Inhibition of the enzymatic activity of papain by peptides and peptide analogues was determined by a colorimetric assay. Papain (Fluka 76218) at a concentration of 1.23 μM was diluted in assay buffer (81 μL) (123 mM phosphate buffer pH 6.2, 1.23 mM EDTA, 1.23 mM DTT), mixed with peptides or peptide analogues (16 μL) (30 mM in 50% DMSO, 50 mM phosphate buffer pH 6.2) and incubated for 10 min at room temperature. The final concentration of papain during the experiment was 0.9 μM. The chromogenic papain substrate Nα-benzoyl-dl-arginine-4-nitroanilide hydrochloride (3 μL) (Sigma B4875, 100 mM in DMSO) was added, and the reaction was incubated for 2 h at 37 °C before termination by the addition of glacial acetic acid (10 μL). Abs405 was measured to determine the amount of cleaved substrate. Samples were run in duplicates on two separate occasions.
Purification of SpeB
For purification of SpeB, S. pyogenes strain 5448 was grown for 16 h in Todd–Hewitt broth (TH) (BD Biosciences) at 37 °C; 5% (v/v) CO2. The growth media was cleared by centrifugation and subjected to ammonium sulfate precipitation (50–80% (w/v)). The resulting pellet was dissolved in 1× PBS, dialyzed o/n at 4 °C against 20 mM sodium acetate buffer, pH 5. The sample was further purified on a HiTrap SP FF anion exchange column (GE Healthcare), equilibrated in 20 mM sodium acetate, pH 5. Proteins were eluted by a linear NaCl gradient and SpeB eluted at approximately 0.2 M NaCl. The eluted protein fraction was dialyzed o/n against 1× PBS and kept at −20 °C.
SpeB Activity Assay
SpeB activity was measured essentially as previously described.30 Briefly, SpeB was activated with 2 mM dithiothreitol (DTT) for 30 min at 37 °C and mixed with 150 μL of 1 mM substrate solution, n-benzoyl-proline-phenylalanine-arginine-p-nitroanilide hydrochloride (BPFA) (pH 4) (Sigma), in 0.1 M phosphate buffer (pH 6). The change in absorbance at OD405 was recorded. The final concentration of SpeB during the experiment was 0.28 μM The amount of active SpeB was determined by active-site titration using various amount of the cysteine protease inhibitor E-64 as previously described.31 For inhibition assays, 11 μL of activated SpeB (1.12 μM in PBS) was mixed with 2 μL of peptide or peptide analogue (30 mM in 50% DMSO, 50 mM phosphate buffer pH 7.4) and incubated for 15 min at room temperature prior to the addition of substrate solution. All assays were performed in triplicate.
Acknowledgments
This work was supported by the Swedish Research Council (project numbers 2005-3145 (K.L.) and 2009-4997 (UvPR) and Insamlingsstiftelsen at Umeå University. We thank AstraZeneca R&D Mölndal, Sweden, for financial support of the graduate student fellowship for KB. We also thank Madeleine Åhman, AstraZeneca, Sweden, for help with the pKa measurements.
Glossary
Abbreviations Used
- Boc
tert-butyloxycarbonyl
- Cbz
benzyloxycarbonyl
- Cbz-OSu
N-(benzyloxycarbonyloxy)succinimide
- DMSO
dimethylsulfoxide
- E64
1-[N-[(3-trans-carboxyoxirane-2-carbonyl)-l-leucyl]amino]-4-guanidinobutane
- EDC
N-(3-methylaminopropyl)-N′-ethylcarbodiimide
- ee
enantiomeric excess
- HOBt
hydroxybenzotriazole
- HPLC
high pressure liquid chromatography
- IdeS
immunoglobulin G-degrading enzyme of Streptococcus pyogenes
- NMR
nuclear magnetic resonance
- pip
the piperidine moiety replacing glycine residues
- rac
racemic
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SpeB
streptococcal pyogenic exotoxin B
- TFA
trifluoroacetic acid
- TLCK
tosyl lysyl chloromethyl ketone
- TPCK
tosyl phenylalanyl chloromethyl ketone
- Z-LVG CHN2
N-benzyloxycarbonyl-leucyl-valyl-glycine diazomethylketone
Supporting Information Available
Synthetic procedures and characterization of compounds shown in Schemes 3–6; NMR spectral assignments of (R)-1, (R)-2, (R)-3, (R)-4, (R)-7, and (R)-10; 1H and 13C NMR spectra of tested compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Rawlings N. D.; Tolle D. P.; Barrett A. J. MEROPS: the peptidase database. Nucleic Acids Res. 2004, 32, D160–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M. W. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 2000, 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis J. R.; Steer A. C.; Mulholland E. K.; Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005, 5, 685–694. [DOI] [PubMed] [Google Scholar]
- Telfer N. R.; Chalmers R. J. G.; Whale K.; Colman G. The role of streptococcal infection in the initiation of guttate psoriasis. Arch. Dermatol. 1992, 128, 39–42. [PubMed] [Google Scholar]
- Elliott S. D. A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J. Exp. Med. 1945, 81, 573–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Pawel-Rammingen U.; Johansson B. P.; Björck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 2002, 21, 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenig K.; Chatwell L.; von Pawel-Rammingen U.; Björck L.; Huber R.; Sondermann P. Structure of the streptococcal endopeptidase IdeS, a cysteine proteinase with strict specificity for IgG. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17371–17376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agniswamy J.; Nagiec M. J.; Liu M. Y.; Schuck P.; Musser J. M.; Sun P. D. Crystal structure of group A Streptococcus Mac-1: insight into dimer-mediated specificity for recognition of human IgG. Structure 2006, 14, 225–235. [DOI] [PubMed] [Google Scholar]
- Kagawa T. F.; Cooney J. C.; Baker H. M.; McSweeney S.; Liu M. Y.; Gubba S.; Musser J. M.; Baker E. N. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: An integrin-binding cysteine protease. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. G.; Dagil R.; Niclasen L. M.; Sørensen O. E.; Kragelund B. B. Structure of the mature streptococcal cysteine protease exotoxin mSpeB in its active dimeric form. J. Mol. Biol. 2009, 393, 693–703. [DOI] [PubMed] [Google Scholar]
- Collin M.; Olsén A. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun. 2001, 69, 7187–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincents B.; von Pawel-Rammingen U.; Björck L.; Abrahamson M. Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry 2004, 43, 15540–15549. [DOI] [PubMed] [Google Scholar]
- Burton D. R. Immunoglobulin-G—functional sites. Mol. Immunol. 1985, 22, 161–206. [DOI] [PubMed] [Google Scholar]
- Rasmussen M.; Björck L. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 2002, 43, 537–544. [DOI] [PubMed] [Google Scholar]
- Agniswamy J.; Lei B. F.; Musser J. M.; Sun P. D. Insight of host immune evasion mediated by two variants of group A Streptococcus Mac protein. J. Biol. Chem. 2004, 279, 52789–52796. [DOI] [PubMed] [Google Scholar]
- Potempa J.; Dubin A.; Korzus G.; Travis J. Degradation of elastin by a cysteine proteinase from Staphylococcus aureus. J. Biol. Chem. 1988, 263, 2664–2667. [PubMed] [Google Scholar]
- Johansson B. P.; Shannon O.; Björck L. Ides: A bacterial proteolytic enzyme with therapeutic potential. PLoS ONE 2008, 3, e1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar K. S.; Johansson B. P.; Björck L.; Holmdahl R. Blocking of experimental arthritis by cleavage of IgG antibodies in vivo. Arthritis Rheum. 2007, 56, 3253–3260. [DOI] [PubMed] [Google Scholar]
- Yang R.; Otten M. A.; Hellmark T.; Collin M.; Björck L.; Zhao M.-H.; Daha M. R.; Segelmark M. Successful treatment of experimental glomerulonephritis with IdeS and EndoS, IgG-degrading streptococcal enzymes. Nephrol., Dial., Transplant. 2010, 25, 2479–2486. [DOI] [PubMed] [Google Scholar]
- Berggren K.; Johansson B.; Fex T.; Kihlberg J.; Björck L.; Luthman K. Synthesis and biological evaluation of reversible inhibitors of IdeS, a bacterial cysteine protease and virulence determinant. Bioorg. Med. Chem. 2009, 17, 3463–3470. [DOI] [PubMed] [Google Scholar]
- Filira F.; Biondi L.; Gobbo M.; Rocchi R. N-Alkylation of amino acids during hydrogenolytic deprotection. Tetrahedron Lett. 1991, 32, 7463–7464. [Google Scholar]
- Hamid M.; Slatford P. A.; Williams J. M. J. Borrowing hydrogen in the activation of alcohols. Adv. Synth. Catalysis 2007, 349, 1555–1575. [Google Scholar]
- Berggren K.; . et al. , unpublished results. [Google Scholar]
- The peptides were purchased from CASLO Laboratory ApS, Lyngby, Denmark.
- Åkesson P.; Moritz L.; Truedsson M.; Christensson B.; von Pawel-Rammingen U. IdeS, a highly specific immunoglobulin G (IgG)-cleaving enzyme from Streptococcus pyogenes, is inhibited by specific IgG antibodies generated during infection. Infect. Immun. 2006, 74, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L.; Groom C. R.; Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discovery Today 2004, 9, 430–431. [DOI] [PubMed] [Google Scholar]
- Wan H.; Holmén A. G.; Wang Y. D.; Lindberg W.; Englund M.; Någård M. B.; Thompson R. A. High-throughput screening of pKa values of pharmaceuticals by pressure-assisted capillary electrophoresis and mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2639–2648. [DOI] [PubMed] [Google Scholar]
- Meloun M.; Bordovska S. Benchmarking and validating algorithms that estimate pK(a) values of drugs based on their molecular structures. Analyt. Bioanalyt. Chem. 2007, 389, 1267–1281. [DOI] [PubMed] [Google Scholar]
- Bogomolovas J.; Simon B.; Sattler M.; Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expression Purif. 2009, 64, 16–23. [DOI] [PubMed] [Google Scholar]
- North M. J. Cysteine endopeptidases of parasitic protozoa. Methods Enzymol. 1994, 523–539. [DOI] [PubMed] [Google Scholar]
- Nyberg P.; Rasmussen M.; von Pawel-Rammingen U.; Björck L. SpeB modulates fibronectin-dependent internalization of Streptococcus pyogenes by efficient proteolysis of cell-wall-anchored protein F1. Microbiology 2004, 150, 1559–1569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.