Abstract
One of the main motivations to study amphioxus is its potential for understanding the last common ancestor of chordates, which notably gave rise to the vertebrates. An important feature in this respect is the slow evolutionary rate that seems to have characterized the cephalochordate lineage, making amphioxus an interesting proxy for the chordate ancestor, as well as a key lineage to include in comparative studies. Whereas slow evolution was first noticed at the phenotypic level, it has also been described at the genomic level. Here, we examine whether the amphioxus genome is indeed a good proxy for the genome of the chordate ancestor, with a focus on protein-coding genes. We investigate genome features, such as synteny, gene duplication and gene loss, and contrast the amphioxus genome with those of other deuterostomes that are used in comparative studies, such as Ciona, Oikopleura and urchin.
Keywords: deuterostomes, evolutionary rates, gene duplication, gene loss, orthology, synteny
INTRODUCTION
Amphioxus (cephalochordates), and especially the model amphioxus Branchiostoma floridae, are often used as proxies for the ancestor of chordates, notably in molecular studies [1–7], and more recently in genomics [8, 9].
Although there is an interest in reconstructing ancestral genomic features in many comparative studies, the amphioxus stands out as an organism, which is strongly studied as an ancestor proxy [7]. Among article abstracts present in PubMed, 27% of those that include the word ‘amphioxus’ also include some variation of the root ‘ancest*’ (e.g. ancestor, ancestral). This figure is only 11% for ‘Ciona’, the most studied invertebrate chordate and 3% for ‘Hydra’. The effect is even stronger in amphioxus-related articles highlighted in Faculty of 1000 of which 69% contain the root ‘ancest*’ only 37% for Ciona.
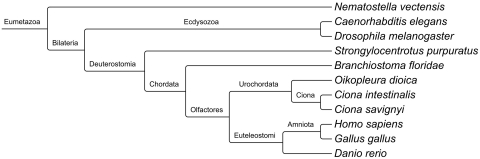
This raises the following question: how good a proxy for the chordate ancestor is amphioxus? Here, we investigate this question from a genomic perspective: how good a proxy for the ancestral chordate genome is the available amphioxus genome? We compare the relevance of the amphioxus genome with those of other invertebrate deuterostomes, whose sequenced genomes are also potentially useful to reconstruct the chordate ancestor: Ciona [10, 11], sea urchin [12] and Oikopleura [13] (Table 1 and Figure 1), plus sea anemone [14].
Table 1:
Representative bilaterian genomes
| Species | Genome size (Gb) | Protein-coding gene count | Assembly (N50) | BRBHs to Nematostella |
|---|---|---|---|---|
| Branchiostoma floridae | 0.52 | 34 717 | Scaffolds (2.6 Mb) | 8139 |
| Danio rerio | 1.5 | 26 095 | Chromosomes | 8134 |
| Strongylocentrotus purpuratus | 0.92 | 29 129 | Scaffolds (0.065 Mb) | 7475 |
| Gallus gallus | 1.1 | 16 736 | Chromosomes | 7300 |
| Homo sapiens | 3.2 | 21 558 | Chromosomes | 7243 |
| Ciona intestinalis | 0.17 | 14 180 | Chromosomesa | 6054 |
| Drosophila melanogaster | 0.17 | 13 781 | Chromosomes | 6022 |
| Ciona savignyi | 0.18 | 11 604 | Scaffolds (1.8 Mb) | 5848 |
| Caenorhabditis elegans | 0.10 | 20 289 | Chromosomes | 5533 |
| Oikopleura dioica | 0.072 | 18 119 | Scaffolds (0.4 Mb) | 5395 |
aOnly 54% of the scaffold length mapped to chromosomes.
Note: Nematostella vectensis is a sea anemone, which is an outgroup to all the bilaterian species presented in the table. The species are ordered according to their number of BRBHs to Nematostella.
BRBH, best reciprocal BlastP hit.
Figure 1:
Phylogenetic relationships between species used in this study. The 11 animals diverged from a common eumetazoan ancestor more than 500 million years ago. Branch lengths are not to scale. Nematostella vectensis: sea anemone; Caenorhabditis elegans: nematode; Drosophila melanogaster: fruit fly; Strongylocentrotus purpuratus: sea urchin; Branchiostoma floridae: amphioxus; Oikopleura dioica: tunicate; Ciona intestinalis and Ciona savignyi: sea squirts; Homo sapiens: human; Gallus gallus: chicken; Danio rerio: zebrafish.
LINEAGE-SPECIFIC DUPLICATED GENES
One of the ways in which a modern-day genome can diverge from the ancestral state is through secondary gene or genome duplications. First, duplication can lead to an increase in the number of genes in some functional classes, relative to others. Second, duplicate genes can diverge in function, leading to greater functional diversity inside the genome [15].
Notably, all jawed vertebrate genomes share at least two rounds of whole-genome duplication [9, 16], and up to three in the ancestry of teleost fishes. Such events are followed by biased gene loss. Thus, a long-term consequence of whole-genome duplication is that the genome is enriched in certain functional categories, such as transcription factors, or in genes expressed in late development, relative to the ancestor [9, 17–19].
Under this metric, one could expect any other deuterostome genome to be a better representative of the ancestor, rather than any vertebrate. However, small scale duplications can also be an abundant source of divergence, and ‘lineage-specific’ explosive duplications of different gene families in different species appear widespread [20]. Indeed, examples of such lineage-specific duplications have been found in all deuterostome genomes. For example, Oikopleura, which has the smallest chordate genome, has 266 homeobox genes, resulting from 87 amplification events [13]. Interestingly, innate immunity genes have expanded independently in the amphioxus and sea urchin genomes [8]. Although such examples can be repeatedly listed [21], a systematic view is required in order to quantify divergence from the ancestor more accurately.
An important technical problem is that many of these genomes are assembled with a lesser quality than those of the vertebrate model organisms, such as human or mouse. Moreover, the assembly is often based on a mixture of haplotypes, from populations with very high levels of polymorphism [discussed in 9, 12, 13]. As a result, it can be difficult to diagnose lineage-specific duplications in an automatic manner, with an acceptable false positive rate.
To gain some insight into the extent of duplication, we have measured the number of homologs descending from one chordate ancestral gene in each genome, using the following procedure: (i) gene trees from Ensembl [22] that contain at least one vertebrate gene and at least one gene outside vertebrates were used to reconstruct the ancestral chordate complement of genes (15 040 genes); (ii) an all-against-all BlastP comparison was performed between Metazoa (sea urchin, oikopleura, sea anemone, amphioxus) absent from the Ensembl data set and representatives of the latter (human, chicken, zebrafish, drosophila, nematode, Ciona intestinalis and Ciona savignyi); (iii) all best reciprocal hits were used to insert the new genes in Ensembl trees according to the species phylogeny. This procedure is very conservative, as fast evolving duplicates will not be identified, but several paralogs per genome can still be identified, if they are best reciprocal hits to different genes of the gene family. The advantage of this procedure is that we remove most false positives, while using a consistent definition that allows a comparison between genomes. It is biased against discovering new lineage-specific duplicates, especially for gene families that are single copy in all Ensembl genomes. We consider this risk of false negatives in amphioxus and other nonmodel organisms to be preferable to a high level of false positives. Thus the results should not be taken as indicative of the absolute level of duplication, but rather of the relative amount of duplication in different genomes.
With this procedure, we find the highest number of duplications in zebrafish, followed by the two vertebrates investigated (Table 2). This shows that whole-genome duplications were the main factor in generating paralogs in chordate genomes (at least those that are sufficiently conserved in sequence to be detected by our approach). Oikopleura, which has the smallest chordate genome, also has fewer duplications, consistent with its general properties of reductive history, whereas amphioxus and Ciona show intermediate levels of duplicate gene retention.
Table 2:
Number of descendants of ancestral chordate genes
| Species | Modern genes of chordate origin | 1 copy, n (%) | ≥1 copies, n (%) | Lost genes, n (%) |
|---|---|---|---|---|
| Branchiostoma floridae | 11 436 | 8089 (54) | 1460 (10) | 5491 (37) |
| Homo sapiens | 15 944 | 6888 (46) | 3202 (21) | 4950 (33) |
| Gallus gallus | 13 291 | 6254 (42) | 2652 (18) | 6134 (41) |
| Oikopleura dioica | 7576 | 6202 (41) | 640 (4) | 8198 (55) |
| Danio rerio | 22 255 | 5557 (37) | 4418 (29) | 5065 (34) |
| Ciona intestinalis | 10 614 | 5418 (36) | 1869 (12) | 7753 (52) |
Note: For each of the 15 040 genes inferred to have existed in the ancestral chordate, the number of descendents (‘modern genes’) that are best reciprocal blast hits was calculated. In a modern genome, one ancestral gene can correspond to: 1 copy, or ≥1 copies or the gene can be lost.
As might be expected, whole-genome duplications have thus had a large impact on vertebrate genomes, suggesting that chordate genomes that did not undergo these duplications are better proxies for the chordate ancestor.
GENE LOSS OR VERY STRONG DIVERGENCE
In Table 1, we present the number of best reciprocal BlastP hits (BRBHs) between an outgroup to bilaterians, the sea anemone and different bilaterian animal genomes. This provides a rough estimate of conserved orthologs between the genomes. If genes were retained in single copy in two species, and did not diverge too much in sequence, then they will be reported. They will also be reported if there were duplications, but one gene copy diverged less than the others, and presumably remained closer to the ancestral function and structure. These are obviously approximations and notably, a recent study failed to support a correlation between sequence and function conservation [23]. We still believe that this provides a useful estimation of the amount of conservation of ancestral genes in each genome. If a genome lost more genes, or if its genes diverged more, then we expect fewer BRBHs between that genome and the outgroup.
The results are striking: amphioxus has the most BRBHs with sea anemone of any species considered. This is despite the fact that the quality of the genome sequences of many of the model species is better, with deeper sequencing, better assembly and better annotation. Supporting the utility of our approximate measure, Oikopleura has the lowest number of BRBHs, consistent with the known pattern of gene loss and gene remodeling in that lineage [13]. The second highest number of BRBHs, very similar to that of amphioxus, is for the zebrafish. Thus, on this measure, these two genomes are the closest to the ancestral genome. But for zebrafish, this should be combined with three rounds of genome duplication, which implies another form of divergence that amphioxus did not experience.
We next considered only the subset of 15 040 genes that were inferred in the ancestor of chordates (Table 2). Of these, amphioxus lost 5491, similar to the 4950 lost in human and much fewer than the 7753 lost in Ciona or 8198 lost in Oikopleura. Moreover, there is a subset of 4629 genes that were repeatedly lost in different lineages (i.e. human, zebrafish, Ciona, amphioxus) (Figure 2). There are only 925 genes that were lost only in amphioxus. This compares with 701 lost only in vertebrates, but 2354 lost only in Ciona. Thus, amphioxus has conserved ancestral genes much more than Ciona, and similarly to vertebrates. Moreover, these results might be biased by the better quality of the human and zebrafish genomes, i.e. there are probably more false negatives in the amphioxus genome.
Figure 2:
Venn diagram of gene loss in different chordates. Each of the four species lost the total number of genes indicated in the last column of Table II. The intersections show the number of genes lost in common between any two, three or four species.
If we combine the results of gene loss and gene duplication, it appears likely that the amphioxus complement of protein-coding genes is close, but not identical, to the ancestral chordate complement. Indeed, it is the only species investigated for which more than half of the ancestral chordate genes are still present in single copy (Table 2) (within the limitations of our reciprocal best hits and of an imperfectly assembled genome).
CONSERVATION OF SYNTENY
Although the conservation of protein-coding genes is an important aspect of evolution, there are many other ways in which a genome can diverge from its ancestral state. An interesting global measure of genome evolution is the conservation of synteny, i.e. of gene order and gene neighborhood. Cases in which exact gene order is functionally important, such as the Hox clusters of vertebrates or of insects [24], appear to be rather exceptional in animals. On the other hand, a more relaxed definition of synteny based on shared gene neighborhood appears to play a functional role in vertebrate genomes [25, 26], and is applicable to the comparison of genomes as distant as human and hydra [14].
Comparative studies of animal genomes have shown a large variability between lineages in the level of synteny conservation. Despite the limitations of the amphioxus genome assembly, and despite a longer divergence time between amphioxus and vertebrates compared with Ciona and vertebrates, the conservation of gene neighborhood with vertebrates is greater for amphioxus than for C. intestinalis [9]. In total, 74% of amphioxus scaffolds have a significant concentration of orthologs from the same human chromosome, as opposed to 9% of Ciona scaffolds. Even less conserved than Ciona, Oikopleura is the only known chordate genome to show no significant conservation of gene neighborhood with other chordates, at a 30 genes neighborhood distance [13]. Even Nematostella (sea anemone) and Caenorhabditis elegans have higher conservation with the chordate gene order than Oikopleura.
A comparative estimation of Deuterostomes plus hydra showed that the lowest rearrangement rates since the ancestral bilaterian were in the lineages leading to urchin and amphioxus [27]. The vertebrate genomes appeared highly impacted by the whole-genome duplications that were followed by intense rearrangements. Yet, the amphioxus genome does not appear very strongly conserved in this analysis, and ‘therefore it cannot be assumed to be uniquely representative of the ancestral chordate genome’ [27].
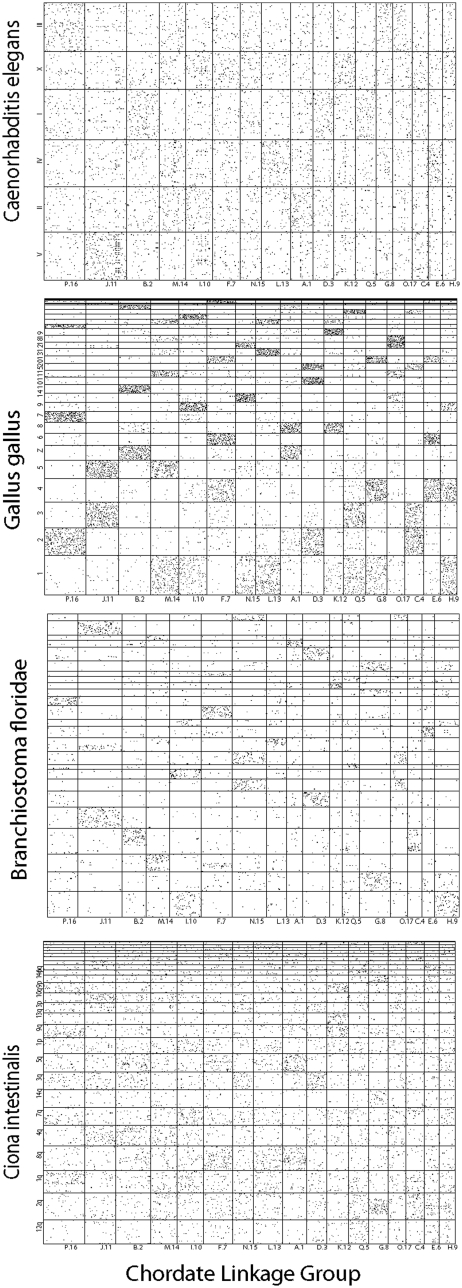
To confirm the extent of synteny conservation between different model genomes and the chordate ancestor, we have used estimated ancestral chordate linkage groups [9]. We plotted the position of these ancestral genes on the amphioxus genome scaffolds, and on scaffolds or chromosomes from other species, ensuring that a similar number of genes were used in each species in order to make comparisons possible. The resulting dot-plots clearly confirm the lack of conservation in Oikopleura, and a similar lack of conservation in Drosophila (data not shown). Some level of synteny conservation is found in C. intestinalis and C. elegans, and still higher for amphioxus (Figure 3). The pattern in sea urchin is not clear, because of a lack of mapped orthologs (data not shown). Despite the post whole-genome duplication rearrangements, the strongest conservation of synteny is found for vertebrates, notably the chicken (Figure 3). There might be a bias in that the estimation of the ancestral linkage groups used more information from the well assembled human genome, than from less well assembled genomes. Of note, the patterns observed show clearly the 4-to-1 homology of chicken to the ancestral chordate, due to two whole-genome duplications. Thus, it seems that either chicken or amphioxus provide the best proxy for the ancestral gene arrangement, depending on the importance of working with a nonduplicated genome (i.e. amphioxus), or the importance of having very well conserved synteny (i.e. chicken).
Figure 3:
Dot plots of chromosomal homology between animal genomes. Columns represent ancestral chordate linkage groups (CLGs) as defined in [9]. Row represent the 25 longest contigs in the Ciona and amphioxus genome assemblies and individual chromosomes in nematode and chicken. Each dot is a gene that was present in a given CLG, and is present in a given contig or chromosome in a modern species. Gene order was shuffled in chicken chromosomes to erase the biases introduced when inferring the CLG from a vertebrate genome in [9]. A random sample of 10 000 chicken genes was selected for the dot-plot so that the number of genes would be equivalent in each species.
Interestingly, comparative synteny and sequence alignments have been used to identify conserved noncoding elements between vertebrates and amphioxus. Such elements were first identified among vertebrates, but not between vertebrates and other species [28], although only the C. intestinalis genome was then available. The amphioxus draft genome allowed the detection of a few conserved noncoding elements, which were shown to be functional, i.e. they drive expression in development [8]. Using conserved synteny with vertebrates, Hufton et al. [29] identified 1299 conserved noncoding elements in amphioxus. All vertebrate genomes had many more such elements. Of those that were tested, about half had enhancer activity in vivo. It seems probable that the 1299 elements in amphioxus are representative of the ancestral state, providing an exciting window into gene regulation in ancestral chordate development.
CONCLUSION
Although the use of the amphioxus as a proxy for the chordate ancestor is frequent in the literature, tests for its appropriateness are much rarer. The identification of many functional conserved noncoding elements in amphioxus [29]—thanks to conserved synteny—is thus particularly interesting, since these elements are not found in other basal chordates, whereas they are highly duplicated in vertebrates. This is consistent with the accumulated evidence from small-scale studies, that gene regulation in the amphioxus is probably much closer to the ancestral state (e.g. in terms of transcription factors), than either the tunicates (because of gene loss and rearrangement) or the vertebrates (because of genome duplication) [8].
The different metrics that we have used paint a picture that is consistent with the conclusions of Hufton et al. [27]: amphioxus is not ‘ancestral’, but has derived from the ancestral chordate in many ways. Yet, it is the less derived of the available species with genomes sequenced, specifically in terms of gene content.
We can reformulate the question as: how useful is amphioxus for reconstructing the ancestral state? If the amphioxus genome is not used as a proxy of the ancestor, but as a data point to reconstruct that ancestor and understand chordate evolution, then it is clear that it is the most useful genome for understanding chordate origins and evolution.
Key Points.
The amphioxus genome has relatively few conserved duplicate genes; in this, it differs from vertebrates, which underwent whole-genome duplications and resembles those of other chordates, such as Ciona or Oikopleura. The amphioxus genome also has few gene losses, comparable with vertebrates, and in contrast to Ciona or Oikopleura. Thus, it has the most genes preserved in single copy since the origin of chordates, of any known genome.
The amphioxus genome has a good conservation of gene neighborhood (synteny), which appears related to conservation of gene regulation. Synteny appears more conserved in chicken or human, but with the added complexity of whole-genome duplication.
The amphioxus genome cannot be taken to represent the ancestral chordate genome but it is the least derived and thus, it is a key element in reconstructing chordate genome evolution.
FUNDING
Research in the Robinson–Rechavi lab is supported by the Swiss National Science Foundation, the Swiss Institute of Bioinformatics, the HP2C program of the Swiss National Supercomputing Center, and Etat de Vaud.
Biographies
Alexandra Louis is a bioinformatics Research Engineer at CNRS, and member of the DYOGEN group at the Institute of Biology of the Ecole Normale Supérieure (IBENS) in Paris. She is working on the reconstruction of ancestral genomes in vertebrates and plants and actively maintains the Genomicus synteny viewer.
Hugues R. Crollius is Research Director at CNRS and Group Leader at the Institute of Biology of the Ecole Normale Supérieure (IBENS) in Paris. He is interested in evolution as a general framework to understand the emergence of genomic and functional properties of organisms through the reconstruction of ancestral genomes.
Marc Robinson-Rechavi is associate Professor at the Department of Ecology and Evolution in the University of Lausanne, and Group Leader at the Swiss Institute of Bioinformatics. His main interest is in the evolution of animal genomes in the context of organismal function and development.
References
- 1.Castro LF, Holland PW. Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol Dev. 2003;5:459–65. doi: 10.1046/j.1525-142x.2003.03052.x. [DOI] [PubMed] [Google Scholar]
- 2.Lacalli TC. Sensory systems in amphioxus: a window on the ancestral chordate condition. Brain Behav Evol. 2004;64:148–62. doi: 10.1159/000079744. [DOI] [PubMed] [Google Scholar]
- 3.Castro LF, Rasmussen SL, Holland PW, et al. A Gbx homeobox gene in amphioxus: insights into ancestry of the ANTP class and evolution of the midbrain/hindbrain boundary. Dev Biol. 2006;295:40–51. doi: 10.1016/j.ydbio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Zmasek CM, Zhang Q, Ye Y, et al. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8:R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Y, Huang G, Zhang Q, et al. The primitive immune system of amphioxus provides insights into the ancestral structure of the vertebrate immune system. Dev Comp Immunol. 2010;34:791–6. doi: 10.1016/j.dci.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Belgacem MR, Escande ML, Escriva H, et al. Amphioxus Tbx6/16 and Tbx20 embryonic expression patterns reveal ancestral functions in chordates. Gene Expr Patterns. 2011;11:239–43. doi: 10.1016/j.gep.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Holland LZ, Sower SA. “Insights of early chordate genomics: endocrinology and development in amphioxus, tunicates and lampreys”: introduction to the symposium. Integr Comp Biol. 2010;50:17–21. doi: 10.1093/icb/icq039. [DOI] [PubMed] [Google Scholar]
- 8.Holland LZ, Albalat R, Azumi K, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–11. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putnam NH, Butts T, Ferrier DEK, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–71. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 10.Dehal P, Satou Y, Campbell RK, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–67. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 11.Small KS, Brudno M, Hill MM, et al. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007;8:R41. doi: 10.1186/gb-2007-8-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sea Urchin Genome Sequencing Consortium. Sodergren E, Weinstock GM, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–52. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denoeud F, Henriet S, Mungpakdee S, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–5. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putnam NH, Srivastava M, Hellsten U, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 15.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–50. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 16.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 17.Brunet FG, Roest Crollius H, Paris M, et al. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–16. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- 18.Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux J, Robinson-Rechavi M. Developmental constraints on vertebrate genome evolution. PLoS Genet. 2008;4:e1000311. doi: 10.1371/journal.pgen.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lespinet O, Wolf YI, Koonin EV, et al. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–59. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minguillon C, Ferrier DE, Cebrian C, et al. Gene duplications in the prototypical cephalochordate amphioxus. Gene. 2002;287:121–8. doi: 10.1016/s0378-1119(01)00828-9. [DOI] [PubMed] [Google Scholar]
- 22.Flicek P, Amode MR, Barrell D, et al. Ensembl 2012. Nucleic Acids Res. 2011;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nehrt NL, Clark WT, Radivojac P, et al. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput Biol. 2011;7:e1002073. doi: 10.1371/journal.pcbi.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemons D, McGinnis W. Genomic evolution of hox gene clusters. Science. 2006;313:1918–22. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 25.Makino T, McLysaght A. Interacting gene clusters and the evolution of the vertebrate immune system. Mol Biol Evol. 2008;25:1855–62. doi: 10.1093/molbev/msn137. [DOI] [PubMed] [Google Scholar]
- 26.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Nl Acad Sci USA. 2010;107:9270–4. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hufton AL, Groth D, Vingron M, et al. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008;18:1582–91. doi: 10.1101/gr.080119.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolfe A, Goodson M, Goode DK, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hufton AL, Mathia S, Braun H, et al. Deeply conserved chordate non-coding sequences preserve genome synteny but do not drive gene duplicate retention. Genome Res. 2009;19:2036–51. doi: 10.1101/gr.093237.109. [DOI] [PMC free article] [PubMed] [Google Scholar]