Abstract
Immune systems evolve as essential strategies to maintain homeostasis with the environment, prevent microbial assault and recycle damaged host tissues. The immune system is composed of two components, innate and adaptive immunity. The former is common to all animals while the latter consists of a vertebrate-specific system that relies on somatically derived lymphocytes and is associated with near limitless genetic diversity as well as long-term memory. Deuterostome invertebrates provide a view of immune repertoires in phyla that immediately predate the origins of vertebrates. Genomic studies in amphioxus, a cephalochordate, have revealed homologs of genes encoding most innate immune receptors found in vertebrates; however, many of the gene families have undergone dramatic expansions, greatly increasing the innate immune repertoire. In addition, domain-swapping accounts for the innovation of new predicted pathways of receptor function. In both amphioxus and Ciona, a urochordate, the VCBPs (variable region containing chitin-binding proteins), which consist of immunoglobulin V (variable) and chitin binding domains, mediate recognition through the V domains. The V domains of VCBPs in amphioxus exhibit high levels of allelic complexity that presumably relate to functional specificity. Various features of the amphioxus immune repertoire reflect novel selective pressures, which likely have resulted in innovative strategies. Functional genomic studies underscore the value of amphioxus as a model for studying innate immunity and may help reveal how unique relationships between innate immune receptors and both pathogens and symbionts factored in the evolution of adaptive immune systems.
Keywords: innate immunity, Toll-like receptors, expanded immune repertoire, allelic complexity, gut immunity
INTRODUCTION
The capacity to maintain the integrity of self is a hallmark characteristic of all metazoan species. The continuum of microbial encounters, which occurs in the lifetime of individuals, creates enormous selective pressure that in turn has driven the development of a wide range of highly integrated immune defenses that also recognize and eliminate dying cells and cancer. Several decades of studies, focused initially on jawed vertebrates, have revealed two clearly demarcated lines of defense termed innate and adaptive immunity. The innate immune system includes a wide range of molecules that exhibit varying levels of polymorphism and are inherited in a simple Mendelian manner. The adaptive immune system, which presently is thought to be confined only to the vertebrates, relies on genetic recombination that takes place in individual somatic cells belonging to the lymphoid lineage. The receptors are displayed on the surfaces of individual lymphocytes and subsequent exposure of cells harboring receptors that are specific to foreign stimuli results in clonal expansion and in the case of immunoglobulin receptors, their eventual secretion as antibody molecules. Approximately 107–108 structurally different receptors, which mediate cellular (T-cell receptor) and humoral (immunoglobulin antibody) immunity, respectively, are created uniquely in an individual human. Much about the evolution of innate and adaptive immunity has been inferred in the absence of data from species that diverged in phylogeny prior to the vertebrate radiations (Figure 1). The resolution of whole genomes from species, such as amphioxus (a cephalochordate), sea urchin (an echinoderm) and sea squirt (a urochordate) has allowed us to examine the phylogeny of integral components of immune function outside of the intrinsic bias of the narrow window of evolution represented by mammals and to a lesser degree other vertebrate forms [1].
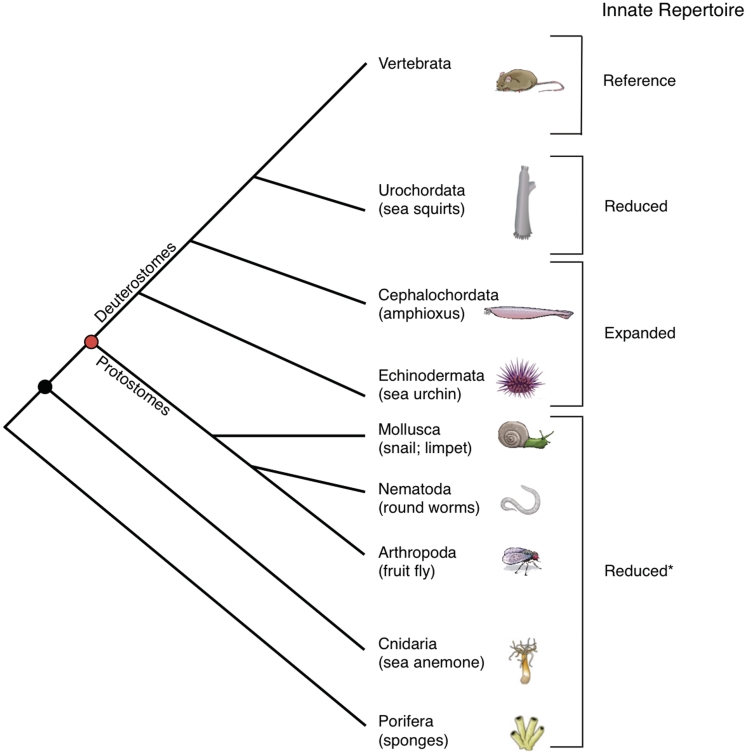
Figure 1:
Simplified phylogeny of extant animal phyla. Complete genomes have been defined for members of the representative phyla shown. The black circle denotes the bilaterian split; the red circle denotes protostome and deuterostome divergence. Overall trends in the complexity of mediators of innate immunity are implied (Innate Repertoire column) relative to a mammalian reference (mouse). *Although reduced numbers of innate immune receptors, relative to those found in mammals, are noted for both urochordates and protostomes, immune mechanisms are present in these species that include (but are not limited to) antimicrobial peptides, prophenoloxidase-associated melanization and various PRRs.
INNATE IMMUNITY
Innate immune mechanisms include: barrier defenses (i.e. host epithelium) and the associated non-specific secretory components (e.g. antibacterial peptides), pattern recognition receptors (PRRs) on phagocytes and other host cells, various phagocyte effector mechanisms (e.g. reactive oxygen species) and different enzymatically catalyzed cascades involved in clotting, melanization and complement activation [2, 3]. In vertebrates, the number of PRRs, which recognize microbial-associated molecular patterns (MAMPs), is relatively small. The Toll-like receptors (TLRs), composed of an intracellular Toll/interleukin-1 receptor (TIR) domain combined with extracellular leucine-rich repeats (LRRs), are the most extensively characterized PRR and include both extracellular and intracellular forms [4, 5]. First described in Drosophila, Toll plays an integral role in development. Later, it was shown to effect immunity after PRR detection of pathogens (e.g. fungal) and subsequent protease cleavage product activation of Toll signaling [6]. Soon thereafter, it was demonstrated that related molecules in mammals, such as TLR4, function directly in immune recognition [7, 8]. PRRs, such as TLRs, effect immune protection across a particularly broad range of metazoans [9–11]. Nucleotide-binding oligomerization domain-like receptors (NLRs), which are intracellular PRRs, consist of a NACHT domain (nucleotide triphosphatase, NTPase, domain; named after different genes) and LRRs. NLRs, which serve as intracellular sensors of danger, are distributed widely and play integral roles in innate immunity [12, 13]. Innate immunity, at least in part through the function of PRRs, shapes and/or influences adaptive immune responses [14, 15].
Both epithelial barriers and innate immune-related products act as a first line of defense in protostomes and deuterostomes. Furthermore, immunocyte mobility has factored significantly in the evolution of innate defenses, as most invertebrates possess open vascular systems. Pathogens that pass the epithelial layers in flies rapidly encounter humoral and cellular responses in the hemolymph of the body cavity, where they can be engulfed or encapsulated by phagocytic cells [16]. Humoral reactions that typically occur in local tissue spaces spontaneously activate enzymatic cascades that can immobilize intruders via melanization (prophenoloxidase, proPO) and/or terminal coagulation. Products and/or by-products of these humoral reactions activate PRRs, such as peptidoglycan recognition proteins (PGRPs) that in turn activate Toll (and associated intracellular signaling), which can stimulate the secretion of antibacterial peptides in immune-related tissues, such as the fat body and hemocytes to mediate direct killing [16, 17].
Activation of Toll receptors in flies and other insects is indirect, i.e. in the receptor itself does not bind MAMPs. In marked contrast, TLR and NLR activation in deuterostomes results from direct interaction with MAMPs, which not only is relevant in innate immune recognition and defense, but is central to activation and regulation of adaptive immunity [3, 5], as well as in governing homeostasis of some symbiotic relationships [18, 19]. Insect Toll and vertebrate TLR-mediated responses are remarkably similar despite the fundamental differences in their recognition potential as well as associated signaling pathways. It is likely that in deuterostomes interactions with MAMPs, as well as a lack of developmental constraints, have contributed to these functional differences and to the evolution of diverse TLR types. Additional conserved effectors of innate and/or mucosal immunity include phagocytosis [20] and encapsulation [21] as well as: (i) the complement system (discussed further below), which represents the predominant enzymatically driven cascade for eliminating potential pathogens [22, 23]; (ii) wound repair which has ancient origins and likely has experienced repeated co-option by the innate immune system [24, 25]; and (iii) melanization reactions that are associated with immune function in some deuterostome invertebrates [26]. Although protostome invertebrates lack many of the innate receptors that have expanded in deuterostomes and have homologous counterparts in vertebrates, they possess an expanded array of alternative mediators, which include complex melanization pathways as well as a massively diverse repertoire of antibacterial peptides [16, 17]. It is difficult to weigh their relative contributions; however, PRR detection of MAMPs as well as immobilization of potential pathogens via catalytic hemolymph reactions or enzymatic cascades are ancient and vital players in innate immune defenses across a remarkably wide phylogenetic spectrum. The mechanisms summarized above rely on inflammation and/or phylogenetically ancient inflammatory mediators as major effectors of innate immunity [27–29] and are not exclusive to the interstitial and mucosal environments.
Deuterostome invertebrates, owing to their phylogenetic position relative to vertebrates, have considerable potential as models for studies of immune phylogeny (Figure 1). Collectively, the deuterostome invertebrates (echinoderms and protochordates) possess multiple homologs of different vertebrate innate immune effectors; the numbers and potential reactivity of these genes suggest that their functional relevance (Table 1). Protochordates (which include urochordates and cephalochordates) share certain developmental features with vertebrates and are the most recently diverged common invertebrate forms that lack adaptive immunity. The massive expansions of TLRs, other PRRs, NLRs and scavenger receptors that have been revealed through large-scale genome analyses represent the most distinguishing differences among the respective innate immune systems of cephalochordates and echinoderms relative to those of vertebrates [30, 31]. Notably, parallel expansions are not seen in the urochordate, Ciona intestinalis [32]. Since most of these gene families arose through expansions from lineage-specific duplications (i.e. paralogous), very little orthology exists among deuterostome invertebrates and/or among related vertebrate counterparts. It is likely that novel evolutionary constraints may have driven the expansion and functional specialization of the aforementioned receptors [33, 34]. Unrelated functional constraints also may have contributed to parallel or convergent evolution [35]. Although the structures of these innate immune receptors and their underlying mechanisms of function are diverse, they likely share conserved immune regulatory and signaling pathways [36, 37]. In-depth genome analysis will likely offer unique insight into these mechanisms. Amphioxus is critical in understanding major transitions in innate immunity and the proposed development of interrelationships with the adaptive immune system.
Table 1:
Relative complexity of innate immune repertoires
| Amphioxusa | Sea squirtb | Sea urchinc | Humand | |
|---|---|---|---|---|
| Complement | ||||
| C3/C4/C5e | 3 | 2 | 4 | 4 |
| Bf/C2f | 2 | 3 | 3 | 2 |
| MASPg/C1r,sh | 5/44i | 4 | 0/2i | 4 |
| TCCj | 9 | 11 | 0 | 6 |
| C1qk-like | 39 | 2 | 4 | 23 |
| LRR-containing | ||||
| TLRl | 72 | 2 | 222 | 10 |
| NLRm | 118 | 0/28n | 203 | 25 |
| LRR-Igo | 125 | NDp | 22 | 30 |
| Other mediators | ||||
| SRCRq | 270 | 5 | 218 | 16 |
| PGRPr | 18 | 0 | 5 | 6 |
| GNBPs | 5 | 3 | 3 | 0 |
| CTLt | 1200/717u | NDP | 104 | 81 |
| VCBPv | 5/10w | 4 | 0 | 0 |
| Cytokines | ||||
| TNFx | 21 | 4 | 4 | 20 |
| TNFRy | 31 | 3 | 9 | 26 |
| IL-17z | 9 | 2 | 30 | 9 |
Numbers are approximations, reflect assembly modeling ambiguities and subjective interpretations, and vary slightly among cited references [30, 31, 39, 42, 48, 53] and the studies Terajima D, Shida K, Takada N, et al. Identification of candidate genes encoding the core components of the cell death machinery in the Ciona intestinalis genome. Cell Death Differ 2003;10:749–53; Parrinello N, Vizzini A, Arizza V, et al. Enhanced expression of a cloned and sequenced Ciona intestinalis TNFalpha-like (CiTNF alpha) gene during the LPS-induced inflammatory response. Cell Tissue Res 2008;334:305–17. Little information exists regarding the function of specific (predicted) genes in the non-vertebrate species shown.
aBranchiostoma floridae.
bCiona intestinalis.
cStrongylocentrotus purpuratus.
dHomo sapien.
eComplement components.
fFactor B-like protease.
gMannose-associated serine protease.
hComplement C1 (initiator) protease components.
iIncludes MASP-like sequences.
jTerminal complement components.
kC1 q subcomponent (complement initiator).
lToll-like receptor.
mNucleotide-binding oligomerization domain-like receptor.
nPredicted models with NACHT domains.
oLeucine-rich repeat immunoglobulin domain-containing molecule.
pNot described.
qScavenger receptor with cysteine-rich domains.
rPeptidoglycan recognition protein.
sGram-negative recognition protein.
tC-type lectin.
uConfirmed by RT-PCR.
vVariable region-containing chitin-binding protein.
wHaplotype-specific multiple paralogs found (# varies).
xTumor necrosis factor.
yTumor necrosis factor receptor.
zInterleukin-17.
AMPHIOXUS GENOME AND IMMUNE-RELATED NOVELTIES
Due to the limited number of physiologically relevant approaches for examining specific immune reactions in amphioxus, resolution of its genome has given us a particularly distinct view of ‘innate immunity’ that is based almost completely on molecular homology [31, 38]. Genome mining as an experimental approach presents rich opportunities for predicting total transcriptome content, identifying homologous gene families, and revealing genome novelties and innovations. Studies in amphioxus have achieved notable success in all three areas.
Computational approaches, as well as a limited number of experimental approaches that have been applied in amphioxus, support predictions of major increases in the number of genes in amphioxus that function in innate immunity. Specifically, as many as 134 TIR domains and approximately 72 models for TLR-like genes have been predicted [31, 39]. Most significantly, 28 of these predictions create high confidence models [31]. In contrast, approximately 222 TLR gene models can be predicted confidently in the sea urchin genome; however, the relatively fewer TLR models in amphioxus are still at least six times more complex than what is found in the human genome. Differences in the number, length and sequence of the LRR domains of the TLRs suggest that variability in form and likely (additional) function of the receptors exceeds that found in other organisms. Despite the apparent complexity of these genes, only a single adaptor molecule encoded by a sterile alpha armadillo motif (SARM)-like gene, is predicted to regulate the TLRs; however, approximately 10 copies of SARM-like genes can be modeled [40, 41]. Amphioxus, like the sea urchin, exhibits extensive expansions of other intracellular PRRs, including the NLRs, where approximately 118 genes can be identified. Furthermore, 270 gene models for scavenger receptors, more than 1200 C-type lectins and more than 1300 LRR-containing gene models, as well as several other innate immunity genes have been predicted from the amphioxus genome [39] (Table 1). Compared to the sea urchin, amphioxus exhibits considerably larger increase (approximately 240 models) in the numbers of LRR-Ig-containing proteins (LRRIGs), C-type lectin receptors (CLRs) and fibrinogen genes (approximately 340 models) [39, 42]. Both amphioxus and sea urchin exhibit similar expansions of scavenger receptors with cysteine-rich repeats (SRCRs). In marked contrast, the numbers of TLR genes in Ciona is markedly decreased from that seen in mammals [43]. However, some conserved functionality of TLRs in Ciona is revealed in the interactions with MAMPs, such as double-stranded RNA and bacterial cell wall components as well as the subsequent activation of NF-kB signaling pathways [44].
Multiple similarities have been identified among the complement components of protochordates and vertebrates [45]. In marked contrast to Ciona and the sea urchin, more than 39 copies of complement C1q-like genes, of which 25 are encoded on a single scaffold, have been identified in the amphioxus genome and may be associated with an expanded recognition repertoire [31]. A functional role for complement is seen in the findings of vertebrate-type lytic activity in amphioxus [46, 47]. Immune challenges in amphioxus and the subsequent analyses of transcription products indicate that complement may be a major component of mucosal immunity in the gut, where tight regulation of other PRRs, including TLRs, has been demonstrated [42]. Notably, some key elements that regulate complement activation as well as terminal pathway components, appear to be missing from the sea urchin genome, which likely lacks a lytic component [48]. Complement and TLRs interact dynamically in vertebrates, and their respective roles in mucosal tissue recognition and homeostasis, including symbiotic interactions with the resident microbial ecosystems, have been established [22, 23, 49]. This relationship will be important to evaluate in amphioxus.
The oxidative system, petidoglycan recognition proteins (PGRPs), Gram-negative binding proteins (GNBPs), chitin binding proteins (CBPs), lysozymes and defensins also have been shown to be major effectors in amphioxus gut mucosal immunity [42]. Neither IL-1, which is a major cytokine in vertebrates, nor other vertebrate-like cytokines have been identified in the amphioxus genome. However, domain architectures similar to IL-1 receptors as well as both tumor necrosis factor (TNF) and associated receptor (TNFR) have been identified and their respective gene families are associated with modest expansions relative to vertebrate genomes [31]. IL-17, which functions in mucosal immunity, phagocyte responses and inflammatory reactions in vertebrates [50], has been modeled from the sea urchin genome [30] (J.P. Rast, unpublished observation). An IL-17 homolog has been shown to be expressed in response to bacterial challenge in amphioxus [51]. Although clear orthologs of IL-17 have been difficult to identify in the Drosophila genome [52], they have been identified in a nematode and mollusc [51], which are both protostomes, and thereby implicate this cytokine as an ancient and likely relevant, proinflammatory molecule.
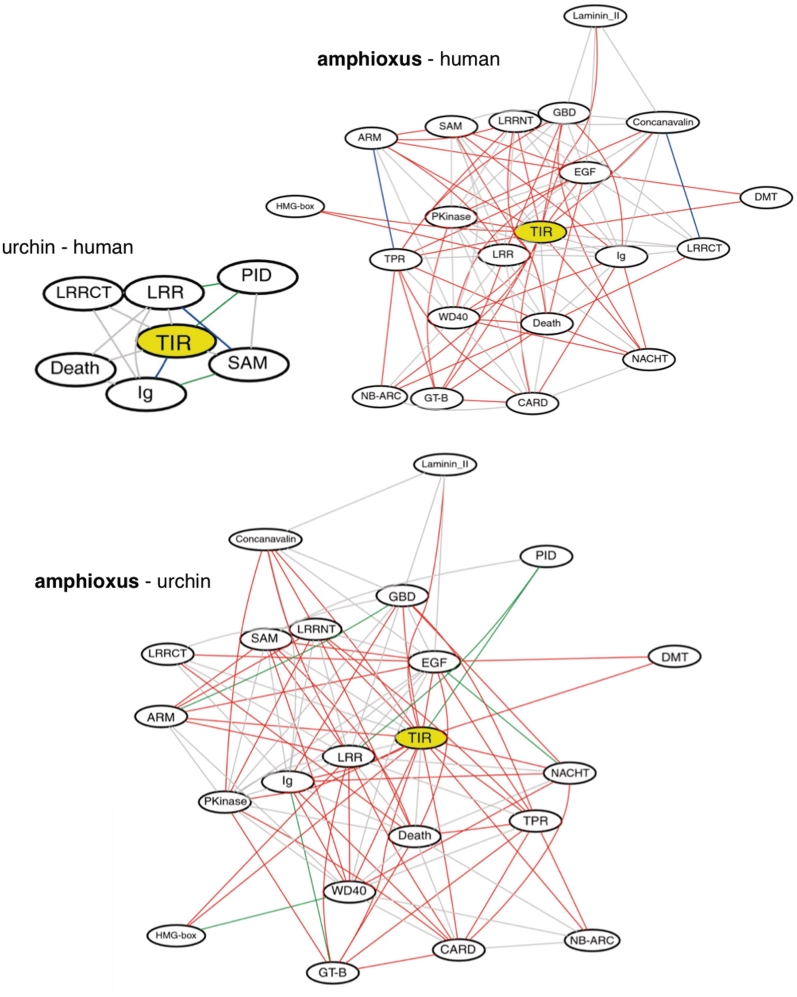
Additional novel protein types have likely arisen in amphioxus through extensive domain expansion and shuffling, which have factored in the expansion of the immune repertoire and creation of putative shortcut pathways [53]. Novel protein architectures have been modeled from these domain rearrangements and include those predicted to mediate immune functions through alternative activation, signaling and network integration routes [33] (Figure 2). However, we currently only can speculate on function. Pathway topology likely is affected by changes in the combinations of different domains (e.g. NACHT domains) as well as by shuffling of novel paralogs of specific domain expansions (e.g. TIR domains, death domain superfamily, others) [53]. These architectures could be associated with novel regulatory connections and/or activation pathways in the innate immune systems [33, 53]. It is likely that constrained selective pressures to expand on a limited supply of domain architectures can result in independently evolved proteins with related domains that lack any true orthologous relationships. Assuming that these novel pathway topologies impart selective advantages, appropriately designed functional studies could help reveal intricate and/or heretofore unrecognized features of innate immune pathways that may factor in adaptive immunity in contemporary vertebrates [33, 34].
Figure 2:
Complexity of domain interaction networks based on TIR domain. Predicted interaction networks relative to the TIR domain. Extensive complexity is indicated among amphioxus, sea urchin and human topologies of innate protein interactions. Common domain combinations between the selected genomes are shown in gray; amphioxus-specific combinations are shown in red; human-specific combinations are shown in blue and sea urchin-specific combinations are shown in green. It is predicted that novel domain combinations result in functionally relevant, innate immune topology for inflammatory pathways in amphioxus. Figure 2 is reproduced from [53], an open access article.
In addition to the significant expansion of the innate immune repertoire in amphioxus defined above, a variety of Ig-containing gene models can be predicted, some of which are V-type. The identification of the V-type domains in amphioxus is of potential interest as jawed vertebrates derive diverse V domains during the somatic development and expansion of antigen-specific lymphocytes in response to antigen. The rearrangement mechanism that gave rise to somatic V diversity in vertebrates involves recombination activating genes (RAG) 1 and 2, which are tightly linked. A homologous, closely linked RAG1 and 2-like gene pair has been identified in the purple sea urchin genome; however, its function is unknown [54]. Both RAG 1 and RAG 2 are required for somatic rearrangement of immunoglobulin and T-cell receptor loci; however, only a RAG1-like segment can be predicted in the amphioxus genome. Despite the absence of several other core molecules that govern the rearrangement process, amphioxus homologs of a variety of other gene products, recruited largely from ancient DNA damage repair pathways, which function in the diversification of immune receptors in jawed vertebrates [55, 56], are found in both amphioxus [31] and sea urchin [30].
THE VARIABLE REGION-CONTAINING CHITIN-BINDING PROTEINS—A DIVERSIFIED FAMILY OF IMMUNE RECEPTORS
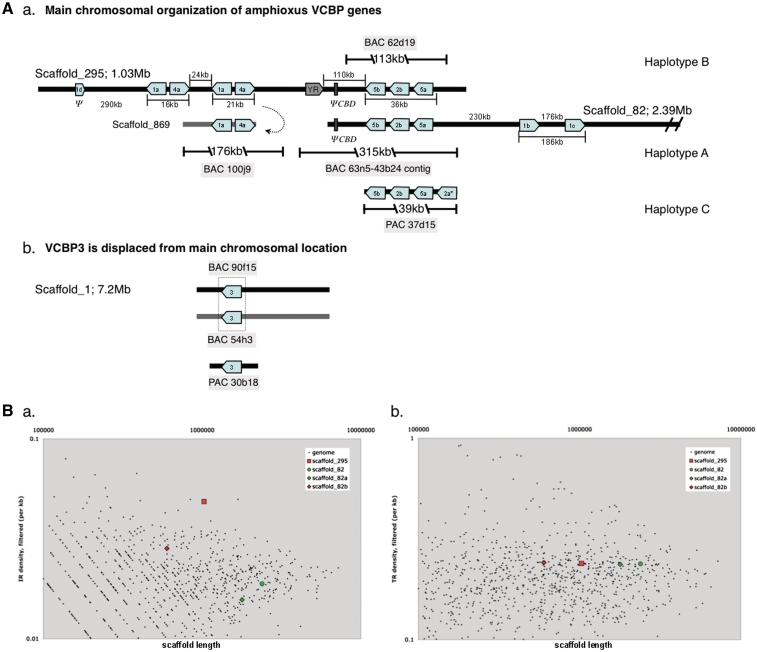
Although genome screening has revealed no evidence for rearranging antigen binding receptors in amphioxus, a family of immune-type genes, which are composed of two Ig V regions and a C-terminal chitin-binding domain (VCBP) [57], have been characterized in both amphioxus and Ciona. Based on the presence of highly polymorphic alleles encoding immunoglobulin domains as well as their expression patterns, it has been hypothesized that VCBPs represent a novel gut-associated form of innate immune proteins in protochordates [57, 58]. VCBPs were discovered prior to the resolution of amphioxus genome [57]; however, the availability of the whole genome, traces, partial assemblies and reference bacterial artificial chromosome (BAC) libraries has permitted an in-depth characterization of the haplotype complexity of VCBPs [59]. VCBPs1, 2, 4 and 5 are linked in a continuous chromosomal region; VCBP3 maps to a separate unrelated scaffold [59] (Figure 3A). VCBP2 and 5 represent paralogous gene sets in which hyperpolymorphism is localized to the N-terminal regions of both the V1 and V2 domains. Folding of the molecule creates a unique hypervariable interface that is predicted to form the binding site of the receptor [60]. VCBPs are organized in a tightly linked cluster and copy number variation among haplotypes has been described. Certain haplotypes of the VCBP2 and 5 cluster demonstrate extensive inverted repeat density, frequent indel (both large and small) polymorphism and an elevated variation in repeat type and density. These chromosomal features, particularly the inverted repeats, are distributed more densely within the VCBP loci than elsewhere in the genome (Figure 3B) and may be associated with transcriptional diversity [59]. Recently, we have shown that VCBPs play an integral role in gut immunity through recognition of bacterial surfaces by the V regions [61].
Figure 3:
(A) Genomic organization of amphioxus VCBPs. (a) Overlapping scaffolds_295 and 82 represent VCBP haplotypes B and A, respectively; assemblies are based on Brafl1, which was modified by sequencing additional genomic libraries, BACs and P1 artificial chromosomes (for details, see [59]). Shaded box (labeled YR) represents a Ngaro-like tyrosine recombinase retroposon gene. Pseudogenes (Y) are indicated. Intergeneic distances are in kilobase (1000 base pairs) and are not to scale. Transcriptional orientation is indicated by an arrow; a and b designations reflect paralogous relationships. (b) VCBP3 is located on an unrelated scaffold from the VCBP1/4 and VCBP2/5 clusters. CHEF analyses (not shown) and extensive BAC library screening support assignments. (B) Genome-wide comparison of repeat densities. (a) Inverted repeats (IRs) among VCBP2/5 clusters. The density of IR distributions in scaffold_295 (and scaffold_82, or the corresponding region of scaffold_82 only, shown as _82b) was compared to the distribution of IRs across the entire amphioxus genome. The density of IRs in Scaffold_295 (haplotype B) is higher than what is seen in most other scaffolds of related size (and larger) in the amphioxus genome, including scaffold_82 (haplotype A). IRs are clustered at high density in the VCBP-associated regions of scaffold _82 (_82b). (b) The distribution and density of tandem repeats has been compared by another method. Distributions of tandem repeats in the VCBP loci are consistent with other areas of the genome and is not elevated across haplotypes A and B. Figure 3A and B are reproduced from [59].
Based on the identification of VCBPs in amphioxus, we have screened available genome databases in the interest of determining the phylogeny of Ig and CBD domain-containing molecules (R. Haire, unpublished data). Several Ig-CBD (i.e. VCBP-like) predicted transcripts can be predicted in Capitella, a protostome representative (polychaete annelid). A 3.3-kb genomic segment is predicted to encode six or seven exons and produce a 1.2–1.5 kb transcript encoding two V-type Ig domains and two CBDs: V1V2(v3)CBD1CBD2 or V-V-v-C-C (lower case v represents an incomplete domain). A second 3.2-kb genomic segment is related closely to the 3.3-kb segment and is predicted to transcribe a 1.4 kb transcript. A third region is encoded in seven exons (4.4 kb) and can be modeled to encode a 3.4 kb transcript (V-V-X-C-C, where x is an unknown domain). In Lottia, a gastropod mollusk representing another protostome, a 2.5-kb genomic region can be modeled from seven exons to encode a 1 kb transcript (X-V-X-C). A single example of proximal V and CBD domains separated by a 1 kb repeat is found in Drosophila. Whereas the various predicted molecules have not been confirmed experimentally, it appears that Ig-CBD proteins may not be unique to deuterostome invertebrate innate immunity.
CONCLUDING REMARKS
Our interests are focused on developing protochordates as physiologically relevant systems in which to explore the functional significance of innate immune innovations. In addition to the significance of the phylogenetic position that amphioxus occupies in chordate phylogeny, this species presents a unique opportunity to assess the functional consequences of expansions in multigene families encoding several different forms of molecules that we presently attribute to innate immunity. Specifically, this model will permit studies of how novel gene duplications are acquired and become integrated in pre-existing pathways or are adapted in newly evolving topologies that regulate or effect innate immune responses (Figure 2). Whole animal experimentation will reveal how selective pressures, likely effected through various microbial interactions, have driven the incorporation of these novel protein architectures into the well conserved and highly adapted innate immune repertoire. Protochordates open a window into the study of innate immunity in integrated systems that are devoid of interactions with the highly derived adaptive immune system of the vertebrates.
Studies of innate immunity in higher vertebrates have emphasized the major role of the gut in immune stasis. Filter-feeding invertebrates, such as amphioxus and Ciona, in which the gut is in direct and continuous contact with a vast range of microbiota, will serve as powerful models in which basic questions such as the roles of gene novelty and diversity can be explored directly. Furthermore, the dynamic interactions between host and microbial communities include many symbiotic relationships resulting in a continuum of selective pressures that directly influence innate immune innovation. It is likely that a more comprehensive understanding of the complex dynamics of host and microbial dialog across innate immune receptors will emerge from studies in both cephalochordates and urochordates, possibly revealing how microbial communities influence innate immunity and have contributed to the origins of adaptive immunity in vertebrates [62, 63].
Key points.
Amphioxus is a protochordate model of immune evolution.
The amphioxus genome demonstrates extensive polymorphism and certain innate immune genes in amphioxus are highly polymorphic.
The innate immune repertoire of amphioxus is extensively expanded; this effect also is seen in sea urchin.
The expanded innate repertoire demonstrates innovation.
Immune gene families in amphioxus likely reflect novel adaptations with microbiota.
FUNDING
This work was supported by the National Institutes of Health (AI23338 to G.W.L.) and the All Children's Hospital Foundation and the H. Lee Moffitt Cancer Center and Research Institute (to L.J.D.).
Acknowledgements
Authors thank M. Gail Mueller and Natasha Gwatney for continuing technical assistance and Ronda T. Litman for sequence analysis. Authors also thank Barb Pryor for editorial assistance.
Biographies
Larry J. Dishaw is an Assistant Professor in the Department of Pediatrics at the University of South Florida, Children's Research Institute. His research is focused on the evolution of gut immunity with special emphasis on the role of gut microbiota.
Robert N. Haire is an Associate Professor in the Department of Pediatrics at the University of South Florida, Children's Research Institute. His research is focused on the evolution of the immunoglobulin domain.
Gary W. Litman is the Hines Professor and Vice Chairman of the Department of Pediatrics at the University of South Florida. His studies are focused on alternative forms of immune recognition and the evolution of immune defense reactions.
References
- 1.Rast JP, Messier-Solek C. Marine invertebrate genome sequences and our evolving understanding of animal immunity. Biol Bull. 2008;214:274–83. doi: 10.2307/25470669. [DOI] [PubMed] [Google Scholar]
- 2.Maldonado-Contreras AL, McCormick BA. Intestinal epithelial cells and their role in innate mucosal immunity. Cell Tissue Res. 2011;343:5–12. doi: 10.1007/s00441-010-1082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavelle EC, Murphy C, O'Neill LA, et al. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 2010;3:17–28. doi: 10.1038/mi.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medzhitov R, Janeway CA., Jr An ancient system of host defense. Curr Opin Immunol. 1998;10:12–15. doi: 10.1016/s0952-7915(98)80024-1. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre B, Nicolas E, Michaut L, et al. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–83. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 7.Medzhitov R, Preston-Hurlburt P, Janeway CAJ. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 8.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 9.Boraschi D, Tagliabue A. The interleukin-1 receptor family. Vitam Horm. 2006;74:229–54. doi: 10.1016/S0083-6729(06)74009-2. [DOI] [PubMed] [Google Scholar]
- 10.Werts C, Girardin SE, Philpott DJ. TIR, CARD and PYRIN: three domains for an antimicrobial triad. Cell Death Differ. 2006;13:798–815. doi: 10.1038/sj.cdd.4401890. [DOI] [PubMed] [Google Scholar]
- 11.Delbridge LM, O'Riordan MX. Innate recognition of intracellular bacteria. Curr Opin Immunol. 2007;19:10–16. doi: 10.1016/j.coi.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–54. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Inohara N, Chamaillard M, McDonald C, et al. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Ann Rev Biochem. 2005;74:355–83. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 14.Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–3. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- 15.Medzhitov R, Janeway CAJ. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 16.Kurata S. Fly immunity: recognition of pathogens and induction of immune responses. Adv Exp Med Biol. 2010;708:205–17. doi: 10.1007/978-1-4419-8059-5_11. [DOI] [PubMed] [Google Scholar]
- 17.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 18.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–44. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 20.Jutras I, Desjardins M. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu Rev Cell Dev Biol. 2005;21:511–27. doi: 10.1146/annurev.cellbio.20.010403.102755. [DOI] [PubMed] [Google Scholar]
- 21.Ratner S, Vinson SB. Phagocytosis and encapsulation: cellular immune responses in arthropoda. Am Zool. 1983;23:185–94. [Google Scholar]
- 22.Trouw LA, Daha MR. Role of complement in innate immunity and host defense. Immunol Lett. 2011;138:35–7. doi: 10.1016/j.imlet.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Ehrnthaller C, Ignatius A, Gebhard F, et al. New insights of an old defense system: structure, function, and clinical relevance of the complement system. Mol Med. 2011;17:317–29. doi: 10.2119/molmed.2010.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massberg S, Grahl L, von Bruehl ML, et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 2010;16:887–96. doi: 10.1038/nm.2184. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Wilhelmsson C, Hyrsl P, et al. Pathogen entrapment by transglutaminase—a conserved early innate immune mechanism. PLoS Pathog. 2010;6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cammarata M, Arizza V, Cianciolo C, et al. The prophenoloxidase system is activated during the tunic inflammatory reaction of Ciona intestinalis. Cell Tissue Res. 2008;333:481–92. doi: 10.1007/s00441-008-0649-x. [DOI] [PubMed] [Google Scholar]
- 27.Doolittle RF. Coagulation in vertebrates with a focus on evolution and inflammation. J Innate Immun. 2011;3:9–16. doi: 10.1159/000321005. [DOI] [PubMed] [Google Scholar]
- 28.Aller MA, Arias JL, Sanchez-Patan F, et al. The inflammatory response: an efficient way of life. Med Sci Monit. 2006;12:RA225–34. [PubMed] [Google Scholar]
- 29.Campbell EL, Serhan CN, Colgan SP. Antimicrobial aspects of inflammatory resolution in the mucosa: a role for proresolving mediators. J Immunol. 2011;187:3475–81. doi: 10.4049/jimmunol.1100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rast JP, Smith LC, Loza-Coll M, et al. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–6. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holland LZ, Albalat R, Azumi K, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–11. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehal P, Satou Y, Campbell RK, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–60. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Zmasek CM, Godzik A. Domain architecture evolution of pattern-recognition receptors. Immunogenetics. 2010;62:263–72. doi: 10.1007/s00251-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capra JA, Pollard KS, Singh M. Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biol. 2010;11:R127. doi: 10.1186/gb-2010-11-12-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leulier F, Lemaitre B. Toll-like receptors—taking an evolutionary approach. Nat Rev Genet. 2008;9:165–78. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 36.Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223–44. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wullaert A. Role of NF-kappaB activation in intestinal immune homeostasis. Int J Med Microbiol. 2010;300:49–56. doi: 10.1016/j.ijmm.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Putnam NH, Butts T, Ferrier DE, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–71. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 39.Huang S, Yuan S, Guo L, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–26. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan S, Wu K, Yang M, et al. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J Immunol. 2010;184:6874–81. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Zmasek CM, Cai X, et al. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev Comp Immunol. 2011;35:461–8. doi: 10.1016/j.dci.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Wang X, Yan Q, et al. The evolution and regulation of the mucosal immune complexity in the basal chordate amphioxus. J Immunol. 2011;186:2042–55. doi: 10.4049/jimmunol.1001824. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki N, Ogasawara M, Sekiguchi T, et al. Toll-like receptors of the ascidian Ciona intestinalis: prototypes with hybrid functionalities of vertebrate Toll-like receptors. J Biol Chem. 2009;284:27336–43. doi: 10.1074/jbc.M109.032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satake H, Sasaki N. Comparative overview of toll-like receptors in lower animals. Zoolog Sci. 2010;27:154–61. doi: 10.2108/zsj.27.154. [DOI] [PubMed] [Google Scholar]
- 45.Nonaka M, Kimura A. Genomic view of the evolution of the complement system. Immunogenetics. 2006;58:701–13. doi: 10.1007/s00251-006-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Zhang S, Wang C, et al. Complement-mediated killing of Vibrio species by the humoral fluids of amphioxus Branchiostoma belcheri: implications for a dual role of O-antigens in the resistance to bactericidal activity. Fish Shellfish Immunol. 2008;24:215–22. doi: 10.1016/j.fsi.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S, Wang C, Wang Y, et al. Presence and characterization of complement-like activity in the amphioxus Branchiostoma belcheri tsingtauense. Zoolog Sci. 2003;20:1207–14. doi: 10.2108/zsj.20.1207. [DOI] [PubMed] [Google Scholar]
- 48.Hibino T, Loza-Coll M, Messier C, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–65. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 49.Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010;31:154–63. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pappu R, Ramirez-Carrozzi V, Sambandam A. The interleukin-17 cytokine family: critical players in host defence and inflammatory diseases. Immunology. 2011;134:8–16. doi: 10.1111/j.1365-2567.2011.03465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu B, Jin M, Gong J, et al. Dynamic evolution of CIKS (TRAF3IP2/Act1) in metazoans. Dev Comp Immunol. 2011;35:1186–92. doi: 10.1016/j.dci.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 52.Aggarwal S, Gurney AL. IL-17: prototype member of an emerging cytokine family. J Leukoc Biol. 2002;71:1–8. [PubMed] [Google Scholar]
- 53.Zhang Q, Zmasek CM, Dishaw LJ, et al. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9:R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fugmann SD, Messier C, Novack LA, et al. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA. 2006;103:3728–33. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung D, Alt FW. Unraveling V(D)J recombination; insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- 56.Litman GW, Cannon JP, Rast JP. New insights into mechanisms of immune receptor diversification. Adv Immunol. 2005;87:209–36. doi: 10.1016/S0065-2776(05)87006-3. [DOI] [PubMed] [Google Scholar]
- 57.Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–7. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- 58.Cannon JP, Haire RN, Schnitker N, et al. Individual protochordates possess unique immune-type receptor repertoires. Curr Biol. 2004;14:R465–R466. doi: 10.1016/j.cub.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Dishaw LJ, Mueller MG, Gwatney N, et al. Genomic complexity of the variable region-containing chitin-binding proteins in amphioxus. BMC Genomics. 2008;9:78. doi: 10.1186/1471-2156-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hernandez Prada JA, Haire RN, Allaire M, et al. Ancient evolutionary origin of diversified variable regions revealed by crystal structures of an immune-type receptor in amphioxus. Nat Immunol. 2006;7:875–82. doi: 10.1038/ni1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dishaw LJ, Giacomelli S, Melillo D, et al. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut-bacteria interactions. Proc Natl Acad Sci USA. 2011;108:16747–52. doi: 10.1073/pnas.1109687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 63.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–73. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]