Abstract
We report an enzymatic end-point modification and immobilization of recombinant human thrombomodulin (TM), a cofactor for activation of anticoagulant protein C pathway via thrombin. First, a truncated TM mutant consisting of epidermal growth factor-like domains 4–6 (TM456) with a conserved pentapeptide LPETG motif at its C-terminal was expressed and purified in E. coli. Next, the truncated TM456 derivative was site-specifically modified with N-terminal diglycine containing molecules such as biotin and the fluorescent probe dansyl via sortase A (SrtA) mediated ligation (SML). The successful ligations were confirmed by SDS-PAGE and fluorescence imaging. Finally, the truncated TM456 was immobilized onto N-terminal diglycine-functionalized glass slide surface via SML directly. Alternatively, the truncated TM456 was biotinylated via SML and then immobilized onto streptavidin-functionalized glass slide surface indirectly. The successful immobilizations were confirmed by fluorescence imaging. The bioactivity of the immobilized truncated TM456 was further confirmed by protein C activation assay, in which enhanced activation of protein C by immobilized recombinant TM was observed. The sortase A-catalyzed surface ligation took place under mild conditions and is rapid occurring in a single step without prior chemical modification of the target protein. This site-specific covalent modification leads to molecules being arranged in a definitively ordered fashion and facilitating the preservation of the protein’s biological activity.
Introduction
Immobilization of protein onto solid surfaces is widely used in the preparation of functional protein microarrays, in the development of protein-based bioassays, biosensors, and for medical devices surface functionalization, and various other applications.1 The success of all these technologies relies on the immobilization technique employed to attach a functional protein to the corresponding surface. Non-specific physical adsorption or chemical cross-linking with appropriate surfaces results in the immobilization of the protein in random orientations and with reduced activity. Thrombomodulin (TM) is an endothelial integral membrane protein and acts as a major cofactor in the protein C anticoagulant pathway.2 When bound to TM on the endothelial surface, thrombin is unable to activate fibrin generation or activate platelets. Instead, it becomes a potent activator of protein C. Subsequently, the activated form of protein C (APC), an anticoagulant protease, selectively inactivates coagulation factors Va and VIIIa, providing an essential feedback mechanism in preventing excessive coagulation. Therefore, TM serves as an excellent target for the engineering of antithrombogenic surfaces for cardiovascular biomaterials by immobilizing TM.3 In the past decade, several groups have covalently immobilized TM onto polymeric surfaces for generating antithrombogenic materials. Kishida et al. conjugated recombinant human thrombomodulin to both aminated and carboxylated surfaces, including poly(vinyl amine) and poly(acrylic acid) surface-grafted polyethylene, and a surface-hydrolyzed poly(ether urethaneurea).4–6 Similarly, Vasilets et al. immobilized TM onto poly(acrylic acid) surface-grafted PTFE.7 Alternatively, Cutler et al. physically adsorbed and crosslinked soluble human TM onto small-caliber ePTFE grafts and reported promising short-term antithrombotic results in vivo.8 Other investigators, such as Sperling et al. and Han et al. have tethered TM to surfaces by using polyethylene glycol spacers.9,10 However, many of these methods may negatively impact activity of the TM due to harsh conjugation conditions possibly leading to protein denaturation and random-site conjugation leaving limited accessibility to the active region of the protein, both potentially hampering the desired bioactivity. The latter issue resulting from common conjugation techniques has been reported in studies where the cofactor activity of human TM for protein C activation decreases upon immobilization due to the potential conformation variation caused by the random covalent immobilization.10
In order to generate the stable, functional, homogeneous, and oriented protein immobilization, attention has focused on methods that enable the efficient and site-specific covalent immobilization of recombinant proteins. Recently, a short TM construct containing epidermal growth factor (EGF)-like domains 4–6 with an azide-functionalized methionine analogue as a C-terminal linker was synthesized in E. coli for site-specific immobilization of the recombinant TM derivative at the C-terminus through click chemistry11 and Staudinger ligation.12 Incorporation of azide-functionalized amino acids into proteins in vivo provides opportunities for protein modification under native conditions and selective labeling of proteins in the intracellular environment. However, the expression yield for recombinant protein with non-natural amino acid incorporation is generally too low for protein modification and practical applications. Also, the incorporation of non-natural amino acids involves complicated and time-consuming molecular biology procedures.
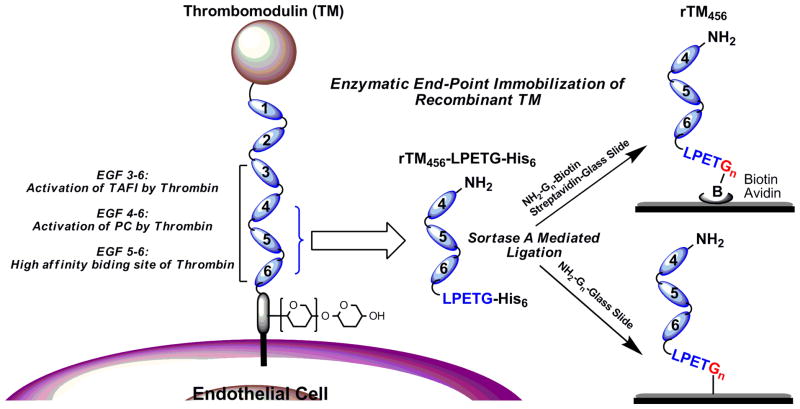
Enzymatic post-translational modifications have recently provided robust tools for covalent protein conjugations. For example, sortase A (SrtA) has been studied extensively for protein ligation applications.13 Sortase A, a transpeptidase from S. aureus, is present on the plasma membrane and catalyzes a cell wall sorting reaction that attaches surface proteins to the cell wall envelope.14 SrtA recognizes substrates containing a pentapeptide LPXTG motif (X being any amino acid) and cleaves the amide bond between the Thr and Gly residues with an active site cysteine to generate a covalent acyl-enzyme intermediate. The carboxyl group of Thr of the thioester intermediate is then attacked by the amino terminus of an oligoglycine nucleophile, resulting in the formation of a native covalent peptide bond.15–18 The advantage of sortase-mediated ligation (SML) is that the reaction takes place in a site-specific manner under mild conditions and occurs rapidly in a single step without prior chemical modification of the target protein. Employing such a strategy, we report an enzymatic end-point modification and immobilization of recombinant TM via SML. Briefly, a truncated TM mutant consisting of epidermal growth factor-like domains 4–6 (rTM456) with a conserved pentapeptide LPETG motif at its C-terminal was expressed and purified in E. coli, which is for SML with N-terminal oligoglycine-containing molecules. It has been shown that LPETG is the optimal isoform of the LPXTG motif.19 Two strategies for site-directed immobilization of recombinant TM via their C-terminus to different solid supports were demonstrated (Figure 1). First, the rTM456 was biotinylated via SML and then immobilized onto streptavidin-functionalized glass slide surfaces. Second, the rTM456 was immobilized onto N-terminal diglycine-functionalized glass slide surfaces via SML directly. The bioactivity of immobilized rTM456 was further confirmed by protein C activation assay, which showed enhanced protein C activation. Overall, sortase A-catalyzed surface ligation is mild, rapid, and occurs in a single step without prior chemical modification of the target protein. Site-specific covalent attachment leads to molecules being arranged in a definite, orderly fashion and allows the use of spacers and linkers to help minimize steric hindrances between the protein and the surface thus facilitating the protein’s stability and biological activity. Since TM is a transmembrane glycoprotein of the endothelium of blood and lymphatic vessels and the basis of a major natural anticoagulant system, the reported strategies should provide an ideal approach for the engineering of antithrombogenic surfaces for cardiovascular biomaterials, minimizing incompatibility and thrombosis.
Figure 1.
Illustration of endothelial thrombomodulin (TM) and site-specific end-point immobilization of LPETG-tagged recombinant thrombomodulin (rTM456) through Sortase A-Mediated Ligation (SML). EGF: Epidermal Growth Factor-like domain.
Materials and methods
Materials
All solvents and reagents were purchased from commercial sources and were used unless otherwise noted. Human protein C, human thrombin and human antithrombin III were obtained from Haematologic Technologies Inc., Chromogenic thrombin substrate BIOPHEN-CS01 was obtained from Aniara, and the DNA fragments encoding the TM derivative and sortase A were synthesized by Genscript Inc..
Synthesis of diGly-Biotin and diGly-Dansyl
A solution of Biotin ethylenediamine (36 mg, 0.129 mmol, Sigma) in 1 mL dry DMF was treated with triethylamine (0.1 mL, 0.71 mmol) and allowed to stir at room temperature for 30 min. The solution was then added to a solution of Z-Gly-Gly-p-nitrophenyl ester (100 mg, 0.258 mmol, Sigma) in dry DMF (4 mL) via syringe. The reaction mixture was stirred for 24 h at room temperature under Ar(g) and then concentrated under vacuum to give a residue, which was then dissolved in MeOH (10 mL) to which 50 mg of Pd/C (10% wt.) was added. The reaction solution was degassed under vacuum and then backfilled with hydrogen gas via a hydrogen gas balloon and stirred for 6 h at room temperature. Finally, the reaction solution was filtered to remove the Pd/C (10% wt.) catalyst and the filtrate was concentrated to give a residue, which was purified by silica gel column using chloroform/methanol (5:1) as elutent to afford diGly-biotin (36 mg, 70%). 1H NMR (D2O) d: 4.39 (m, 1 H, CH-1-Biotin), 4.19 (m, 1 H, CH-4-Biotin), 3.87 (s, 2H, CH2-Gly), 3.73 (s, 2H, CH2-Gly), 3.39 (t, 2H, CH2CH2), 3.13 (t, 2H, CH2CH2), 3.60 (dd, 1 H, CH-Biotin), 2.91 (m, 1H, CH-Biotin), 2.79 (dd, 1 H, CH-Biotin), 2.60 (m 1 H, CH-Biotin), 2.06 (t, 1 H, CH2CO-Biotin), 1.53, 1.43, 1.22 (m, 6 H, (CH2)3-Biotin). Under similar reaction conditions, diGly-Dansyl was synthesized from Z-Gly-Gly-p-nitrophenyl ester (100 mg, 0.258 mmol, Sigma) and Dansyl ethylenediamine (38 mg, 0.129 mmol, Sigma) in 73% (40 mg). 1H NMR (D2O) δ: 8.34 (d, 1H, aromatic), 8.09 (m. 2H, aromatic), 7.55 (m, 2H, aromatic), 7.26 (m, 1H, aromatic), 3.58 (s, 2H, CH2-Gly), 3.37 (s, 2H, CH2-Gly), 3.02 (t, 2H, CH2CH2), 2.91 (t, 2H, CH2CH2), 2.71 (s, 6H, CH3x2).
Synthesis of Gly-Gly-modified glass slides
Several pre-cut pieces of the amine-coated glass slide were placed in a small recystallization dish. To the dish was added 10 mL of an incubation solution of ~ 20 mM Z-GG-p-nitrophenyl ester in dry DMF and then 5 drops of triethylamine (Et3N) was added. The dish was then immediately covered and placed on a mechanical shaker to shake gently overnight at room temperature. After 24 h, the slides were removed from the incubation solution and sequentially rinsed with EtOH, doubly distilled H2O, EtOH again and then blown dry with argon gas.
To deprotect the Z-group at the N-terminus of the diglycine residues, the newly functionalized glass slides were gently placed into a 100 mL round bottom flask turned on its side. To the flask was added 8 mL of a 3:1 Methanol/H2O solution and ~ 5 mg of Pd/C (10% wt). A 3-way stopcock with a hydrogen balloon was then attached to the flask. Prior to introducing H2 gas into the vessel, the air was slowly evacuated while slowly flushing with argon gas over a period of ~ 5 min. After the air was removed, H2 gas was introduced into the system and the flask was set on a mechanical shaker to shake gently for 5 h at room temperature performing hydrogenolysis of the benzyl carbamate-protecting group. Then, the vessel was flushed with argon to remove the excess H2 and the slides were then removed and rinsed with a 1:1 methanol/H2O solution. The slides were then blown dry with argon gas and set into a plastic container to be stored in a 4–8 °C refrigerator until further use.
Expression and purification of TM456 fusion protein
The gene encoding the EGF domain 4–6 of human thrombomodulin (corresponding to residues 345–473) with a C-terminal LPETG motif and an N-terminal human Factor Xa cleavage site (IEGRS) was designed and synthesized (Genscript Inc.). In order to prevent oxidation and reduce proteolytic susceptibility, three residues of TM were mutated (M388L, R456G and H457Q) as described before20 (See Supporting Information for protein sequence). The gene fragment was then inserted into the NcoI-BamHI sites of the pET39b vector to get the new expression plasmid, pET39b-TM456. This new construct was transformed in E. coli BL21 (DE3) and grown at 37°C to an OD (600 nM) of 0.8. Protein expression was induced by the addition of IPTG to a final concentration of 1 mM, and the E. coli culture was incubated overnight at 25°C. The bacteria were then centrifuged at 6,000 g for 10 min and the pellet was then resuspended in 40 mL of lysis buffer (20 mM Tris and 150 mM NaCl, pH 8.0) and following lysis by sonication, the extract was then loaded onto a 5 mL HiTrap Chelating column (GE Healthcare) charged with Ni2+ ions and eluted with 20 mM Tris, 0.5 M NaCl and 250 mM imidazole, pH 8.0. The chromatographic fractions were analyzed by 12% gradient SDS-PAGE gel electrophoresis and visualized by western blot analysis using mouse monoclonal antibody towards human TM. The nitrocellulose membrane was developed using the ECL plus western blotting detection kit (Amersham Biosciences, U.K.). Pooled fractions containing recombinant TM were collected and then dialyzed against 20 mM Tris and 150 mM NaCl, pH8.0, and lyophilized to afford DsbA/LPETG-tagged TM456 fusion protein (dlTM456F). FXa cleavage removed the fusion DsbA tag and generated the target LPETG-tagged TM456 fusion protein (lTM456F).
Expression and purification of soluble SrtA
Soluble SrtA with a C-terminal His6-tag was expressed and purified from E. coli BL21, briefly, the gene encoding S. aureus COL SrtA-ΔN59 (residues 60–206) was inserted into NcoI-XhoI sites of pET28b vector. The plasmid pET28b-SrtA was transformed into E. coli BL21 (DE3). The cells were grown in LB medium to an OD (600 nm) of 0.8 at 37°C, protein expression was induced by the addition of IPTG to a final concentration of 0.5 mM. After growth for an additional 4 h at 37°C, the cells were harvested by centrifugation and resuspended in 40 mL lysis buffer containing 20 mM Tris and 250 mM NaCl, pH 8.0, and lysed by sonication. The extract was centrifuged at 20,000 g for 20 min, and the supernatant was applied onto a 5 mL HiTrap Chelating column (GE Healthcare) charged with Ni2+ ions and eluted with 20 mM Tris, 0.5 M NaCl and 250 mM imidazole, pH8.0. The pooled fractions containing rSrtA were collected and then dialyzed against 20 mM Tris and 150 mM NaCl, pH 8.0, and stored at −80°C until use.
C-terminal covalent modification of TM456 fusion protein via SML
dlTM456F (10 μM) was mixed with SrtA (5 μM) and biotin/dansyl modified diglycine nucleophiles (100 μM) in reaction buffer (20 mM Tris, 150 mM NaCl and 5 mM CaCl2, pH 8.0), the reaction mixture was then incubated at 37°C for 6 h. For further immobilization reactions, unreacted diglycine nucleophiles were removed by dialysis against 20 mM Tris and 150 mM NaCl, pH 8.0. In order to detect the TM456 conjugates, the reaction mixtures were loaded on SDS-PAGE and analyzed by fluorescence imaging or under UV light.
OG488 labeling of TM456 fusion protein
dlTM456F was labeled with Oregon Green 488 (OG488) succinimidyl ester (Molecular Probes, Inc). Briefly, to a solution of 150 μL of carbonate-bicarbonate buffer (pH 9.0), 50 μL of OG488 solution (10 mg/mL in DMSO) was added, followed by 150 μL aqueous dlTM456F solution (2 mg/mL). The mixture was gently vortexed at room temperature in the dark for 2 h. After the coupling reaction, the unreacted OG488 was removed by dialysis using centrifuge filter devices with a cutoff molecular weight of 10,000 Da to afford OG488-labeled dlTM456F.
End-point immobilization of TM456 fusion protein onto solid supports
For immobilization of biotinylated dlTM456F, the mixture containing biotinylated OG488-dlTM456F from above was incubated with Streptavidin coated glass slides (1×2 cm, Xenopore, Hawthorne, NJ) at 37°C for 1 h, the glass slides were then rinsed with PBS (pH 7.4) for 1 h to remove any unbounded proteins. Surface-bound OG488-labeled dlTM456F was further detected by fluorescence image at EX/EM = 480/520.
For direct immobilization of dlTM456F via SML, OG488-labeled dlTM456F (2 μM) was co-incubated with SrtA (1 μM) and diglycine-coated glass slides (1×2 cm) in reaction buffer (20 mM Tris, 150 mM NaCl and 5 mM CaCl2, pH 8.0) for 16 h at room temperature. Then the glass slides were removed from reaction buffer and were washed with the PBS (pH 7.4) for 1 hr to remove unreacted dlTM456F prior to measuring TM activity. The successful immobilization was confirmed upon examining the fluorescence image of OG488-labeled dlTM456F (EX/EM = 480/520 nm)
Protein C activation activity assay of immobilized TM456 fusion protein
The cofactor activity of immobilized dlTM456F was assessed by protein C activation assay as previously described with some modification.10 The dlTM456F-immobilized glass slide (1×2 cm) was placed in a 6-well plate and immersed by 3 mL of assay buffer (20 mM Tris, 150 mM NaCl and 5 mM CaCl2, pH 8.0) containing 300 nM of Human protein C, reaction was initiated with the addition of human α-thrombin to a final concentration of 10 nM. After incubation for 2 hrs at 37°C with shaking, the protein C activation was terminated by addition of 30 μL human antithrombin III (1 mg/mL) and 2 μL heparin (10 U/mL) and further incubated for 5 min at 37°C. The enzymatic activity of activated protein C was measured by the addition of chromogenic substrate H-D-Phe-Pip-Arg-pNa, 2HCl (0.5 mM), after 20 min incubation, the change in absorbance at 405 nm was measured.
Results and Discussion
Expression of TM456 fusion protein and recombinant SrtA
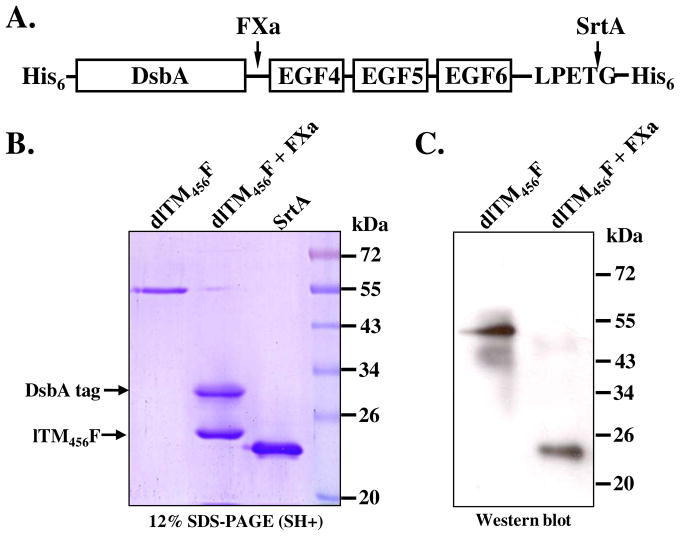
TM is membrane protein and is composed of five distinct domains: an NH2-terminal domain, a domain containing six epidermal growth factor (EGF)-like structures, an O-glycosylation site-rich domain, a transmembrane domain, and a cytoplasmic domain. It has been reported that the region including the fourth, fifth, and sixth EGF-like structures (EGF 4–6) is the minimum domain required for protein C-activating cofactor activity and the protein’s subsequent anticoagulant activity21,22 In this study, a fragment containing the last three consecutive EGF-like domains 4–6 of human TM (TM456) was chosen as the target protein to test the enzyme-mediated ligation using various substrates, such as biotinylation, fluorophore labeling and immobilization. To closely mimic the TM structure as it appears at the cell surface and consequently preserve its antithrombotic bioactivity, we investigated the site-specific immobilization of the recombinant TM at the C-terminus via SML (Scheme 1). In order to reduce steric effects and increase the flexibility of C-terminal LPETG motif, eight hydrophilic amino acids at the C-terminal of EGF4–6 of the native TM (KVDGGDSG, corresponding residue 465–472) were retained (Figure 2A). Also, a LPETG-His6 tag was genetically fused to the C-terminal of TM456. The gene construct was synthesized and inserted into expression vector pET39b. Vector pET39b-TM456 was constructed for expression of an N-terminal DsbA/LPETG-tagged TM456 derivative with two His6 tags at both termini (dlTM456F) (Figure 2A). Protein expression was conducted in the E. coli culture overnight at 25°C. Purified dlTM456F (~55 kDa) was analyzed by SDS-PAGE (Figure 2B), and further confirmed by western blot (Fig. 2C) and protein C activation assay (Figure 4B and Figure 5B). Treatment with Factor Xa removed the DsbA fusion tag (~33 kDa) and generated the target lTM456F (~22 kDa, Figure 2B and 2C). Since the SrtA reaction is highly specific, the reaction can take place even in the presence of impure protein, with the exception of impure protein containing N-terminal oligoglycine, thus the use of the impure target TM456 after fusion tag removal was acceptable. In this study, dlTM456F was used for the reason that there are 21 lysine residues in DsbA-tagged TM456 which can be more efficiently labeled by OG488 succinimidyl ester compared with only one lysine residue in lTM456.
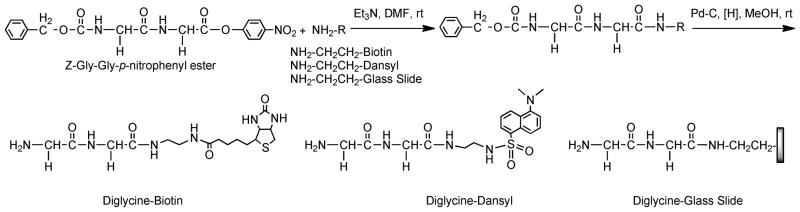
Scheme 1.
Synthesis of diGlycine containing Biotin, Dansyl and glass slide conjugates.
Figure 2.
Structure of DsbA/LPETG-tagged TM456 fusion protein (dlTM456F) (A), its SDS-PAGE characterization (B): Lane 1. purified dlTM456F, Lane 2. dlTM456F incubated with human Factor Xa, Lane 3. purified SrtA-ΔN59, Lane 4. molecular makers, and Western blot analysis (C): Lane 1. purified dlTM456F, Lane 2. dlTM456F incubated with human Factor Xa.
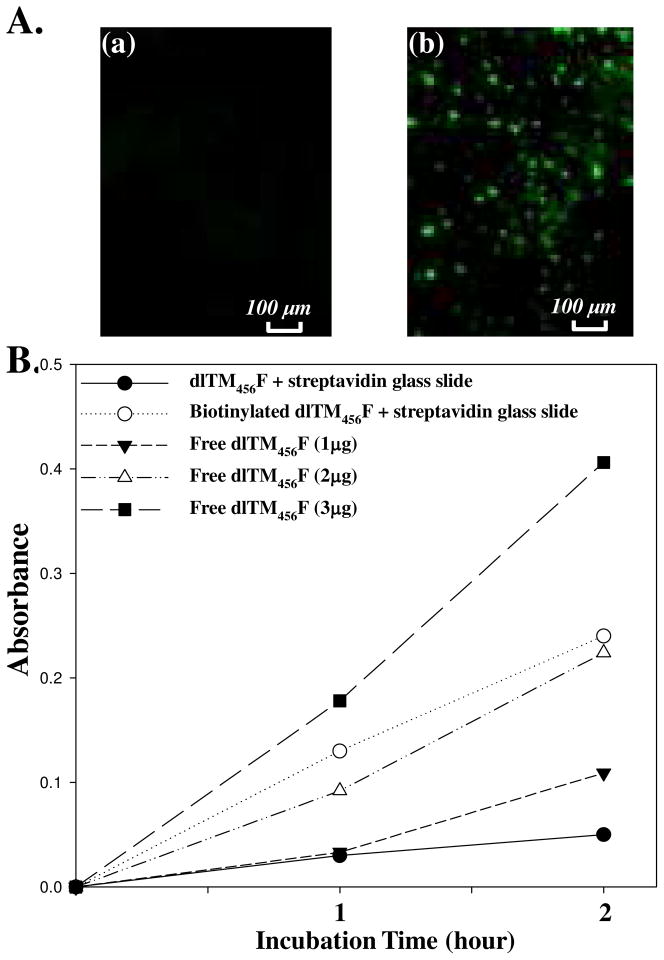
Figure 4.
A. Fluorescence images of Streptavidin-coated glass slides incubated with (a) OG488-labeled dlTM456F and (b) biotinylated OG488-labeled dlTM456F (Ex/Em=480/520). B. Protein C activation assay for (●) streptavidin-glass slide pretreated with dlTM456F in the presence of SrtA; (○) streptavidin-glass slide pretreated with biotinylated dlTM456F in the presence of SrtA; (▼) 1 μg of free dlTM456F; (△)2 μg of free dlTM456F and(■)3 μg of free dlTM456F measured in the same assay condition for immobilized dlTM456F.
Figure 5.
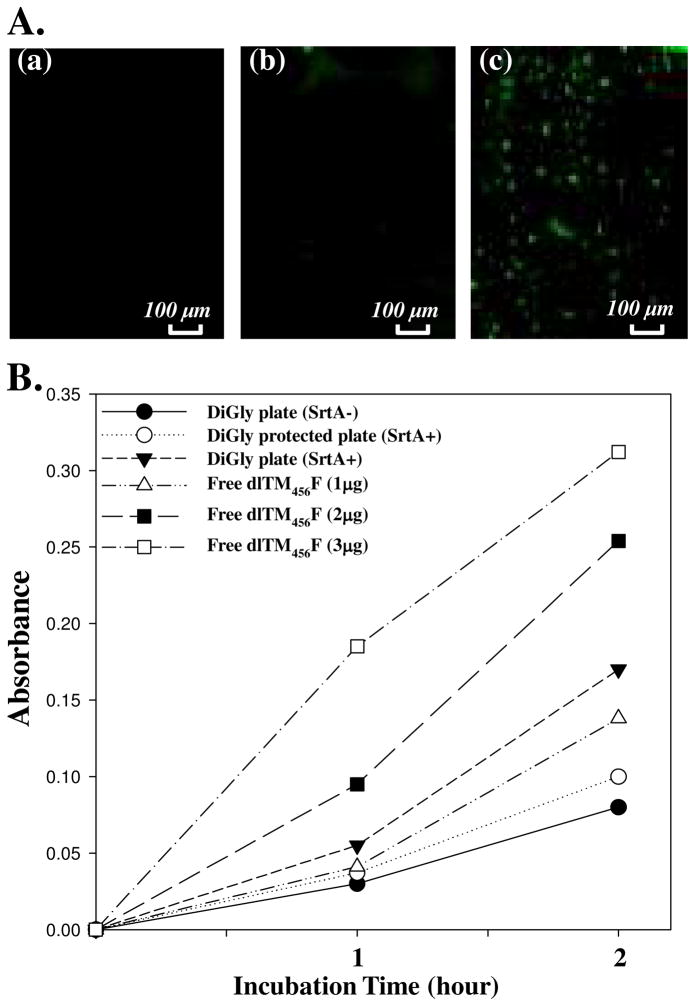
A. Direct immobilization of dlTM456F via SML. OG488-labeled dlTM456F in 20 mM Tris, 150 mM NaCl and 5 mM Ca2+, pH8.0 was incubated with (a) diglycine plate without SrtA; (b) Z-group protected diglycine plate and SrtA, and (c) diglycine plate and SrtA, respectively. OG488 was detected by fluorescence image (EX/EM=480/520). B. Protein C activation assay of (●) diglycine plate pretreated with dlTM456F without SrtA; (○) Z-group protected diglycine plate pretreated with dlTM456F and SrtA; (▼)diglycine plate pretreated with dlTM456F and SrtA; (△)1 μg of free dlTM456F;(■)2 μg of free dlTM456F, and (□) 3 μg of free dlTM456F measured in the same assay condition for immobilized dlTM456F.
Wild type SrtA is a polypeptide of 206 amino acids with an N-terminal membrane-spanning region, therefore the full-length SrtA is difficult to purify in sufficient amounts. However, truncated SrtA protein with 59 N-terminal amino acids deleted (SrtA-ΔN59), which encode outer-membrane region of wild type SrtA, was readily expressed and purified as a soluble protein. SrtA-ΔN59 have been expressed and used in previous studies since it retains the same transpeptidation activity as the full-length SrtA enzyme.15,17 Expression vector pET28b-SrtA was constructed for expression of soluble SrtA with a His6 tag at its C-terminal. The proteins were expressed in E. coli cells and in one step, purified in large amounts. The transpeptidation activity of SrtA was verified by penta-glycine and sortase substrate Abz-Leu-Pro-Glu-Thr-Gly-Lys(Dnp)-NH2 (Anaspec) (Supporting Information).
Site-specific modification of TM456 fusion protein with diGly-biotin and diGly-dansyl via SML
With dlTM456F and SrtA in hand, site-specific modification of dlTM456F with N-terminal oligoglycine-containing molecules via SML was investigated. Although single glycine at the N-terminus of nucleophile is sufficient for sortase-catalyzed transpeptidation, it has been reported that maximum reaction efficiency is generally obtained with substrates in which two or more glycines are incorporated, the addition of any more glycines having no significant effect on the reaction rate15 Therefore, diglycine was chosen as the nucleophile in our study. Two kinds of diglycine-containing molecules, diGly-Biotin and diGly-Dansyl, were prepared for site-specific modification of dlTM456F via SML (Figure 3). DiGly-Biotin and diGly-Dansyl were synthesized in two steps, first amidation of Biotin-ethylenediamine and Dansylethylenediamine with commercially available Z-Gly-Gly-p-nitrophenyl ester (Sigma) followed by removal of the Z-protection group under hydrogenation conditions, respectively (Scheme 1). The resulting compounds were characterized by 1H NMR and FTIR spectroscopy. In the same reaction process, diglycine-functionalized glass slides were also prepared from amine functionalized glass slides (Xenopore, Hawthorne, NJ) for dlTM456F immobilization via SML as depicted below (Scheme 1).
Figure 3.
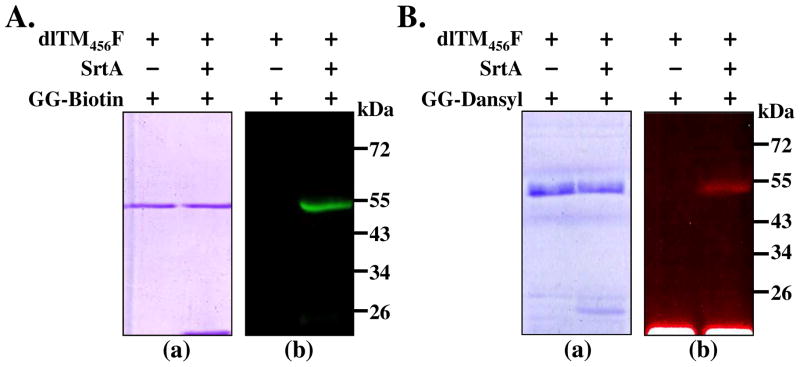
SrtA mediated site-specific biotinylation and dansylation of dlTM456F: A. reactions between dlTM456F (10 μM) and GG-Biotin(100 μM) in the presence or absence of SrtA (5 μM) analyzed by SDS-PAGE (a) and fluorescence scanning imaging of biotinylated dlTM456F confirmed by incubation of SDS-PAGE gel with Streptavidin-Bodipy (EX/EM=480/520) (b); B. Reactions between dlTM456F and GG-Dansyl in the presence or absence of SrtA, Reaction mixtures were analyzed by SDS-PAGE (a) and its photo image for Dansyl-labeled dlTM456F taken under UV light (b).
To investigate whether the LPETG moiety in the dlTM456F was reactive toward diglycine, dlTM456F was mixed with SrtA and diGly-Biotin in the presence of Ca2+ following the conditions mentioned in methods section of this paper. The products were analyzed by SDS-PAGE. Within the gel, it was very difficult to observe the band migration of the biotinylated product due to the minute change in molecular weight of dlTM456F conjugate (less than 1 kDa). Therefore, the gel was incubated with a Bodipy-labeled streptavidin (Sigma) solution, allowing the biotinylated dlTM456F to be visualized by fluorescence scanning imaging, whereas no unbiotinylated dlTM456F was observed in the absence of SrtA in the control reaction (Figure 3Ab). Furthermore, dansylation of the dlTM456F via SML was conducted successfully under the same conditions as biotinylation. Protein sample was separated by electrophoresis, and dansyl labeling was verified by visualizing the fluorescent bioconjugate under UV light (Figure 3Bb). Overall, these results indicated that the LPETG moiety at the C-terminal of dlTM456F can be site-specifically modified with diglycine-containing molecules via SML.
End-point immobilization of TM456 fusion protein via SML
Streptavidin/biotin-based protein immobilization has been widely used for functional surface fabrication.23 In this study, immobilization of biotinylated TM456 onto a streptavidin-covered glass slide surfaces was investigated first. dlTM456F was pre-labeled with Oregon green 488 (OG488) before biotinylation. The reaction mixture containing biotinylated dlTM456F from above was incubated with streptavidin-coated glass slides (Xenopore, Hawthorne, NJ) in PBS (pH 7.4) buffer for 1 h at room temperature. Then the glass slides were removed from the reaction solution and washed with PBS buffer for 1 h to remove unreacted dlTM456F prior to measuring TM activity. The successful immobilization was confirmed by the fluorescence image of OG488 labeled dlTM456F (Figure 4Ab), while no fluorescence image was observed with dlTM456F without biotinylation (Figure 4Aa). Next, the biological activity of the immobilized dlTM456F was accessed via the activation of human protein C. Figure 4B showed the absorbance variation of the chromogenic substrate at 405 nm for different glass slides. In order to compare the relative activity of immobilized dlTM456F with free dlTM456F, activities of various concentrations of free dlTM456F were measured under the same conditions, respectively. As a result, the dlTM456F-immobilized glass slides exhibited a protein C activation-promoting capability comparable to that of 2–3 μg of free dlTM456F in this analysis with the surface areas of the covered glass substrates being ~2 cm2. By contrast, the precursor streptavidin-glass slide, which was pre-incubated with a mixture of dlTM456F without biotinylation, did not enhance the protein C activation to any degree above that of 1 μg of free dlTM456F. These results indicated that biotinylated dlTM456F from a mixture of proteins can be efficiently and selectively immobilized onto streptavidin or avidin coated solid supports. The biotin-mediated immobilization strategy has its advantages and disadvantages. For example, the protein biotinylation reaction is more effective than direct protein-surface reaction due to the small molecule weight of diGly-biotin with a less steric hindrance. However, streptavidin-biotinylated protein interaction might be hindered due to the protein size. Also, streptavidin immobilization is costly and thus is not ideal for practical applications for applications in the future.
Finally, direct site-specific immobilization of dlTM456F onto diglycine derivatized glass slides via SML was investigated. The diglycine functionalized glass slides obtained above was used for the SML. In this experiment, Z-group protected diglycine-glass slides (Figure 5Ab) were used as control. The successful immobilization was confirmed by fluorescence image (Figure 5Ac). The protein C activation activity of immobilized dlTM456F was then further evaluated. The absorbance variation of the cleaved chromogenic substrate at 405 nm is shown in Figure 5B. dlTM456F-immobilized glass slides generated a tremendous amount of APC, compared to the control diGly slides treated without SrtA and Z-protected diglycine glass slides (Figure 5B). The relative activity of dlTM456F immobilized glass slides was approximately that 1–2 μg of free dlTM456F as determined under the same conditions as above. The results confirmed that in the presence of SrtA, dlTM456F can be site-specifically immobilized onto oligoglycine derivatized glass slides directly and exhibit protein C activation-promoting capabilities approximate to that of 1–2 μg of free dlTM456F. Overall, the SrtA-mediated surface ligation was capable of being performed under mild conditions, occurring rapidly in a single step without any prior chemical modification of the target protein. Importantly, the site-specific covalent attachment leads to molecules being arranged in a definitively ordered fashion and thus facilitates protein’s biological activity.
Conclusion
We have designed and expressed a truncated TM mutant consisting of epidermal growth factor-like domains 4–6 (EGF 4–6) with a conserved pentapeptide LPETG motif at its C-terminal for end-point immobilization via SML. The SML provides a convenient and mild approach to the covalent linkage of LEPTG tagged TM to small molecules or solid supports. The TM456 was successfully immobilized onto a diglycine modified glass slide surface by a single-step reaction scheme via SML, and biotinylated TM456 was successfully immobilized via streptavidin-biotin interaction. The sortase-mediated immobilization method presented here has several advantages over traditional methods. First, chemo-and bio-orthogonal immobilization provides a simple and convenient route to uniform protein immobilization in short times creating dense surface coverages. Second, mild immobilization conditions and even low temperatures minimize chances of protein denaturation. Finally, direct immobilization of recombinant proteins from crude cell lysates makes it feasible to fabricate protein arrays in high throughput fashion by eliminating time-consuming and costly purification steps. As TM is a transmembrane protein of the endothelium of blood and lymphatic vessels and the basis of a major natural anticoagulant system, the reported strategies provide an ideal mimetic approach for the engineering of antithrombogenic surfaces for cardiovascular biomaterials minimizing incompatibility and thrombosis.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant 1R01HL102604-2 and Ohio Research Scholar Program and National Science Foundation MRI Grant (CHE-1126384). Thanks to Dr. Dale Ray at Case NMR Center for NMR study.
Footnotes
Supporting Information available: 1H NMR and IR spectra of diGly-biotin and diGly-dansyl, full amino acid sequence of TM456 fusion protein and enzymatic activity assay of recombinant SrtA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wong LS, Khan F, Micklefield J. Selective Covalent Protein Immobilization: Strategies and Applications. Chem Rev. 2009;109:4025–4053. doi: 10.1021/cr8004668. [DOI] [PubMed] [Google Scholar]
- 2.Esmon CT. Thrombomodulin as a model of molecular mechanisms that modulate protease specificity and function at the vessel surface. FASEB J. 1995;9:946–955. doi: 10.1096/fasebj.9.10.7615164. [DOI] [PubMed] [Google Scholar]
- 3.Tseng P, Jordan SW, Sun X-L, Chaikof EL. Catalytic efficiency of a thrombomodulin-functionalized membrane-mimetic film in a flow model. Biomaterials. 2006;27:2768–2775. doi: 10.1016/j.biomaterials.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 4.Kishida A, Ueno Y, Fukudome N, Yashima E, Maruyama I, Akashi M. Immobilization of human thrombomodulin onto poly(ether urethane urea) for developing antithrombogenic blood-contacting materials. Biomaterials. 1994;15:848–852. doi: 10.1016/0142-9612(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 5.Kishida A, Ueno Y, Maruyama I, Akashi M. Immobilization of human thrombomodulin on biomaterials: evaluation of the activity of immobilized human thrombomodulin. Biomaterials. 1994;15:1170–1174. doi: 10.1016/0142-9612(94)90238-0. [DOI] [PubMed] [Google Scholar]
- 6.Kishida A, Ueno Y, Maruyama I, Akashi M. Immobilization of human thrombomodulin onto biomaterials. Comparison of immobilization methods and evaluation of antithrombogenicity. ASAIO J. 1994;40:840–845. doi: 10.1097/00002480-199407000-00116. [DOI] [PubMed] [Google Scholar]
- 7.Vasilets VN, Hermel G, König U, Werner C, Müller M, Simon F, Grundke K, Ikada Y, Jacobasch HJ. Microwave CO2 plasma-initiated vapour phase graft polymerization of acrylic acid onto polytetrafluoroethylene for immobilization of human thrombomodulin. Biomaterials. 1997;18:1139–1145. doi: 10.1016/s0142-9612(97)00045-8. [DOI] [PubMed] [Google Scholar]
- 8.Li JM, Singh MJ, Nelson PR, Hendricks GM, Itani M, Rohrer MJ, Cutler BS. Immobilization of human thrombomodulin to expanded polytetrafluoroethylene. J Surg Res. 2002;105:200–208. doi: 10.1006/jsre.2002.6381. [DOI] [PubMed] [Google Scholar]
- 9.Sperling C, Salchert K, Streller U, Werner C. Covalently immobilized thrombomodulin inhibits coagulation and complement activation of artificial surfaces in vitro. Biomaterials. 2004;25:5101–5113. doi: 10.1016/j.biomaterials.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Han HS, Yang SL, Yeh HY, Lin JC, Wu HL, Shi GY. Studies of a novel human thrombomodulin immobilized substrate: surface characterization and anticoagulation activity evaluation. J Biomater Sci Polym Ed. 2001;12:1075–1089. doi: 10.1163/15685620152691869. [DOI] [PubMed] [Google Scholar]
- 11.Sun X-L, Stabler CL, Cazalis CS, Chaikof EL. Carbohydrate and protein immobilization onto solid surfaces by sequential Diels-Alder and azide-alkyne cycloadditions. Bioconjugate Chem. 2006;17:52–57. doi: 10.1021/bc0502311. [DOI] [PubMed] [Google Scholar]
- 12.Stabler C, Sun X-L, Cui W, Wilson J, Haller C, Chaikof EL. Surface re-engineering of pancreatic islets. Bioconjugate Chem. 2007;18:1713–1715. doi: 10.1021/bc7002814. [DOI] [PubMed] [Google Scholar]
- 13.Proft T. Sortase-mediated protein ligation: an emerging biotechnology tool for protein modification and immobilisation. Biotechnol Lett. 2010;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- 14.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 15.Tsukiji S, Nagamune T. Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. ChemBioChem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 16.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C, Zhu J, Wang Y, Umeda A, Cowmeadow RB, Lai E, Moreno GN, Person MD, Zhang Z. Staphylococcus aureus sortase A exists as a dimeric protein in vitro. Biochemistry. 2007;46:9346–9354. doi: 10.1021/bi700519w. [DOI] [PubMed] [Google Scholar]
- 18.Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci USA. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruger RG, Otvos B, Frankel BA, Bentley M, Dostal P, McCafferty DG. Analysis of the substrate specificity of the Staphylococcus aureus sortase transpeptidase SrtA. Biochemistry. 2004;43:1541–1551. doi: 10.1021/bi035920j. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Adams TE, Nangalia J, Esmon CT, Huntington JA. Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex. Proc Natl Acad Sci USA. 2008;105:4661–4666. doi: 10.1073/pnas.0711055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zushi M, Gomi K, Yamamoto S, Maruyama I, Hayashi T, Suzuki K. The last three consecutive epidermal growth factor-like structures of human thrombomodulin comprise the minimum functional domain for protein C-activating cofactor activity and anticoagulant activity. J Biol Chem. 1989;264:10351–10353. [PubMed] [Google Scholar]
- 22.Fuentes-Prior P, Iwanaga Y, Huber R, Pagila R, Rumennik G, Seto M, Morser J, Light DR, Bode W. Structural basis for the anticoagulant activity of the thrombin-thrombomodulin complex. Nature. 2000;404:518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 23.Rusmini F, Zhong Z, Feijen J. Protein immobilization strategies for protein biochips. Biomacromolecules. 2007;8:1775–1789. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.