Abstract
Hepatocellular carcinoma (HCC) is characterized by a propensity for multifocality, growth by local spread, and dysregulation of multiple signaling pathways. These features may be determined by the tumoral microenvironment. The potential of tumor cells to modulate HCC growth and behavior by secreted proteins has been extensively studied. In contrast the potential for genetic modulation is poorly understood. We investigated the role and involvement of tumor derived nanovesicles capable of altering gene expression, and characterized their ability to modulate cell signaling and biological effects in other cells. We show that HCC cells can produce nanovesicles, exosomes, that differ in both RNA and protein content from their cells of origin. These can be taken up and internalized by other cells, and can transmit a functional transgene. The microRNA content of these exosomes was examined, and a subset that is highly enriched within exosomes was identified. A combinatorial approach to identify potential targets identified transforming growth factor β activated kinase-1 (TAK1) as the most likely candidate pathway that could be modulated by these miRNA. Loss of TAK1 has been implicated in hepatocarcinogenesis and is a biologically plausible target for inter-cellular modulation. We showed that HCC cell derived exosomes can modulate TAK1 expression and associated signaling and enhance transformed cell growth in recipient cells. Conclusion: Exosome mediated miRNA transfer is an important mechanism of inter-cellular communication in HCC cells. These observations identify a unique inter-cellular mechanism that could potentially contribute to local spread, intrahepatic metastases or multifocal growth in HCC.
Keywords: Liver cancer, exosomes, gene expression, TAK1, vesicle
Inter-cellular communications are an essential part of the relationship between cells that enable normal cellular function and maintain tissue homeostasis. Cells use a variety of approaches to communicate with each other such as direct membrane-to-membrane contact, or release of soluble mediators (1). Small vesicles shed from cells have also been shown to play important roles in cell-to-cell communication. These membrane bound vesicles contain membrane proteins similar to those of the donor cell and contain protein and RNA derived from their donor cell cytoplasm (2). They can be taken up and transfer their content to modulate cellular activities in recipient cells. These vesicles have been shown to be secreted into the medium from a variety of normal or tumor cells in culture (5–11). They are also found in biological fluids such as blood, urine and ascites (10, 12–14). Thus, they have the ability to signal and transfer their molecular content within the local microenvironment as well as at a distance.
At least two types of vesicles are recognized based on the size and presumed mechanism of their formation. Exosomes are vesicles with 50–100 nm in diameter secreted from intracellular multivesicular endosomes (3). These vesicles are unrelated to the RNA exosome, an RNA processing intercellular complex. Membrane vesicles which are also referred to as microvesicles, or microparticles have a diameter of 100–1000 nm in diameter and are presumed to form by budding or shedding from plasma membrane (4).
Studies of the functional contributions of these vesicles to inter-cellular communication have focused on understanding the role of their membrane and cytoplasmic protein content. A role in the modulation of immune function has emerged; for example dendritic cells can secrete exosomes that contain immune molecules such as MHC-I, -II which can regulate T-cell responses; Interleukin-15 receptor α and Natural Killer Group 2 member D ligand which can activate NK cells (15–17). The presence of RNA within these vesicles can also enable genetic inter-cellular communication. Studies have showed that mRNA transferred by vesicles from glioma or mast cells can be translated thereby raising the potential for epigenetic modulation by this mechanism (7, 10).
The cellular microenvironment is a critical determinant of tumor progression and development. Hepatocellular cancer (HCC) is characterized by dysregulation of multiple signaling pathways that mediate tumor behavior, local spread and a propensity for multifocal tumor development (18). All of these can potentially be modulated by a maladaptive inter-cellular signal which promotes cellular signaling and responses that enable clonal proliferation, anchorage independent growth and tumor spread. Therefore, epigenetic modulation by inter-cellular signaling could represent an important mechanism contributing to hepatocarcinogenesis. However the involvement and role of this mechanism in HCC is unknown. Understanding the role of these mechanisms and their relevance to HCC offers the potential for new insights into tumor growth and interventions to modulate tumor formation and progression. Thus, we sought to evaluate the ability of HCC cells to release vesicles capable of modulating gene expression, and to investigate their capability to modulate cell signaling and biological effects.
Experimental procedures
Cell lines and culture
Human HCC cell lines, Hep3B, HepG2 and PLC/PRF/5 were obtained from the American Type Culture Collection (Manassas, VA) and cultures were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen Corp., Carlsbad, CA) containing 10% fetal bovine serum and 1% antibiotic-antimycotic (Invitrogen) at 37ºC with 5%CO2. For all studies, vesicle depleted (VD) medium was prepared by centrifuging cell-culture medium at 100,000g over night to spin down any pre-existing vesicular content. Luciferase-expressing PLC/PRF/5 (PLC-luc) generated by stable transfection with phCMV plasmid expressing firefly luciferase cDNA were kindly provided by Dr Ching-Shih Chen (Columbus, OH).
Isolation of cellular nanovesicles
HCC cells (1×106) were plated in 11 ml of VD medium on collagen-coated 10 cm dishes. After 3–4 days, the medium was collected and sequential centrifugation performed (19). The medium was first centrifuged at 300g for 10 min and then at 2,000g for 20 min in 4ºC to remove cells. The supernatant was then centrifuged at 10,000g for 70 min at 4ºC. The supernatant was further ultracentrifuged at 100,000g for 70 min at 4ºC to pellet cellular nanovesicles, which were then washed by resuspending in PBS and ultracentrifuged at 100,000g for 70 min in 4ºC. The final pellet comprising of cellular nanovesicles was used for experiments or resuspended with 50–100 μl of PBS and stored at −80ºC. The protein yield was measured using BCA Protein Assay Kit, (Pierce Biotechnology Inc., Rockford, IL). Electron microscopy was performed using an EM208S transmission electron microscope (Philips, Eindhoven, The Netherlands). Using a particle sizer, ~10% of nanovesicles were noted to range in size between 100μM and 150μM suggesting the presence of large exosome aggregates, large exosomes or microvesicles.
RNA extraction and analysis
Total RNA was extracted from nanovesicles or donor HCC cells using Trizol (Invitrogen, Carlsbad, CA) with an overnight precipitation at −20ºC to increase the yield of RNA. RNA concentration was measured using NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE) and RNA content was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, CA). RNase degradation studies were performed using 100 μg/ml RNase A (Qiagen Inc., Valencia, CA).
Real-time Quantitative RT-PCR
cDNA was transcribed from a total of 600ng of DNase I-treated RNA using the cDNA reverse transcription kit and random primers (Invitrogen, Carlsbad, CA). Real-time quantitative RT-PCR (qRT-PCR) was performed using a Mx3000p System (Stratagene, La Jolla, CA) to detect firefly luciferase (Fluc) mRNA, 18S ribosomal RNA (rRNA) and small nucleolar RNA (snoRNA) U43 with SYBR green I (SYBR® Advantage® qPCR Premix, Clontech Laboratories, Inc., Mountain View, CA). The following PCR primers were used: Fluc primers, forward: 5′-AGGTCTTCCCGACGATGA-3′, reverse: 5′-GTCTTTCCGTGCTCCAAAAC-3′, 18S rRNA primers, forward: 5′-GTAACCCGTTGAACCCCATT-3′, reverse: 5′-CCATCCAATCGGTAGTAGCG-3′, snoRNA U43, forward: 5′-CACAGATGATGAACTTATTGACG-3′, reverse: 5′-CAGAACGTGACAATCAGCAC-3′.
Isolation and detection of protein in cellular vesicles
Hep3B derived nanovesicles were resuspended in 30 μl of Complete Lysis-M buffer (Roche Diagnostics GmbH, Mannheim, Germany) and the lysate was centrifuged at 12,000g for 15 min at 4ºC. 15 μg of protein was mixed with NuPAGE LDS Sample Buffer (Invitrogen, Carlsbad, CA) and separated using NuPAGE Novex 4–12% Bis-Tris Gels (Invitrogen, Carlsbad, CA). The gel was stained with SYPRO Ruby Protein Gel Stain (Molecular Probes, Inc. Eugene, OR) and imaged using the Gel-Doc EQ imaging system (Bio-Rad Laboratories, Hercules, CA). The expression of specific proteins was analyzed by flow cytometry. PLC/PRF/5 derived nanovesicles were conjugated with 4 μm-aldehyde/sulfate latex beads (Invitrogen, Carlsbad, CA, USA), washed in PBS/1% BSA, and stained with primary antibodies against CD63 (Santa Cruz Biotechnology, Santa Cruz, CA), COX-IV (Abcam, Cambridge, MA), Calnexin (Abcam), PMP70 (Abcam) or iso-type controls followed by FITC- or PE-labeled secondary antibodies. Analysis was performed using an Accuri C6 flow cytometer (Accuri Cytometers, Inc., Ann Arbor, MI).
Cellular internalization of Hep3B derived nanovesicles
Hep3B derived nanovesicles were labeled with PKH67 (Sigma-Aldrich, St. Louis, MO) as follows. Two micro liter of PKH67 was added to 25 μg of Hep3B derived nanovesicles in a total 1 ml of diluent and incubated for 15 min at room temperature. A mixture without nanovesicles was used as a control for detecting any carry over of PKH67 dye. Labeling was stopped by adding 1 ml of 1% of BSA and the mixture was added into 18 ml of PBS and was centrifuged at 120,000g for 2 hours in 4ºC. The supernatant was removed and the pellet was resuspended in 20 ml of PBS and centrifuged at 120,000g for 2 hours in 4ºC. The pellet containing PKH67-labeled nanovesicles was resuspended in 2.5 ml of NPD medium. HepG2 cells were cultured in a 4-chamber slide with NPD medium to 80% confluency. The medium was replaced with NPD medium containing PKH67-labeled nanovesicles (0.5 ml per chamber) and cells were incubated for 24 hours in 37ºC, 5%CO2. After incubation, cells were washed twice with PBS and fixed in pure methanol for 10 min in −20ºC. The slide was mounted with ProLong® Gold Antifade Reagents with DAPI (Molecular Probes, Inc., Eugene, OR) and internalization of nanovesicles was examined by fluorescence microscopy.
Transfer of firefly luciferase mRNA by nanovesicles
PLC-luc derived nanovesicles were collected and RNA was isolated. An equivalent amount of RNA from PLC-luc derived nanovesicles or their donor cells was reverse transcribed and qRT-PCR was performed to detect Fluc mRNA. Transfer of Fluc mRNA by nanovesicles was examined by treating PLC/PRF/5 (recipient cells) with PLC-luc derived nanovesicles. PLC/PRF/5 cells were seeded on a 6-well plate with NPD medium and were incubated with 15 μg/ml PLC-luc derived nanovesicles. After 16-hours, recipient cells were washed twice with PBS and RNA was isolated. An equivalent amount of RNA was transcribed to cDNA and Fluc mRNA was detected by qRT-PCR. The luciferase activity in recipient cells was examined using luciferase assay system (Promega corp., Madison, WI). PLC/PRF/5 cells (15,000 cells/well) were plated in 0.1 ml of NPD medium in a 96-well white plate (BD Biosciences, Rockville, MD). After an overnight incubation, the medium was replaced with NPD medium containing various concentrations of PLC-luc derived nanovesicles. After 16-hour incubation the recipient cells were lysed with 20 μl of cell lysis buffer and luminescence in each well was measured using a luminometer (FLUOstar Omega, BMG LABTECH GmbH, Offenburg, Germany) immediately after adding 100 μl of luciferase assay reagent.
MicroRNA profiling by quantitative RT-PCR
Expression profiling of 424 human mature miRNAs was performed using an Applied Biosystems 7900HT real-time PCR instrument equipped with a 384-well reaction plate as previously described (20). Briefly, RNA samples from nanovesicles or donor cells (n = 4 per each cell line) were treated with DNase I (QIAGEN Inc., Valencia, CA). 500 ng of DNase-treated RNA was reverse transcribed using miRNA specific primers (TaqMan® MicroRNA Assays, Applied Biosystems, Foster City, CA). Primers for snoRNA U38B, snoRNA U43, 18S rRNA, and snRNA U6 as internal controls were included in the mix of primers. Real-time PCR was performed and the cycle number at which the reaction crossed a threshold (CT) was determined for each gene. The expression level of miRNAs was evaluated by a comparative CT method using global median normalization. There are no genes that are known to be expressed with the same copy number in both nanovesicle samples and donor cells that could be used as normalization controls. Thus, raw CT values were normalized using a median CT value (ΔCT = CTmiRNA − CTmedian) and the relative amount of each miRNA in nanovesicles relative to donor cells (fold change) was described using the equation 2−ΔΔCT where ΔΔCT =ΔCTnanovesicle − ΔCTdonor cell. For miR-16 expression studies, total RNA was obtained from PLC/PRF/5 cells incubated with GW4869 (Sigma-Aldrich, St. Louis, MO) for 3 days. 5 nM cel-miR-39 (Qiagen) was added as a spike-in control. The RNA was transcribed using miRNA specific stem loop primers (TaqMan® MicroRNA Assays, Applied Biosystems, Foster City, CA) and real-time PCR was performed to detect miR-16 and cel-miR-39. The expression of miR-16 was evaluated by a comparative CT method.
Statistical analysis
Data were analyzed by ANOVA followed by Fisher’s PLSD test. Results were considered to be statistically significant when p < 0.05. Data were expressed as the mean and standard error.
Additional Experimental procedures are described in the supplementary material.
Results
Can tumor cell derived nanovesicles be isolated?
In order to study the potential of tumor cell derived nanovesicles in tumor growth, we first optimized conditions for their isolation. The approach used was based on their differential sedimentation properties and used sequential ultracentrifugation for their isolation from culture supernatant from HCC cells in culture. Electron microscopy showing membrane limited particles that were homogeneous in appearance and ranging from 40–100 nm in size, (Figure 1). By flow cytometry, the isolated particles expressed markers associated with exosomes (CD63), but not those associated with mitochondria (COX IV), peroxisomes (PMP70), or endoplasmic reticulum (calnexin) (Supplementary Figure 1). The yield was confirmed by measuring protein content. The yield (mean ± SE of eight separate isolations) from Hep3B cells was 0.84 ± 0.05 μg/106 cells/day, whereas the yield from PLC/PRF/5 cells was 0.88 ± 0.05 μg/106 cells/day. Thus, these nanovesicles have characteristics of exosomes and could be isolated in a consistent manner.
Figure 1. Characterization of isolated vesicles.

(A) Electron microscopy was performed on a whole mount of particles isolated from PLC/PRF/5 cells following multistage differential centrifugation. A homogeneous population of particles was obtained. (B) Flow cytometry was performed on isolated particles conjugated with 4 μm-aldehyde/sulfate latex beads using primary antibodies to CD63 or isotype controls followed by a phycoerythrin labeled secondary antibodies.
Is the cellular content of exosomes similar to the cells of origin?
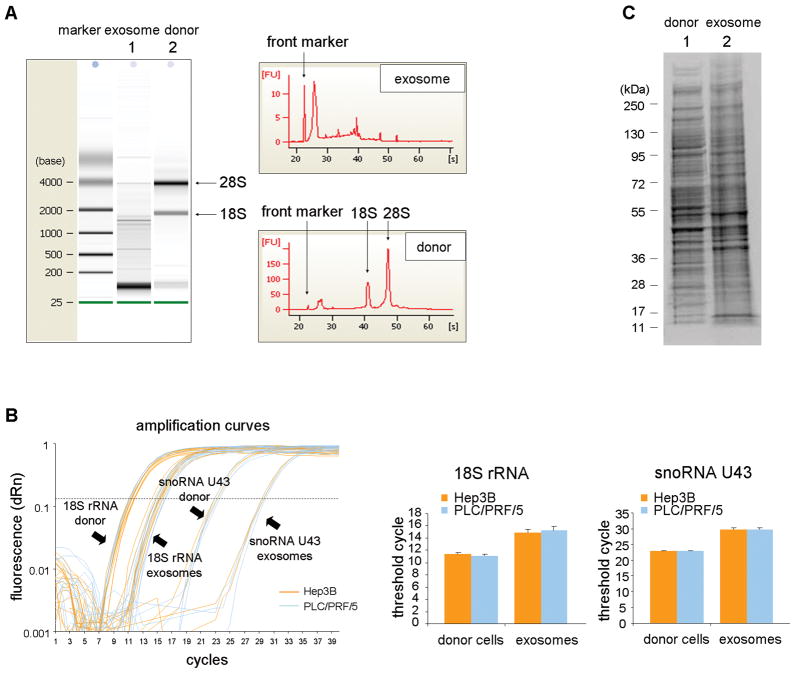
Next, we sought to determine whether the cellular constituents of exosomes were similar to those of the cells of origin. First, we evaluated the profile of total RNA extracted from exosomes by capillary electrophoresis (Figure 2A). Compared to the donor cells, RNA extracted from exosomes did not show clear bands of 18S and 28S ribosomal RNA. However, a distinguishable band was detected below 200 bases suggesting that the RNA content of exosomes is selectively enhanced for small RNAs such as microRNAs. Next, we examined the expression of 18S rRNA and snoRNA U43 by qRT-PCR using equivalent amount of RNA from exosomes and donor cells. These are commonly used as an internal control for small RNA quantification in mammalian cells (Figure 2B). Compared to their expression in either Hep3B or PLC/PRF/5 cells, the expression of these 2 genes was reduced in exosomes derived from these cells. RNA degradation and the yield of RNA obtained was not reduced by RNase treatment compared to controls indicating that the RNA was within the isolated particles (Supplementary figure 2). We next examined the protein expression profile in exosomes. Equivalent amount of proteins extracted from Hep3B-derived exosomes or from their donor cells were separated by SDS-PAGE and stained with SYPRO Ruby (Figure 2C). The protein from exosomes had a different profile showing several distinct bands. Thus, both the RNA and protein content of exosomes is different from that of their cells of origin.
Figure 2. RNA and protein content of cellular exosomes from HCC cells.
(A) RNA was extracted from Hep3B derived exosomes (lane 1) or their corresponding donor cells (lane 2) and analyzed by capillary electrophoresis (Bioanalyzer, Agilent). The RNA content is strikingly different, with the majority of RNA in Hep3B derived exosomes below 2 kb in size and with a very low fraction of 18S ribosomal RNA (rRNA) and 28S rRNA compared to RNA from donor cells. (B) An equivalent amount (600 ng) of RNA from Hep3B or PLC/PRF/5 donor cells, or exosomes obtained from these cells was transcribed to cDNA and the expression of 18S ribosomal RNA (18S rRNA) and small nucleolar RNA U43 (snoRNA U43) examined by real-time PCR. Amplification curves and the mean value ± SEM from four independent samples of threshold cycles for 18S rRNA and snoRNA U43 are shown. Both 18S rRNA and snoRNA U43 show higher CT values in exosomes than in their corresponding donor cells. (C) Protein was isolated from Hep3B derived exosomes or their donor cells and 15 μg of protein was separated on a Bis-Tris gel and stained with SYPRO Ruby (lane 1, donor cells; lane 2, exosomes). The protein content in exosomes was different from that in their corresponding donor cells.
Can cellular exosomes be taken up and internalized by other cells?
To examine the potential for uptake and internalization by other cells, we labeled exosomes derived from Hep3B with the fluorescent dye PKH67 as described in the Methods section. PKH67-labeled exosomes were incubated with HepG2 cells for 24 hours and localization of exosomes was examined by fluorescent microscopy (Figure 3). We observed internalization of PKH67-labeled exosomes as endosome-like vesicles in the cytoplasm of HepG2 cells. These studies indicate that tumor cell derived exosomes can be taken up by other cells.
Figure 3. Internalization of Hep3B-derived exosomes into HepG2 cells.
HepG2 cells in culture were incubated with Hep3B-derived exosomes that were labeled with PKH67 (green). HepG2 cells were also incubated with PKH67 dye without exosomes as a control to detecting any carry-over of the dye (exosomes (−)). Cells are fixed with cold methanol at −20ºC and mounted with ProLong® Gold Antifade Reagent with DAPI as described in Experimental procedures. High magnification images of HepG2 cells incubated with exosomes (A–C), or low magnification images of HepG2 cells incubated with exosomes (D–F), or controls without PKH67 (G–I) are shown. Hep3B derived exosomes are shown to be internalized into the cytoplasm of HepG2 cells.
Can exosomes transfer a functional transgene to other HCC cells?
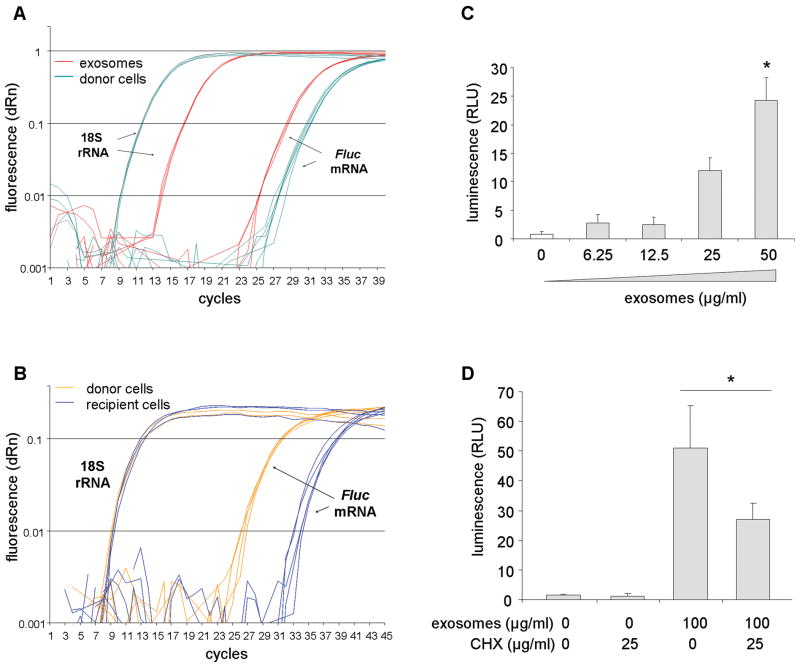
We examined whether exosomes can deliver functional mRNA of a transgene to other HCC cells. Exosomes were collected from PLC-luc cells, which are stably transfected to express a functional luciferase expressing construct. The presence of Fluc mRNA in exosome was verified by qRT-PCR; CT values (mean ± SE) of Fluc mRNA were 29.0 ± 0.1 in exosomes and 31.5 ± 0.2 in donor cells (Figure 4A). PLC/PRF/5 cells were then incubated with various concentrations of PLC-luc-derived exosomes for 16 hours. Fluc mRNA expression and luciferase activity was then assessed in the recipient PLC/PRF/5 cells. We detected Fluc mRNA in the recipient PLC/PRF/5 cells with ΔCT values (CTFluc mRNA − CT18S rRNA, mean ± SE) of 24.7 ± 0.2 compared to 18.0 ± 0.1 in donor PLC-luc cells (Figure 4B). In addition, a concentration-dependent increase in luciferase activity was detected in recipient cells incubated with PLC-luc derived exosomes consistent with a gene-dosing effect (Figure 4C). A reduction of luciferase activity was noted with PLC-luc derived exosomes incubated in recipient cells pre-treated with cycloheximide compared to controls, indicating a requirement for new protein translation for luciferase activity (Figure 4D). These data show that exosomes can deliver a functionally active Fluc mRNA to other cells.
Figure 4. Cell-to-cell transfer of firefly luciferase by exosomes.
(A) Real-tine PCR was performed on an cDNA transcribed from an equivalent amount (600 ng) of RNA from PLC-luc derived exosomes or their donor cells (n = 3, each in duplicate). PCR amplification curves for Fluc mRNA and 18S rRNA are shown. (B) PLC/PRF/5 cells were incubated with 15 μg/ml of PLC-luc derived exosomes for 16 hours. RNA was isolated and equivalent amount of RNA (300 ng) was transcribed to cDNA (n = 3). Amplification curves by quantitative real-time PCR for Fluc mRNA and 18S rRNA in PLC/PRF/5 (recipient cells) and PLC-luc (donor cells) are shown. (C) PLC/PRF/5 cells in a 96-well plate were incubated with various concentrations of PLC-luc derived exosomes, and luciferase activity was assessed in these cells after 16 hours. Bars express the mean value of luminescence ± SEM of four separate determinations. *, p < 0.05. (D) PLC/PRF/5 cells were pretreated with 25 μg/ml of cycloheximide for 2 hours to inhibit de novo protein synthesis. Cells were washed with PBS and incubated with 100 μg/ml of PLC-luc derived exosomes. Luciferase activity was assessed in these cells after 6 hours. Bars express the mean value of luminescence ± SEM of five separate determinations. *, p < 0.05.
Do exosomes contain microRNAs?
Since HCC cell-derived exosomes contain an enriched fraction of small RNAs (Figure 1), we hypothesized that exosomes contain selected miRNAs that could contribute to inter-cellular communication. To examine this possibility, we performed microRNA expression profiling in both Hep3B and PLC/PRF/5 HCC cells and exosomes derived from these cells. Four independent samples were used for each cell line/exosome pair. The expression of total 424 miRNAs and the internal control genes (18S rRNA, snRNA U6, snoRNA U38B and snoRNA U43) were measured by qRT-PCR. The expression level of individual miRNAs in exosomes and donor cells were expressed as the relative expression to global median expression of all miRNA since no internal control genes are available for exosomes. The raw CT values of 18S rRNA and snoRNA U43 vary between donor cells and their exosomes (Figure 2B), and the ability for spiked controls to be expressed in exosomes is unknown.
Of the miRNAs examined, only 134 miRNAs were identified in exosomes isolated from Hep3B cells. Of these, 55 miRNAs were differentially expressed in exosomes more than 4-fold compared to their expression in their donor cells; 25 miRNAs of those were enriched (up to 166-fold) and 30 miRNAs were decreased (up to 113-fold). Notably, 11 miRNAs were detected exclusively in exosomes indicating a very high enrichment in exosomes compared to donor cells (Figure 5A). Similar observations were made in PLC/PRF/5-derived exosomes. 140 miRNAs were identified in PLC/PRF/5-derived exosomes of which 74 miRNAs were differentially distributed in exosomes more than 4-fold compared to the donor cells. Of these, 28 miRNAs were enriched (up to 71-fold), with 20 miRNA detected exclusively in exosomes and 45 miRNAs were decreased (up to 255-fold). There was a moderate correlation in levels of miRNAs contained in exosomes isolated from Hep3B and PLC/PRF/5 (Figure 5B), indicating the existence of a common mechanism of selective enrichment of exosomes with specific miRNAs. The detailed lists of miRNAs and expression levels are available in Supplemental table 1.
Figure 5. Identification of microRNAs in HCC derived exosomes.
Profiling of miRNAs in exosomes or their donor cells was performed from four independent replicates as described in Experimental procedures. (A) The mean values of fold change of expression of miRNA detected in exosomes relative to that in donor cells is shown (n = 4) and the numbers of miRNA that were exclusively detected in either donor cells or exosomes are depicted. 134 miRNAs were identified in Hep3B-derived exosomes, and 140 miRNAs in PLC/PRF/5 derived exosomes. Some miRNAs were predominantly expressed in exosomes compared to their donor cells. (B) Correlation of miRNA expression in Hep3B derived exosomes and in PLC/PRF/5 derived exosomes are shown. A total of 97 miRNAs were detected in exosomes from both Hep3B and PLC/PRF/5. (C) PLC/PRF/5 cells were incubated for 3 days with GW4869, a neutral sphingomyelinase inhibitor which can inhibit ceramide biosynthesis. The cellular expression of miR-16 in donor cells was unchanged whereas expression of miR-16 in exosomes obtained from these cells was reduced following incubation with GW4869 compared to controls. Thus, miRNA release into exosomes occurs via a ceramide dependent pathway. Bars express mean ± SEM from three independent experiments. *, p < 0.05.
The release of miRNA into membrane vesicles can occur via a ceramide dependent manner (21). To evaluate the potential role of this pathway, we treated cells with an nSMase inhibitor, GW4869, which is known to inhibit ceramide biosynthesis and examined the expression of miR-16, a microRNA that is expressed in both donor cells and in exosomes. The cellular expression of miR-16 was unchanged whereas the extracellular expression of miR-16 in exosomes was reduced following incubation with GW4869 compared to controls. Thus, miRNA release into exosomes occurs via a ceramide dependent pathway.
What are potential targets of miRNA enriched in exosomes?
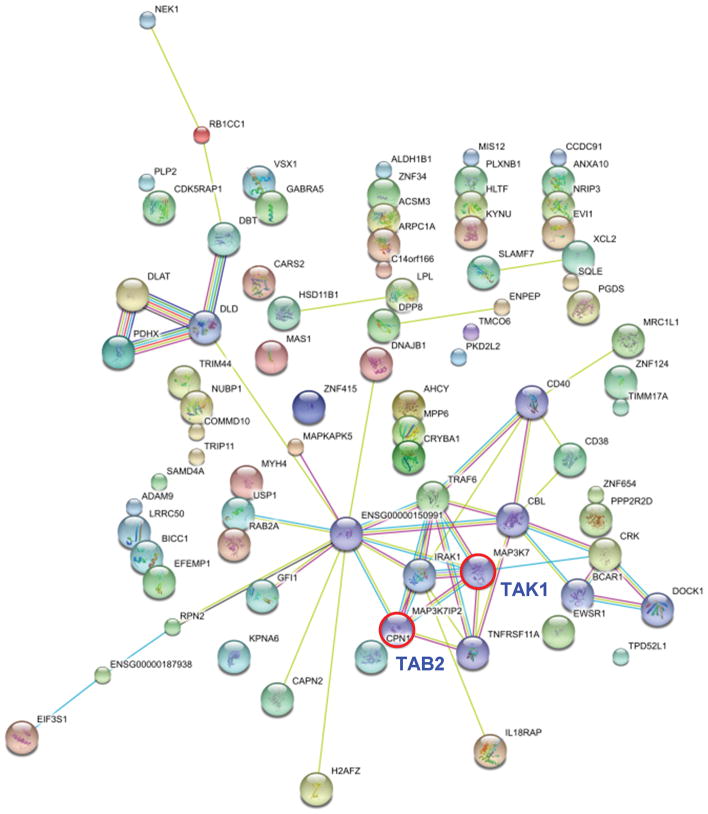
To identify potential roles of exosome-derived miRNA in cell to cell communication, we focused on the 11 miRNAs that were exclusively detected, and hence highly enriched, in Hep3B-derived exosomes (Table 1). We postulated that these miRNA would coordinate regulation of a set of genes, and used a combinatorial approach to analyze potential targets of this set of miRNA in combination. Using the miRror algorithm which integrates data from a dozen miRNA target prediction programs (22) (http://www.proto.cs.huji.ac.il/mirror/), 108 genes were predicted as the targets of these 11 miRNAs (Supplemental table 2). Network analysis of these 108 genes using the String 8.3 program (23) indicated the central involvement of transforming growth factor-β activated kinase-1 (TAK1) signaling (Figure 6). This analysis predicted the TAK1 pathway as the most likely candidate pathway that would be modulated by the selected group of miRNA acting in concert. TAK1 is an upstream member of the mitogen-activated protein kinase kinase kinase (MAP3K) family and has been implicated as an essential component of cellular homeostasis and tumorigenesis in the liver (24, 25).
Table 1.
microRNAs expressed exclusively in exosomes derived from Hep3B human HCC cells.
| microRNA | Expression indexξ |
|---|---|
| miR-584 | 165.8 |
| miR-517c | 39.8 |
| miR-378 | 38.2 |
| miR-520f | 36.4 |
| miR-142-5p | 20.3 |
| miR-451 | 18.4 |
| miR-518d | 13.1 |
| miR-215 | 12.4 |
| miR-376a* | 12.4 |
| miR-133b | 8.3 |
| miR-367 | 7.4 |
miRNA expression in exosomes calculated relative to expression in Hep3B donor cells using an assigned CT value of 40 for donor cell expression.
Figure 6. Bioinformatics analysis of miRNA target genes.
The set of target genes that could be regulated by the 11 microRNAs predominantly expressed in Hep3B derived exosomes were analyzed using the miRror program. 108 genes were predicted by combinatorial analysis of these microRNAs. Network analysis of these genes using String 8.3 software indicated the central involvement of TAK1 signaling.
Can exosomes modulate TAK1 expression and signaling in recipient cells?
The involvement of TAK1 in cell responses to environmental changes and a demonstrated role in HCC formation and growth make this kinase a highly attractive and biologically plausible target for inter-cellular modulation (26). TAK1 can be activated by cytokines and stress stimuli such as transforming growth factor (TGF)-β, tumor necrosis factor (TNF)-α, IL-1β and lipopolysaccharides. TAK1 forms a complex with TNF receptor-associated factor and TAK-1 binding protein (TAB)1, -2, and -3, which is necessary for the recruitment and activation of TAK1. TAK1 can subsequently activate c-Jun NH2-terminal kinase (JNK)/p38 MAPK and nuclear factor (NF)-κB. To examine the potential of exosomes to modulate the TAK1 pathway, Hep3B cells were incubated with Hep3B-derived exosomes (10 μg/ml). The expression of TAK1 and TAB2 proteins and the activation by phosphorylation of downstream JNK and p38 MAP kinases was examined by immunoblot analysis (Figure 7). A reduction in TAK1 of upto 45% and of TAB2 of upto 42% was noted after 72 hours. Concomitantly, a decrease in constitutive active-site phosphorylation of JNK1, JNK2/3 and p38 MAPK were also noted after 72 hours. Thus, exosomes can modulate the constitutive expression of TAK1 and modulate downstream signaling associated with TAK1.
Figure 7. Modulation of TAK1 expression and downstream signaling by exosomes.
Hep3B cells were incubated with 10 μg/ml of exosomes for 24–72 hours in VD medium. Cell were lysed, and an equal amount of protein from each sample was examined by immunoblotting using specific antibodies against TAK1, TAB2, phosphorylated (p-) JNK, total JNK, p-p38 or total p38. (A) A representative immunoblot is shown along with (B) quantitative data by densitometry from three independent experiments. Expression of phosphorylated protein bands are reported as ratio of phosphorylated protein bands to total protein bands. Data represent the mean ± SEM. *, p < 0.05.
Can exosomes modulate transformed cell growth or cell death in target cells?
We began by assessing the effect of exosomes on anchorage-independent growth, a hallmark of transformed cell behavior. HCC cells were seeded in soft agar in the presence or absence of Hep3B derived exosomes and allowed to grow with or without Hep3B-derived exosomes for 7 days. Incubation with Hep3B-derived exosomes increased the number of colonies in soft agar 9.6-fold in Hep3B cells and 1.6-fold in HepG2 cells (Figure 8A) after 7 days. These studies indicate a cell-type dependent yet consistent effect of exosomes on transformed cell growth in vitro. A loss of Tak1 has been associated with spontaneous hepatocyte death. Thus, we determined the effect of exosomes on cell death. Incubation of Hep3B cells with Hep3B-derived exosomes (10 μg/ml) for 24 hrs increased caspase-3/7 activity by 6.1-fold after 24 hours (Figure 8B). This was associated with a 20.5% reduction in cell viability after 72 hours in Hep3B cells incubated with 10 μg/ml Hep3B derived exosomes (Figure 8C). Similar effects were also noted in PLC/PRF/5 cells incubated with Hep3B-derived exosomes. The enhancement in anchorage independent growth in the setting of this modest reduction in cell viability indicate that exosomes have a potent overall effect on transformed cell behavior.
Figure 8. Effect of exosomes on HCC cell behavior.
(A) Transformed cell growth was assessed by determining anchorage-independent growth in soft agar. Cells in culture medium containing 0.4% agarose with or without 15 μg of Hep3B-derived exosomes were plated in a 96-well plate (1000 cells per well) over a base agar layer consisted of culture medium containing 0.6% agarose. After incubation for 7 days, the number of colonies was evaluated fluorometrically and was expressed as fluorescence relative to that in controls without exosomes. Bars represent the mean ± SEM of six separate determinations. *, p < 0.05. (B) Induction of caspase-3/7 activation. Hep3B cells were plated in a 96-well plate (10,000 cells per well) and treated with various concentrations of Hep3B derived exosomes for 24 hours. Caspase-3/7 activity was assessed using a commercial luminometric assay. The data was expressed as relative value of luminescence to control without exosomes. Bars represent the mean ± SEM of three independent experiments. *, p < 0.05. (C) Cell viability assay. Cells (5,000 cells per well) were plated in a 96-well plate with VD medium and incubated with varying concentrations of Hep3B-derived exosomes for 72 hours. Cell viability was assessed using an MTS assay and was expressed as a percentage of control without exosomes. Bars represent the mean ± SEM of 6 separate determinations. *, p < 0.05.
Discussion
Although the role of genetic alterations in oncogenes and tumor suppressor genes has been extensively studied, epigenetic mechanisms contributing to tumor development are less well characterized. The influence of the cellular microenvironment on tumor development and growth is becoming increasingly recognized. In these studies, we show that HCC cells can epigenetically modulate gene expression and cell signaling related to transformed cell growth in vitro through the release of miRNA contained within exosomes. Moreover, we identify selective enhancement of miRNA content within these cellular vesicles, and the potential of exosomal miRNA transfer to effect cellular gene expression and cell behavior in target cells. The significance of these studies is that they identify a mechanism of cell-to-cell communication that may have widespread effects on tissue behavior. The potential of the cellular microenvironment to modulate gene expression and to stimulate cell signaling contributing to tumor behavior could be exploited for therapeutic targeting.
A selective subset of miRNA is present within exosomes which is markedly different from that in their cells of origin. Mature miRNA are released into the cytoplasmic space where they associate with the RISC complex to effect gene expression. The selective enrichment of these miRNA in cellular exosomes, the consistency in expression between different isolations, and the cell-type specificity indicates the presence of a mechanism for their active elaboration within these particles. This may arise from selective transport into a membrane bound exosome, or association and sequestration with proteins that are selectively enriched within the exosome. Alternatively, the possibility exists that these miRNA are rapidly degraded within the cytoplasm but protected from this when they are sequestered in vesicles prior to their elaboration from cells as cellular exosomes. Since the tumor cells studied vary in their cellular behavior, it is not unexpected that some differences were noted between the cell types in miRNA content. These observations regarding cell-type specificity of miRNA content are similar to those made with respect to protein content of exosomes. They emphasize the need to study and interpret data on an individual cell-type specific basis rather than generalizing across cell types.
It is noteworthy that our combinatorial analysis identified TAK1 as a central target of exosome mediated miRNA. TAK1 is an essential inhibitor of hepatocarcinogenesis, and its absence in vivo is associated with the spontaneous development of hepatocellular cancer related to aberrant responses to inflammatory and stress signaling, and associated with hepatocyte injury and apoptosis. TAK1 can also have a direct effect on cancer progression through repression of the telomerase reverse transcriptase gene (27). Thus, the modulation of TAK1 expression and signaling by exosome mediated inter-cellular signaling could represent an important mechanism of tumor progression that may not be dependent on clonal expansion or hyperproliferative effects.
Influences from cellular elements within the tumoral microenvironment such as tumor-associated fibroblasts or soluble mediators such as cytokines, hormones or neurotransmitters are the focus of intense study to understand the influence of this space on tumor behavior. While the autocrine or paracrine effects of proteins that are secreted into the extracellular space has been extensively studied, the involvement of genetic modulation by miRNA adds another layer of complexity and regulation that may be equally as important. We speculate that environmentally mediated epigenetic modulation is likely to be an important determinant of tumor cell growth, and cellular resistance to environmental perturbations.
Inter-cellular communication by exosomes can trigger signal transducing intra-cellular mechanisms that can modulate cell behavior. The potential role of these may be to coordinate cellular responses that are adaptive and can serve to maintain tissue homeostasis. Alternatively, in tumor cells, exosome-mediated intercellular signaling may serve a maladaptive role to promote tumor growth. The current studies demonstrate the potential for tumor cells themselves to exert paracrine or autocrine effects mediated by miRNA as extracellular effectors of cell-to-cell communication. The aberrant expression of selected miRNA in tumor cells and their ability to modulate multiple oncogenic or tumor suppressor genes make them well-suited for such a role. Therefore further studies to understand the mechanisms of cellular exosome production, selective enrichment of RNA genes and other genetic content, and cell-type specificity of response are justified to establish the roles of this mechanism in the development and progression of HCC.
Supplementary Material
Acknowledgments
Financial support
Supported in part by Grant DK069370 from the National Institutes of Health
Abbreviations
- CNV
cellular nanovesicles
- HCC
hepatocellular carcinoma
- miRNA
microRNA
- VD
vesicle-depleted
- TAB
TAK-1 binding protein
- TAK1
transforming growth factor β activated kinase-1
Contributor Information
Takayuki Kogure, Email: kogure.takayuki@mayo.edu.
Wen-Lang Lin, Email: lin.wenlang@mayo.edu.
Irene K. Yan, Email: yan.irene@mayo.edu.
Chiara Braconi, Email: chiara.braconi@osumc.edu.
Tushar Patel, Email: patel.tushar@mayo.edu.
References
- 1.Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci U S A. 2007;104:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 3.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 4.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Saunderson SC, Schuberth PC, Dunn AC, Miller L, Hock BD, MacKay PA, Koch N, et al. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J Immunol. 2008;180:8146–8152. doi: 10.4049/jimmunol.180.12.8146. [DOI] [PubMed] [Google Scholar]
- 6.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 7.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 8.Mallegol J, Van Niel G, Lebreton C, Lepelletier Y, Candalh C, Dugave C, Heath JK, et al. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology. 2007;132:1866–1876. doi: 10.1053/j.gastro.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 9.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutwein P, Stoeck A, Riedle S, Gast D, Runz S, Condon TP, Marme A, et al. Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Clin Cancer Res. 2005;11:2492–2501. doi: 10.1158/1078-0432.CCR-04-1688. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. 2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Cheruvanky A, Hu X, Matsumoto T, Hiramatsu N, Cho ME, Berger A, et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–621. doi: 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 15.Utsugi-Kobukai S, Fujimaki H, Hotta C, Nakazawa M, Minami M. MHC class I-mediated exogenous antigen presentation by exosomes secreted from immature and mature bone marrow derived dendritic cells. Immunol Lett. 2003;89:125–131. doi: 10.1016/s0165-2478(03)00128-7. [DOI] [PubMed] [Google Scholar]
- 16.Luketic L, Delanghe J, Sobol PT, Yang P, Frotten E, Mossman KL, Gauldie J, et al. Antigen presentation by exosomes released from peptide-pulsed dendritic cells is not suppressed by the presence of active CTL. J Immunol. 2007;179:5024–5032. doi: 10.4049/jimmunol.179.8.5024. [DOI] [PubMed] [Google Scholar]
- 17.Viaud S, Terme M, Flament C, Taieb J, Andre F, Novault S, Escudier B, et al. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: a role for NKG2D ligands and IL-15Ralpha. PLoS One. 2009;4:e4942. doi: 10.1371/journal.pone.0004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 19.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, Schmittgen TD, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010 Jun 4;285(23):17442–52. doi: 10.1074/jbc.M110.107821. Epub 2010 Mar 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman Y, Naamati G, Linial M. MiRror: a combinatorial analysis web tool for ensembles of microRNAs and their targets. Bioinformatics. 2010;26:1920–1921. doi: 10.1093/bioinformatics/btq298. [DOI] [PubMed] [Google Scholar]
- 23.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettermann K, Vucur M, Haybaeck J, Koppe C, Janssen J, Heymann F, Weber A, et al. TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell. 2010;17:481–496. doi: 10.1016/j.ccr.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Inokuchi S, Aoyama T, Miura K, Osterreicher CH, Kodama Y, Miyai K, Akira S, et al. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc Natl Acad Sci U S A. 2010;107:844–849. doi: 10.1073/pnas.0909781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malato Y, Willenbring H. The MAP3K TAK1: a chock block to liver cancer formation. Hepatology. 2010;52:1506–1509. doi: 10.1002/hep.23878. [DOI] [PubMed] [Google Scholar]
- 27.Fujiki T, Miura T, Maura M, Shiraishi H, Nishimura S, Imada Y, Uehara N, et al. TAK1 represses transcription of the human telomerase reverse transcriptase gene. Oncogene. 2007;26:5258–5266. doi: 10.1038/sj.onc.1210331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.