Abstract
Sirtuins catalyze the NAD+-dependent deacetylation of target proteins, which are regulated by this reversible lysine modification. During deacetylation, the glycosidic bond of the nicotinamide ribose is cleaved to yield nicotinamide and the ribose accepts the acetyl group from substrate to produce O-acetyl-ADP-ribose (OAADPr), which exists as an ∼50:50 mixture of 2’ and 3’ isomers at neutral pH. Discovery of this metabolite has fueled the idea that OAADPr may play an important role in the biology associated with sirtuins, acting as a signaling molecule and/or an important substrate for downstream enzymatic processes. Evidence for OAADPr-metabolizing enzymes indicates that at least three distinct activities exist that could modulate the cellular levels of this NAD+-derived metabolite. In Saccromyces cerevisiae, NUDIX hydrolase Ysa1 cleaves OAADPr to AMP and 2- and 3-O-acetylribose-5-phosphate, lowering the cellular levels of OAADPr. A buildup of OAADPr and ADPr has been linked to a metabolic shift that lowers endogenous reactive oxygen species and diverts glucose towards preventing oxidative damage. In vitro, the mammalian enzyme ARH3 hydrolyzes OAADPr to acetate and ADPr. A third nuclear-localized activity appears to utilize OAADPr to transfer the acetyl-group to another small molecule, whose identity remains unknown. Recent studies suggest that OAADPr may regulate gene silencing by facilitating the assembly and loading of the Sir2-4 silencing complex onto nucleosomes. In mammalian cells, the Trpm2 cation channel is gated by both OAADPr and ADP-ribose. Binding is mediated by the NUDIX homology (NudT9H) domain found within the intracellular portion of the channel. OAADPr is capable of binding the Macro domain of splice variants from histone protein MacroH2A, which is highly enriched at heterochromatic regions. With recently developed tools, the pace of new discoveries of OAADPr-dependent processes should facilitate new molecular insight into the diverse biological processes modulated by sirtuins.
Introduction
Sirtuins are a conserved family of protein/histone deacetylases found in organisms ranging from bacteria to humans [1–3]. Members of this family share a similar enzymatic core domain of ∼250 amino acids. The founding member Sir2 (Silencing Information Regulator 2) was originally discovered as a gene required for efficient mating in Sacchromyces cerevisiae [4]. Yeast Sir2p is involved in many processes that include gene silencing, ribosomal DNA recombination, DNA repair and longevity[5]. In higher organisms, Sir2 orthologs are also linked to lifespan regulation [6, 7]. Expressing extra copies of Sir2 homologues leads to apparent lifespan extension in both C. elegans and D. melanoganster[2]. Although it is not yet clear if Sir2 homologs modulate lifespan in mammals, mammalian sirtuins are reported to induce positive effects on physiological processes, such as DNA repair, cell survival, stress resistance, metabolic control and insulin sensitivity [2]. Moreover, sirtuins may mediate the beneficial effects of caloric restriction (CR) [7–9], which is the only non-genetic method that extends lifespan in nearly all organisms where this regimen has been investigated.
Discovery of OAADPr
Once yeast Sir2 was identified as an essential factor in gene silencing [4, 10, 11], it was noted that Sir2 shared sequence homology to a bacterial enzyme, CobB that could partially complement a phosphoribosyltransferase defect in cobalamin biosynthesis [12–14]. Initial investigations of Sir2 enzymatic function reported a protein ADP-ribosylation activity, which required NAD+ [15, 16]. However, a number of subsequent reports began to reveal a more robust activity, NAD+-dependent histone deacetylation [17–19]. In 2000, the surprising molecular role of NAD+ in protein deacetylation was revealed [20, 21]. The efficient histone/protein deacetylase reaction is tightly coupled to the formation of a novel acetyl-ADP ribose product O-acetyl-ADP ribose (OAADPr, also abbreviated as AAR and AADPR by some authors). One molecule of NAD+ and acetyl-lysine are readily converted to one molecule of deacetylated lysine, nicotinamide, and OAADPr (Figure 1). The details of the chemical reaction have been reported [22, 23], and will not be reviewed here. However, the idea that some sirtuins mediate protein ADP-ribsoylation, and not deacetylation, remains active. The potential ADP-ribosylating activity of sirtuins stems primarily from two main observations: Some sirtuins exhibit little or no detectable protein deacetylase activity on tested substrates, and low levels of ADPr transfer to protein are often detected under extremely long incubations with NAD+. Several published studies have provided evidence that the reported ADP-ribosylation activity of sirtuins represents a slow side reaction that may not reflect an important physiological function of these enzymes [24–28].
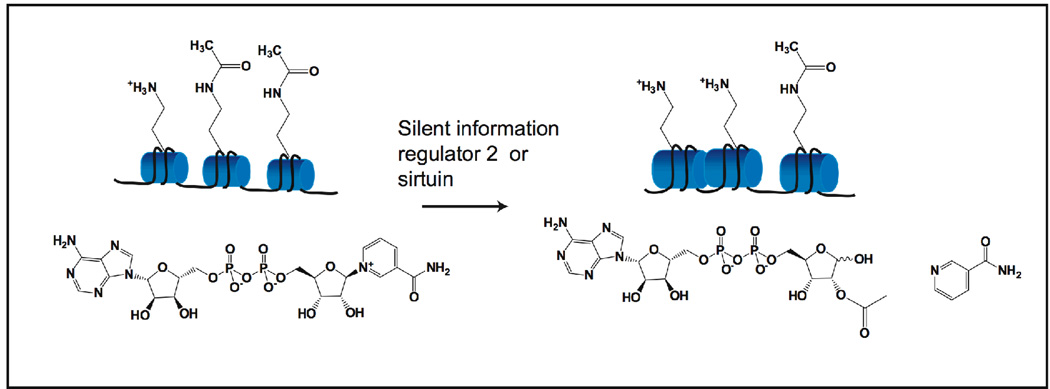
Figure 1. OAADPr production by Sir2/sirtuins deacetylation reaction.
Sir2 and sirtuins catalyzes NAD+ dependent deacetylation of histone tails, or other non-histone acetylated proteins. The reaction transfers the acetyl group from acetylated lysine residues to the ADP-ribose moiety of NAD+, generating deacetylated histone tails, nicotinamide, and the novel metabolite OAADPr.
From several mechanistic studies, the deacetylation product released by sirtuins appears to be the 2'-O-acetyl-ADP-ribose isomer of OAADPr [21, 23, 29]. At neutral pH, 2'-O-acetyl-ADP-ribose and 3'-O-acetyl-ADP-ribose were found to be the solution products, existing in ∼1:1 ratio and generated through a non-enzymatic intramolecular transesterification [23, 29], (Figure 1). The ability of sirtuins to produce OAADPr is well conserved ranging from bacterial CobB to mammalian sirtuins ([3, 19, 27, 30]). It is important to note that the energy released by hydrolysis of NAD+ is 8.2 kcal/mol, in a comparable range for the hydrolysis of ATP to ADP[15, 31]. It is striking that Sir2 enzymes (Class III protein deacetylases) retain this energetically expensive mechanism, compared to the Class I and II deacetylases, which directly produce acetate and do not require NAD+. There is accumulating evidence that the elaborate mechanism of sirtuin catalysis renders OAADPr which can elicit downstream responses that might synergize or antagonize the biological functions of sirtuin genes. In the following sections, we will discuss several studies published over the last few years focusing on the metabolism and cellular functions of OAADPr.
OAADPr metabolism
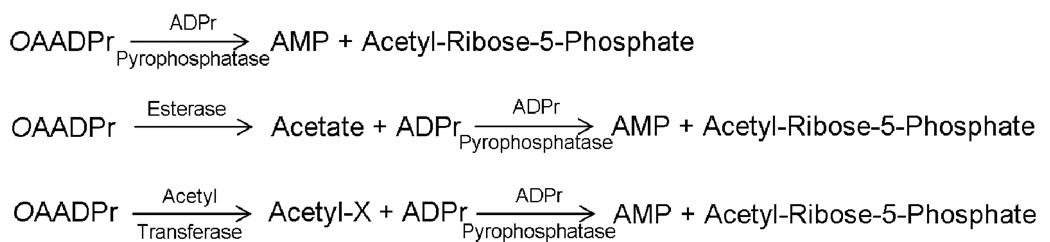
After the discovery of OAADPr, a quantitative microinjection assay of OAADPr into starfish oocytes caused a delay/block in oocyte maturation[32], supporting the hypothesis that OAADPr can evoke biological activity. It is worth noting that microinjection of purified sirtuin enzymes also led to the identical effect. Although the molecular basis behind these observations is unknown, it was reasonable to postulate that the metabolism of OAADPr in vivo might be tightly controlled. Several OAADPr-metabolizing enzymes have been reported (Figure 2). The best characterized are the NUDIX hydrolases (hydrolysis of a nucleoside diphosphate linked to another moiety x), including Ysa1 from yeast and NudT5 from mouse[33]. Recombinant Ysa1 and mNudT5 cleave the pyrophosphate bond of OAADPr, generating 2- and 3-O-acetylribose-5-phosphate and AMP. ARH3 from the human ADP-ribosyl hydrolase (ARH) family hydrolyzes the acetyl group of OAADPr, generating ADPr, which might act as a negative feedback inhibitor of ARH3 [34]. Additionally, two uncharacterized enzyme activities detected in both yeast and human cells have been reported, namely, a cytoplasmic esterase that removes the acetyl group from OAADPr, and a nuclear acetyl-transferase that transfers the acetyl group from OAADPr to an unknown molecule[33]. Future identification and detailed analysis of those activities will likely be important for clarifying the physiological roles of OAADPr.
Figure 2. OAADPr metabolism by the ADPr pyrophosphatase, unknown esterase and acetyl transferase.
The pyrophosphatase cleaves the pyrophosphate bond of OAADPr, generating acetyl-ribose-phosphate and AMP; the esterase removes the acetyl group of OAADPr; the unknown acetyl-transferase transfers the acetyl group from OAADPr to an unknown molecule X.
OAADPr hydrolysis by NUDIX hydrolases
NUDIX hydrolases catalyze the hydrolysis of a nucleoside diphosphate linked to another moiety x, thus the abbreviation “NUDIX”[35, 36]. They are found in more than 250 organisms and characterized by the highly conserved array of amino acids GX5EX7REUXEEXGU, (U represents a bulky, hydrophobic amino acid and X represents any amino acid). The nucleoside diphosphate linkage is common to the broad range of substrates utilized by the family, including NADH, dinucleoside polyphosphates, and nucleotide sugars such as ADPr. NUDIX hydrolases are further divided into sub-families based on their substrate specificity. Comparison among the members of the ADPr pyrophosphatase subfamily revealed a conserved proline, 16 amino acid residues downstream of the NUDIX box, which determines a catalytic preference for ADPr. Considering the structural similarity between OAADPr and ADPr, several members from the ADPr pyrophosphatase subfamily were analyzed for their activity against OAADPr[33]. Ysa1 from Saccromyces cerevisiae[37], NudT5 from mouse[38] and NudT9 from human[39] belong to this family and exhibit ADPrase activity. Ysa1 and mouse NUDT5 (mNUDT5) are able to catalyze hydrolysis of a variety of ADP sugar conjugates with a preference for ADPr. Human NUDT9 (hNUDT9) is highly specific for ADPr and non-physiological IDPR. The shared mechanism among ADPr pyrophosphatases is a nucleophilic attack by water on the pyrophosphate linkage of the substrate, producing AMP and ribose-5-phosphate. Thus, OAADPr hydrolysis yields the corresponding products AMP and 2- and 3-O-acetylribose-5-phosphate ([40–42]).
Steady-state kinetic analyses were performed with Ysa1, mNudT5 and NudT9 using OAADPr and ADPr as substrates (Table I) [33]. Ysa1 hydrolyzes OAADPr with a ∼2 fold lower kcat value (37s−1) and a ∼3.5 fold lower kcat/Km value (4.5 × 104 M−1s−1) compared to ADPr. mNudT5 catalyzes the hydrolysis of both ADPr and OAADPr at a similar maximal rate (0.8–0.9 s−1) and kcat/Km value (1.96 × 104 vs 1.73 × 104, respectively). This kinetic analysis demonstrates that Ysa1 and mNudT5 bind and hydrolyze OAADPr and ADPr with similar catalytic efficiency (Table I). In contrast, NudT9 poorly catalyzes hydrolysis of OAADPr compared with ADPr. The Km value for NudT9 catalysis of OAADPr is ∼500- fold higher than that of ADPr, suggesting that the low catalytic activity results from weak binding affinity [33] (Table I). Consistent with poor binding affinity, OAADPr was unable to inhibit significantly ADPr hydrolysis by NudT9. These results suggest that steric clash from the acetyl group leads to decreased affinity.
Table I.
Kinetic parameters for YSA1, mNudT5, and NUDT9 using ADPr or OAADPr as substrates (Adapted from ref. [33])
| Enzyme (substrate) |
Vmax (µmol·min−1·mg−1) |
kcat (s−1) |
Km (mM) |
kcat/Km (M−1s−1) |
|---|---|---|---|---|
| YSA1 (OAADPr) a | 83.9 ± 3.7 | 36.5 ± 1.6 | 0.081 ± 0.019 | 4.51 × 104 |
| YSA1 (ADPr) a | 175.7 ± 25.1 | 76.4 ± 10.9 | 0.048 ± 0.011 | 1.59 × 105 |
| YSA1 (ADPr)b | 182.3 ± 5.5 | 79.2 ± 2.4 | 0.040 ± 0.009 | 1.98 × 106 |
| mNudT5 (OAADPr) a | 1.95 ± 0.46 | 0.78 ± 0.18 | 0.045 ± 0.009 | 1.73 × 104 |
| mNudT5 (ADPr) a | 2.35 ± 0.48 | 0.94 ± 0.19 | 0.048 ± 0.016 | 1.96 × 104 |
| mNudT5 (ADPr)c | 5.67 ± 0.47 | 0.038 ± 0.005 | ||
| NUDT9 (OAADPr)a,d | 7.9–16.4 | 4.48 ± 0.24 × 102 | ||
| NUDT9 (ADPr)a | 11.0 ± 0.3 | 7.7 ± 0.2 | 0.033 ± 0.006 | 2.33 × 105 |
| NUDT9 (ADPr)e | 11.8 ± 0.3 | 0.100 ± 0.010 |
These data from Ref. [33]
These data from Ref. [37]
These data from Ref. [38]
Due to the inability to saturate, the kcat/Km for NUDT9 with OAADPr was determined from the slope of the line fitted by linear-least squares regression. An estimated range for the Km value was determined by fixing the Vmax at the same rate or 2-fold slower than that obtained with ADPr as substrate (a reasonable assumption based on the results with NudT5 and YSA1, see above) and fitting the data to the Michaelis-Menten equation.
These data from Ref. [76]
Ysa1 is the major endogenous NUDIX hydrolase for both OAADPr and ADPr in yeast [43]. Using an HPLC based assay, cell extracts with endogenous Ysa1 cleaved OAADPr/ADPr to AMP, but cells lacking Ysa1 (Δysa1) exhibited no significant NUDIX activity. Utilizing an OAADPr analog that is resistant to esterase, N-acetyl-ADPr analog was cleaved into AMP by cell extracts containing endogenous Ysa1, supporting the conclusion that OAADPr is directly hydrolyzed by Ysa1[43]. Given that Ysa1 is the major metabolizing enzyme for OAADPr/ADPr in yeast, cellular concentrations of these metabolites under different levels of Ysa1 were determined by LC/MS-MS. Both OAADPr and ADPr levels increase when Ysa1 was deleted [43, 44]. In the Δysa1 strain, OAADPr concentration increased 49% and ADPr concentration increased 50% compared to wild type cells. AMP, the hydrolysis product of Ysa1, decreased 22% in the deletion strain, demonstrating that the increase in OAADPr/ADPr concentration was the direct result of loss of Ysa1 activity. In comparison, whole cell concentrations of NAD+, NADH, and ATP were not significantly affected[43].
ARH3
The human ADP-ribosyl hydrolase (ARH) family contains three members, ARH1-3, which share considerable sequence homology[45]. Based on the available EST (http://www.ncbi.nlm.nih.gov/unigene) sequences, human ARH1 (UGID:142774 UniGene Hs.99884), ARH2 (UGID:142438 UniGene Hs.98669) and ARH3(UGID:133769 UniGene Hs.18021) are widely expressed in multiple tissues including brain, heart, liver, and uterus. ARH1 cleaves the ADP-ribose-arginine bond in the α-ADP-ribosyl-(arginine) protein to generate ADPr and (arginine) protein[46, 47]. Substrates of ARH2 have not been identified[45]. ARH3 binds ADP-ribose but is incapable of hydrolyzing ADP-ribosylarginine[45]. Instead, ARH3 catalyzes the hydrolysis of poly(ADP-ribose). Ono et al.[34] examined the activity of ARH3 against OAADPr. The hydrolyzed product was consistent with ADPr as analyzed by HPLC, high resolution gel electrophoresis and MALDI-TOF. ARH3-dependent OAADPr hydrolysis required Mg2+, similar to the ARH1 catalyzed cleavage of ADP-ribosylarginine and ARH3-catalyzed poly (ADP-ribose) hydrolysis. ARH1 is also capable of hydrolyzing OAADPr, albeit at a 250 fold slower rate. In contrast, ARH2 did not display any activity for OAADPr under the same conditions. ADPr competitively inhibited ARH3 catalyzed OAADPr reactions with a Ki of 2 µM, while NAD+, AMP and ADP showed little or no inhibition. As shown previously, the vicinal carboxylic amino acids Asp60/Asp61 in ARH1 and Glu755/Glu756 in PARG are critical for their catalytic activity. To investigate the importance of similar groups in ARH3, Asp77/Asp78, Glu261/Glu262 and Glu238/Glu239 were mutated. Only the Asp77/Asp78 mutation completely abolished catalytic activity, though their exact roles in OAADPr hydrolysis have not been elucidated [34, 45]. Whether ARH3 metabolizes OAADPr in vivo awaits further clarification. Given these observations, it is possible that ARH3 is the previously observed OAADPr esterase.
OAADPr function
OAADPr in gene silencing
The three regions of the S. cerevisiae genome that are silenced include the two silent mating type loci, the telomeres, and the rRNA-encoding DNA (the rDNA). Silencing at telomere and mating type loci is mediated by a multi-protein nucleosomal binding complex called the Silent Information Regulator (SIR) complex, which contains the Sir2, Sir3, and Sir4 proteins [48]. No obvious homologues to Sir3p and Sir4 have been detected in multi-cellular eukaryotes [49]. Sir2p-dependent deacetylation of histone tails is essential for spreading the Sir2-4 complex along silent chromatin (Figure 3). Considerable evidence indicates that Sir3 and Sir4 only interact with deacetylated histone tails, and that this interaction is required for the establishment and the maintenance of silenced regions [50, 51]. Therefore, in the analysis of Sir2 function in silencing, major attention has been drawn to the deacetylase activity. However, loss of Sir2 activity cannot be rescued by simultaneous deletion of a major histone acetyl transferase, Sas2[52, 53], suggesting that other aspects of Sir2 activity, such as NAD+ hydrolysis and OAADPr synthesis, might function in gene silencing.
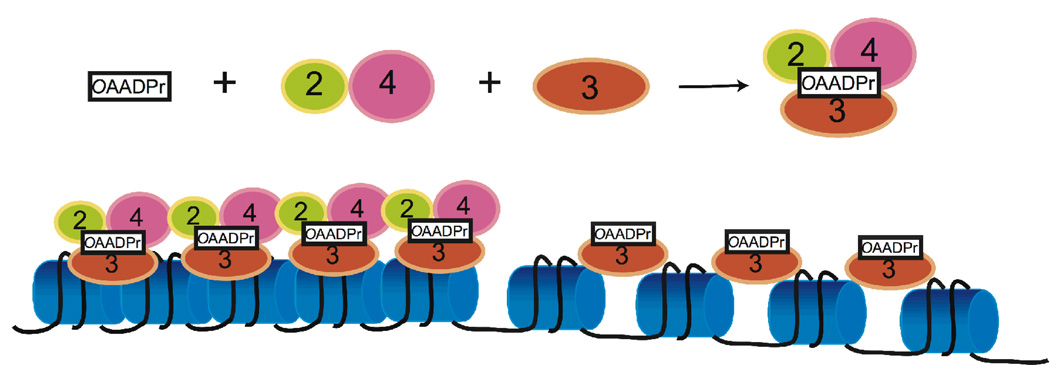
Figure 3. Model for the role of OAADPr in facilitating Sir complex assembly and loading onto the chromatin.
OAADPr interaction with the Sir2-4 complex facilitates loading of the complex onto nucleosomes, probably through binding to Sir2/Sir3. OAADPr might bind Sir3 alone and promote Sir3 loading onto nucleosomes.
OAADPr facilitates SIR complex assembly and loading of SIR complex onto nucleosomes in vitro
The initial model of silent chromatin assembly proposed that the Sir2/Sir4 complex is recruited to the silencer region where Sir2 deacetylates adjacent histone tails. Additional Sir2-4 complexes are then recruited via Sir4/Sir3 interactions, producing subsequent rounds of Sir2 deacetylation and further Sir2-4 recruitment [11, 53–56]. Moazed and colleagues measured the binding affinities of Sir-Sir interactions using full length proteins purified from yeast cells [57]. The Sir2/Sir4 complex bound Sir3 with an apparent disassociation constant (Kd) of ∼3 × 10−11 M, while the Kd for Sir2/Sir3 interaction was ∼6 × 10−8 M and for the Sir3/Sir3 was ∼2 × 10−9 M. Using a number of analytical and biochemical methods, multiple forms of Sir3 oligomers ranging from dimers to decamers were observed ([57, 58] and (unpublished observations, Tong and Denu)). Using TAP-Sir4/HA-Sir2 immobilized on IgG-Sepharose beads, Sir3 bound to the resin and eluted with Sir4/Sir2, suggesting the possibility of forming of a stable SIR complex among the three components in solution [57, 58].
Utilizing an in vitro complex assembly approach, other factors affecting the SIR complex were examined [57, 58]. With different combinations of acetyl-H4-peptide, H4-peptide and NAD+ present, only the addition of both NAD+ and acetyl-H4-peptide lead to an apparent increase of Sir3 bound to the Sir2/Sir4 complex, suggesting the effect was either due to NAD+ hydrolysis or the production of OAADPr. The role of OAADPr in the observed structural rearrangement was shown by adding purified OAADPr. In solution, addition of OAADPr together with unacetylated H4 peptide promoted changes in the stoichiometry and in the appearance of SIR complex, visualized by electron microscopy[57, 58].
Gasser and colleagues investigated the effect of OAADPr on silent chromatin formation at the nucleosomal level[59]. Preincubation of the Sir2/Sir3/Sir4 complex with OAADPr resulted in a marked increase in SIR complex affinity for the chromatin template. In comparison, preincubation with ADPr, ATP, γATP-S or NAD+ had no effect.
Sir3 contains an AAA+ ATPase motif within the C-terminal domain. AAA+ ATPase motif-containing proteins are associated with the assembly, operation, and disassembly of protein complexes[60]. Although missing putative residues for catalysis, the motif in Sir3 appears to retain conserved residues for ATP binding. Considering the structural similarity between OAADPr and ATP, this domain could mediate OAADPr binding with Sir3. Consistent with this hypothesis, preincubating Sir3 with OAADPr led to increased affinity of Sir3 for chromatin [59], indicating that the SIR complex loading efficiency could be mediated by the Sir3/OAADPr interaction (Figure 3). With key catalytic residues missing, no OAADPr hydrolytic activity was detected with the AAA+ ATPase domain of Sir3 [60, 61].
The AAA+ ATPase domain of Sir3 is a potential binding site for OAADPr. Interaction between recombinant purified Sir3 AAA+ domain and OAADPr was detected by photo-crosslinking using [32P]-8N3-OAADPr, though binding evidence from isothermal titration calorimetry was inconclusive (unpublished observations, Tong and Denu). The role of this domain in OAADPr binding and regulated chromatin assembly awaits further evaluation. However, interaction of Sir2 with OAADPr was readily detected by both crosslinking, ITC (Kd of ∼10 µM) and pull down assays ([43] and unpublished observations, Tong and Denu). Collectively, it is quite possible that either Sir2 or Sir3, or both are involved in OAADPr binding and SIR complex assembly and spreading along the chromatin (Figure 3).
While the studies referenced above provided evidence for an important role of OAADPr in silent chromatin assembly in vitro, Gartenberg and colleagues came to a different conclusion based on an engineered protein-chimera employed to examine the role of OAADPr in vivo[62]. Using a Sir3 chimera bearing Hos3, an unrelated NAD+-independent histone deacetylase, silencing was restored in a Δsir2 strain. Even in strains that lack all five sirtuin homologs, the Sir3-Hos3 chimera was able to repress gene expression at the HM loci. The study concluded that OAADPr is not required for silencing in those systems and deacetylation of histone tails is the essential function of Sir2 in HM loci silencing. Nevertheless, these apparently conflicting reports can be rationalized. Considering that OAADPr is not essential for Sir3 to interact with Sir2/Sir4 as determined by SPR (surface plasmon resonance) and in vitro complex assembly [57], silent chromatin can be established in the absence of OAADPr. However, the presence of OAADPr might enhance the efficiency of Sir3 binding to Sir2/Sir4 and chromatin[57, 59], and thus enhance the construction of silent chromatin. Consistent with this hypothesis, Sir3-Hos3 only partially rescued silencing in the strain lacking all five sirtuin homologs, presumably in the absence of any OAADPr [62][60].
OAADPr interaction with macroH2A
In higher eukaryotes, heterochromatin is functionally akin to silent chromatin in yeast. Binding partners like macroH2A1 (mH2A1) also play important roles in chromatin structure and transcriptional activity. MH2A1 is a histone variant that contains a ∼20kD macro domain and is highly enriched in heterochromatin regions such as the female inactive X chromosome, suggesting functions in gene repression[63, 64]. There are currently ±300 proteins in the SMART database [65] that contain a macro domain with diverse functions. A chromatin-remodeling enzyme, ALC1 (Amplified in Liver Cancer 1, also known as CHD1L) interacts with poly(ADP-ribose) (PAR) and catalyzes PARP1-stimulated nucleosome sliding. PAR binding was detected with the macro domain–containing ALC1 proteins but not with a macro-domain mutant. As a consequence, recruitment of ALC1 to DNA damage sites of the macro domain mutant was severely impaired[66]. BAL1 (B-aggressive lymphoma 1) encodes a nuclear protein with N-terminal macro domains and a putative C-terminal poly(ADP-ribose) polymerase (PARP) active site. BAL1 protein represses transcription when tethered to a promoter. A BAL1 proximal N-terminal construct which lacks the macro domains lost transcriptional repression activity. In contrast, the macro domain-containing BAL1 N terminus and the full-length BAL1 protein were equally effective in repressing transcription[67]. Although other macro domain containing proteins can bind poly (ADP-ribose), mH2A1 does not[68]. Interestingly, the gene H2AFY, which expresses mH2A1, generates two isoforms, mH2A1.1 and mH2A1.2. When screening for mH2A1 binding mononucleotides, Kustatscher et al found that the Sir2 metabolite OAADPr binds mH2A1.1 in vitro with a dissociation constant of 2.7 µM and stoichiometry of 1:1 [69]. ADPr binds with similar affinity but with lower binding enthalpy. ADP, AMP, NAD+ and cyclic ADPr were also tested for binding with mH2A1.1, but only ADP showing moderate binding while the other molecules revealed no detectable binding. Thus mH2A 1.1 appears to bind OAADPr and ADPr specifically. However, with the other isoform mH2A1.2, no binding was detected.
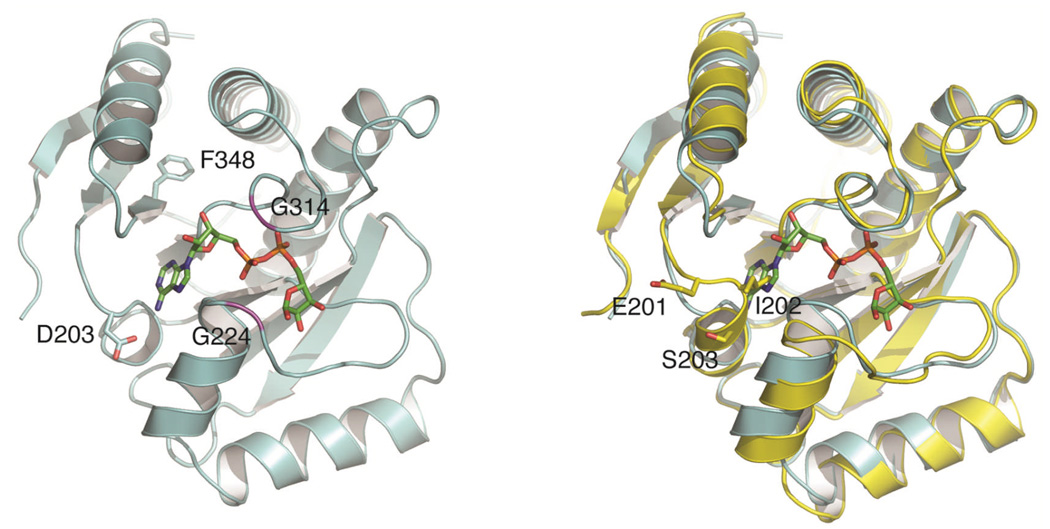
The X-ray crystal structure mH2A 1.1 with OAADPr modeled in (Figure 4) revealed that Phe348 and Asp203 interact with the adenine ring through hydrophobic stacking and hydrogen bonding, respectively, whereas Gly224 and Gly314 were predicted to contact the phosphates of OAADPr [69]. F348A and D203R mutations reduced the binding affinity up to 30-fold, while mutations of residues Gly224 and Gly314 to Glu abolished binding. The crystal structure in combination with mutagenesis predicts that the binding pocket would accommodate the extended confirmation of OAADPr. The major differences between mH2A 1.1 and 1.2 are three residues (EIS) inserted into the adenine binding portion of the mH2A1.1-OAADPr binding site, which flipped the adenine stacking phenylalanine and replaced the phosphate interacting residues Gly223 and Gly224 in mH2A1.1 with larger residues (Lys226 and Asp227) in mH2A1.2. Although these changes are small, they abolished the interaction between mH2A1 and OAADPr (Figure 4).
Figure 4.
Left: model of mH2A 1.1 in complex with ADPR. Model generated by overlaying mH2A 1.1 structure (PDB ID: 2FXK) onto a macro domain-ADPR complex (PDB ID: 2BFQ). Two important glycine residues involved in pyrophosphate binding are colored magenta. F348 and D203 side chains are shown in stick representation. Right: the side chain of the Ile in the EIS insertion clashes with the adenine ring. mH2A 1.1 colored cyan, mH2A 1.2 colored yellow.
Prior to the observation of differential binding to OAADPr, most studies considered macroH2A1.1 and 1.2 to have identical functions. In addition to this difference, the tissue distribution and temporal expression differ noticeably[64]. As mH2A1 represents ∼3 % of total cellular H2A[63], it may compose a considerable amount of chromatin, and presents a novel way for the NAD+ metabolite OAADPr to affect chromatin properties, especially at the heterochromatic regions with enriched macroH2A1.1. SIRT1 has reported roles in heterochromatin establishment[70]. The production of OAADPr by SIRT1 and the potential synergy with chromatin bound mH2A may facilitate the establishment and/or maintenance of heterochromatin.
OAADPr modulation of the TRPM2 ion channel
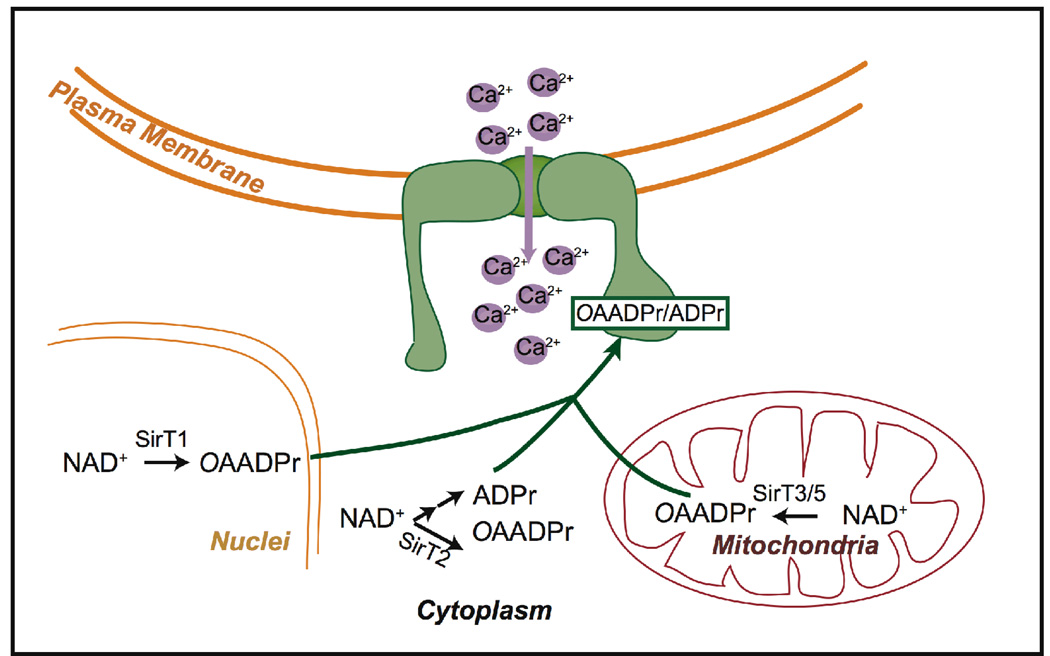
Transient receptor potential malastatin-related channel 2 (TRPM2) is a non-selective cation channel, enriched in brain, gastrointestinal tissues and immune cells[71]. Although its physiology functions are not firmly established, TRPM2 is stimulated by oxidative and nitrative stress that lead to calcium influx into the cell [72, 73]. Though the exact mechanisms are disputed, ADPr binding to NudT9H appears to be required for the well-known oxidative stress activation of the channel, as depletion of ADPr by ADPr hydrolase overexpression rescues cells from TRPM2 over activation [74] In response to oxidative and nitrative stress, ADPr may be generated in large quantities when PARP and PARG become hyperactivated in response to DNA damage [74, 75]. TRPM2 activation is induced by ADPr binding to its C-terminal cytoplasmic domain[39, 76]. This domain, termed NudT9H, shares significant homology to the mitochondrial NUDIX hydrolase NudT9. From electrophysiological experiments, ADPr activates the channel at a low µM levels, a process that requires the NudT9H domain[76, 77]. The NudT9H domain displays no enzymatic activity, suggesting that ADPr binding to this domain is a key step for stress induced TRPM2 channel gating[74] [78].
OAADPr produced by sirtuins induces TRPM2 channel gating [78]. Nicotinamide, a commonly employed inhibitor of sirtuins [79–81], abolished cell death caused by puromycin-induced TRPM2 over activation, consistent with inhibition of OAADPr production and abolished TRPM2 activation. Two mammalian sirtuins, SIRT2 and SIRT3, were examined as possible sources of OAADPR generation that led to TRPM2-dependent cell death induced by the puromycin. RNAi knockdown of SIRT2 and SIRT3 in TRPM2 expressing cells suppressed cell death induced by puromycin treatment. At 20 µM puromycin, loss of SIRT2 or SIRT3 increased cell survival 2–3 fold. With 50 µM of puromycin, 30% of SIRT2 or SIRT3 knocked down cells survived the treatment, while survival of control cells was negligible. Consistent with the hypothesis that OAADPr production is critical for TRMP2 gating, OAADPr induced currents in whole cell patch-clamp experiments in TRPM2 expressing cells, but not when TRPM2 was absent. The kinetics and dose dependence of OAADPr activation were similar to that of ADPr. Hence, OAADPr-dependent gating of the TRPM2 channel is as effective as ADPr and may in fact function as a physiological regulator of TRPM2. OAADPr gating of TRPM2 was also observed in a genetically unmodified cell line CRI-G1, which has a high level of endogenous TRPM2 expression. Consistent with a role for sirtuins, nicotinamide protected CRI-G1 cells from puromycin induced cell death [78].
OAADPr and ADPr bind directly to the NudT9H domain of TRPM2. Using recombinant NudT9H domain, a Kd for ADPr of 130 µM was measured by ITC, whereas no NAD+ binding was detected [78]. A UV crosslinking approach was utilized to examine OAADPr binding to NudT9-H, yielding a Kd of ∼100 µM. These findings provide direct evidence that OAADPr/ADPr bind to the NudT9H domain of the TRPM2 channel, and are likely an important component of the gating mechanism (Figure 5).
Figure 5. OAADPr and ADPr modulates calcium influx through TRPM2 channel.
In mammalian cells, OAADPr produced by sirtuins binds the NudT9H domain of TRPM2 and induces calcium influx into the cells. ADPr, another metabolite of NAD+, is believed to gate TRPM2 in a similar mechanism as OAADPr.
Although OAADPr/ADPr modulate TRPM2 gating in a similar manner, they could accumulate under different circumstances. For instance, ADPr may be generated in large quantities in response to H2O2 stress, when PARP and PARG are activated [74, 75], while OAADPr could accumulate during upregulation of sirtuin activity in response to metabolic adaptation. It is also plausible that OAADPr is rapidly converted to ADPr [33], resulting in a buildup of ADPr, which then mediates TRPM2 gating in vivo (Figure 5).
OAADPr/ADPr modulate cellular REDOX (Reduction-oxidation status)
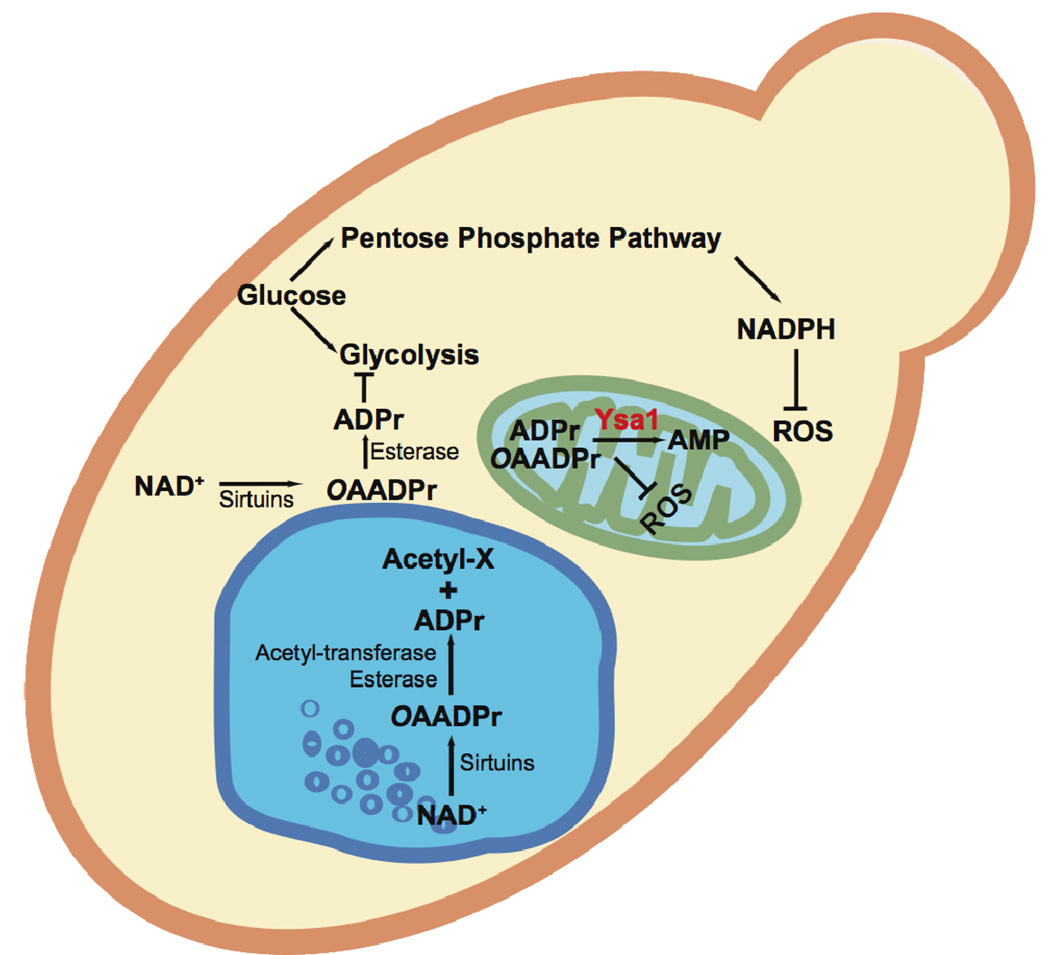
The biological significance of altered OAADPr and ADPr levels in wild type versus Δysa1 yeast strains[44] was examined recently [43]. When cells were treated with H2O2, Δysa1 cells displayed higher resistance to exogenous oxidative stress compared to wild type cells. Similarly, Δysa1 cells were more resistant to endogenous ROS (Reactive Oxygen Species) resulting from treatment with CuSO4, a compound known to increase cellular ROS [82, 83]. Collectively, increased concentration of OAADPr/ADPr appears to endow cells with a higher capacity to resist ROS stress. In addition, basal levels of ROS under normal conditions were 40% lower in the Δysa1 cells compared to wild type cells, providing a strong correlation between lowered ROS with increased levels of OAADPr/ADPr [43]. The mechanism(s) underlying enhanced ROS resistance at higher OAADPr/ADPr levels was explored and two explanations were offered (Figure 6): (i) OAADPr/ADPr inhibit pathway(s) that generate ROS; (ii) OAADPr/ADPr promote pathway(s) that suppress ROS accumulation. The mitochondrial membrane potential ΔΨm in wild type cells was significantly higher than in Δysa1 cells, suggesting that increased OAADPr/ADPr down regulates efficient coupling of respiration, and therefore could suppress electron leakage and ROS generation, which was substantially higher in wild type versus Δysa1 cells [43]. The described biological functions of free ADPr/OAADPr and NUDIX hydrolases are likely conserved among yeast and mammals. Mammalian genomes encode two close homologs of yeast Ysa1, NudT5 and NudT9 [40], both of which display robust catalytic activity toward ADPr in vitro. Consistent with an analogous function in mammalian cells, Formentini et al recently reported the importance of NudT5 and NudT9 in the mitochondrial energy dysfunction caused by PARP-1 hyperactivation [91].
Figure 6. OAADPr metabolism and proposed functions in S. cerevisiae.
OAADPr is produced by sirtuins and can be metabolized by Ysa1, an unknown esterase, and unknown acetyl transferase. Cells lacking Ysa1 display accumulation of ADPr/OAADPr and a corresponding decrease in AMP. Increased ADPr/OAADPr levels might protect cells against ROS via inhibition of the ETC and generation of higher NADPH levels from increased flux through the pentose phosphate pathway.
The observation that OAADPr/ADPr inhibits ETC (electron transport chain)[43] supports the hypothesis that increased OAADPr/ADPr inhibits pathway(s) that generates ROS (Figure 6). Additional data suggests that OAADPr/ADPr might stimulate pathways that suppresses ROS damage and accumulation. To eliminate ROS damage from acute stress of H2O2 and Cu2+, yeast cells reroute glucose metabolism from glycolysis to the NADPH generating pentose phosphate pathway[84–87]. NADPH is utilized to reduce oxidized glutathione and thioredoxin, which are cofactors required by ROS scavenging enzymes[88, 89]. Therefore, rerouting of the metabolic flux in yeast minimizes the damage caused by oxidative stress inducing reagents. With structural similarity to cofactors (ATP, NAD+, etc) utilized by the glycolytic enzymes, OAADPr/ADPr might modulate the activities of key glycolytic enzymes through interacting with the co-enzyme or substrate binding sites. Using an affinity-binding procedure, major proteins interacting with OAADPr/ADPr were identified as phosphoglycerate kinase (PGK), alcohol dehydrogenase (ADH), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which are glycolytic or glycolysis-related enzymes. Steady-state inhibition of GAPDH by ADPr revealed competitive inhibition, with a Ki value of 60 µM. OAADPr inhibits the enzyme with a similar IC50 value [43]. Consistent with the idea that increased levels OAADPr/ADPr induced rerouting of glucose to the pentose phosphate pathway, NADPH levels were substantially higher in the Δysa1 cells (Figure 6). Although mitochondrial proteins were not identified as the major proteins in the OAADPr affinity-binding experiments, it remains to be determined whether the citric acid cycle or other metabolic pathways are regulated directly by ADPr/OAADPr.
Synthesis of OAADPr analogs
To date, evidence supports direct binding of OAADPr to macro H2A, TRPM2, NUDIX hydrolases, ARH3 and possibly Sir2 and Sir3 of the SIR complex. However, the complete picture of OAADPr cellular function and full identification of interacting proteins are likely incomplete. Current analysis of OAADPr function is greatly hindered by its instability and conversion by at least 3 types of enzymatic reactions. ARH3 cleaves the acetyl group on the ribose sugar and NUDIX hydrolases cleave the pyrophosphate linkage. To overcome these limitations in OAADPr research, Comstock et al.[90] developed a synthetic method to produce authentic OAADPr and non-hydrolyzable analogs by substitution of the O-acetyl moiety with an N-acetyl group. N-acetyl analogs at both 2’- and 3’- positions of the ribose sugar were synthesized, which will facilitate the differentiation of the 2’- and 3’- OAADPr isomers by their protein targets. Using macroH2A 1.1, Comstock et al. demonstrated the ability of both analogs to mimic properties of authentic OAADPr. More recently, installment of P-C-P to replace the P-O-P pyrophosphate moiety yields OAADPr analogs that are resistant to cleavage by NUDIX hydrolases (Comstock, Tong, Denu, manuscript in preparation). These synthetic and non-hydrolyzable OAADPr analogs should prove valuable in probing the biological functions of this unique metabolite, derived from the NAD+-consuming activity of Sirtuin protein deacetylases.
CONCLUSIONS AND PERSPECTIVES
Over the last two decades, increasing attention has been paid to newly recognized enzymes that consume NAD+. The sirtuin protein deacetylases represent a novel class that employs NAD+ as a co-substrate to produce nicotinamide, deacetylated product and OAADPr. Although protein deacetylation of targets proteins, like histones, transcription factors and metabolic enzymes, have received the majority of attention among researchers, it will be crucial to account for all the potential functions of the product OAADPr. Given the recent work suggesting OAADPr might regulate gene silencing, ion channel gating, and REDOX regulation, this molecule could play a central role in sirtuin biology, perhaps in synergy with specific protein deacetylation. Because of the structural similarity between OAADPr and ADPr, it is reasonable to suggest that many of OAADPr’s functions and interacting partners might be shared with ADPr. On the other hand, mitochondrial Nudt9 is an important example of an enzyme that displays a 500-fold discrimination between ADPr and OAADPr, favoring ADPr. The rapid conversion of OAADPr into ADPr by several reported enzymes adds another level of complexity to resolving these issues. The non-hydrolyzable OAADPr analogs are thus potentially useful for differentiating the effect of OAADPr and ADPr in future studies.
REFERENCES
- 1.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 2.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 4.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 7.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 9.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 10.Klar AJ, Fogel S, Macleod K. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci U S A. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 13.Rondon MR, Trzebiatowski JR, Escalante-Semerena JC. Biochemistry and molecular genetics of cobalamin biosynthesis. Prog Nucleic Acid Res Mol Biol. 1997;56:347–384. doi: 10.1016/s0079-6603(08)61010-7. [DOI] [PubMed] [Google Scholar]
- 14.Trzebiatowski JR, O'Toole GA, Escalante-Semerena JC. The cobT gene of Salmonella typhimurium encodes the NaMN: 5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-alpha-D-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J Bacteriol. 1994;176:3568–3575. doi: 10.1128/jb.176.12.3568-3575.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moazed D. Enzymatic activities of Sir2 and chromatin silencing. Curr Opin Cell Biol. 2001;13:232–238. doi: 10.1016/s0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 16.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 17.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 18.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 24.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 25.Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J Biol Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- 26.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 27.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 29.Jackson MD, Denu JM. Structural identification of 2'- and 3'-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 30.Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Rowen JW, Kornberg A. The phosphorolysis of nicotinamide riboside. J Biol Chem. 1951;193:497–507. [PubMed] [Google Scholar]
- 32.Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 33.Rafty LA, Schmidt MT, Perraud AL, Scharenberg AM, Denu JM. Analysis of O-acetyl-ADP-ribose as a target for NUDIX ADP-ribose hydrolases. J Biol Chem. 2002;277:47114–47122. doi: 10.1074/jbc.M208997200. [DOI] [PubMed] [Google Scholar]
- 34.Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc Natl Acad Sci U S A. 2006;103:16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or "NUDIX" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 36.McLennan AG. The MutT motif family of nucleotide phosphohydrolases in man and human pathogens (review) Int J Mol Med. 1999;4:79–89. doi: 10.3892/ijmm.4.1.79. [DOI] [PubMed] [Google Scholar]
- 37.Dunn CA, O'Handley SF, Frick DN, Bessman MJ. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J Biol Chem. 1999;274:32318–32324. doi: 10.1074/jbc.274.45.32318. [DOI] [PubMed] [Google Scholar]
- 38.Yang H, Slupska MM, Wei YF, Tai JH, Luther WM, Xia YR, Shih DM, Chiang JH, Baikalov C, Fitz-Gibbon S, Phan IT, Conrad A, Miller JH. Cloning and characterization of a new member of the NUDIX hydrolases from human and mouse. J Biol Chem. 2000;275:8844–8853. doi: 10.1074/jbc.275.12.8844. [DOI] [PubMed] [Google Scholar]
- 39.Shen BW, Perraud AL, Scharenberg A, Stoddard BL. The crystal structure and mutational analysis of human NUDT9. J Mol Biol. 2003;332:385–398. doi: 10.1016/s0022-2836(03)00954-9. [DOI] [PubMed] [Google Scholar]
- 40.McLennan AG. The NUDIX hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabelli SB, Bianchet MA, Ohnishi Y, Ichikawa Y, Bessman MJ, Amzel LM. Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a NUDIX hydrolase. Biochemistry. 2002;41:9279–9285. doi: 10.1021/bi0259296. [DOI] [PubMed] [Google Scholar]
- 42.Gabelli SB, Bianchet MA, Bessman MJ, Amzel LM. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the NUDIX family. Nat Struct Biol. 2001;8:467–472. doi: 10.1038/87647. [DOI] [PubMed] [Google Scholar]
- 43.Tong L, Lee S, Denu JM. Hydrolase regulates NAD+ metabolites and modulates cellular redox. J Biol Chem. 2009;284:11256–11266. doi: 10.1074/jbc.M809790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Tong L, Denu JM. Quantification of endogenous sirtuin metabolite O-acetyl-ADP-ribose. Anal Biochem. 2008;383:174–179. doi: 10.1016/j.ab.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 46.Okazaki IJ, Moss J. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J Biol Chem. 1998;273:23617–23620. doi: 10.1074/jbc.273.37.23617. [DOI] [PubMed] [Google Scholar]
- 47.Moss J, Oppenheimer NJ, West RE, Jr, Stanley SJ. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1986;25:5408–5414. doi: 10.1021/bi00367a010. [DOI] [PubMed] [Google Scholar]
- 48.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 49.Perrod S, Gasser SM. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell Mol Life Sci. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 51.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 52.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 53.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 54.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 55.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 56.Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 58.McBryant SJ, Krause C, Hansen JC. Domain organization and quaternary structure of the Saccharomyces cerevisiae silent information regulator 3 protein, Sir3p. Biochemistry. 2006;45:15941–15948. doi: 10.1021/bi061693k. [DOI] [PubMed] [Google Scholar]
- 59.Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 60.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 61.Buchberger JR, Onishi M, Li G, Seebacher J, Rudner AD, Gygi SP, Moazed D. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28:6903–6918. doi: 10.1128/MCB.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chou CC, Li YC, Gartenberg MR. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell. 2008;31:650–659. doi: 10.1016/j.molcel.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 64.Pehrson JR, Costanzi C, Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J Cell Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 65.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 68.Till S, Ladurner AG. Sensing NAD metabogblites through macro domains. Front Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 69.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 70.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 71.Perraud AL, Schmitz C, Scharenberg AM. TRPM2 Ca2+ permeable cation channels: from gene to biological function. Cell Calcium. 2003;33:519–531. doi: 10.1016/s0143-4160(03)00057-5. [DOI] [PubMed] [Google Scholar]
- 72.Hara Y, Wakamori M, Ishii M, Maeno E, Nishida M, Yoshida T, Yamada H, Shimizu S, Mori E, Kudoh J, Shimizu N, Kurose H, Okada Y, Imoto K, Mori Y. LTRPC2 Ca2+-permeable channel activated by changes in redox status confers susceptibility to cell death. Mol Cell. 2002;9:163–173. doi: 10.1016/s1097-2765(01)00438-5. [DOI] [PubMed] [Google Scholar]
- 73.Wehage E, Eisfeld J, Heiner I, Jungling E, Zitt C, Luckhoff A. Activation of the cation channel long transient receptor potential channel 2 (LTRPC2) by hydrogen peroxide. A splice variant reveals a mode of activation independent of ADP-ribose. J Biol Chem. 2002;277:23150–23156. doi: 10.1074/jbc.M112096200. [DOI] [PubMed] [Google Scholar]
- 74.Perraud AL, Takanishi CL, Shen B, Kang S, Smith MK, Schmitz C, Knowles HM, Ferraris D, Li W, Zhang J, Stoddard BL, Scharenberg AM. Accumulation of free ADP-ribose from mitochondria mediates oxidative stress-induced gating of TRPM2 cation channels. J Biol Chem. 2005;280:6138–6148. doi: 10.1074/jbc.M411446200. [DOI] [PubMed] [Google Scholar]
- 75.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 76.Perraud AL, Fleig A, Dunn CA, Bagley LA, Launay P, Schmitz C, Stokes AJ, Zhu Q, Bessman MJ, Penner R, Kinet JP, Scharenberg AM. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by NUDIX motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- 77.Sano Y, Inamura K, Miyake A, Mochizuki S, Yokoi H, Matsushime H, Furuichi K. Immunocyte Ca2+ influx system mediated by LTRPC2. Science. 2001;293:1327–1330. doi: 10.1126/science.1062473. [DOI] [PubMed] [Google Scholar]
- 78.Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud AL, Scharenberg AM, Denu JM. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J Biol Chem. 2006;281:14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- 79.Rankin PW, Jacobson EL, Benjamin RC, Moss J, Jacobson MK. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J Biol Chem. 1989;264:4312–4317. [PubMed] [Google Scholar]
- 80.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 82.Liang Q, Zhou B. Copper and manganese induce yeast apoptosis via different pathways. Mol Biol Cell. 2007;18:4741–4749. doi: 10.1091/mbc.E07-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 84.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 85.Shenton D, Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, Krobitsch S. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Juhnke H, Krems B, Kotter P, Entian KD. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol Gen Genet. 1996;252:456–464. doi: 10.1007/BF02173011. [DOI] [PubMed] [Google Scholar]
- 88.Winyard PG, Moody CJ, Jacob C. Oxidative activation of antioxidant defence. Trends Biochem Sci. 2005;30:453–461. doi: 10.1016/j.tibs.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 89.Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 90.Comstock LR, Denu JM. Synthesis and biochemical evaluation of O-acetyl-ADP-ribose and N-acetyl analogs. Org Biomol Chem. 2007;5:3087–3091. doi: 10.1039/b710231c. [DOI] [PubMed] [Google Scholar]
- 91.Formentini L, Macchiarulo A, Cipriani G, Camaioni E, Rapizzi E, Pellicciari R, Moroni F, Chiarugi A. Poly(ADP-ribose) catabolism triggers AMP-dependent mitochondrial energy failure. J Biol Chem. 2009;284:17668–17676. doi: 10.1074/jbc.M109.002931. [DOI] [PMC free article] [PubMed] [Google Scholar]