Abstract
Vitamin D3 is critical for the regulation of calcium and phosphate homeostasis. In some individuals, mineral homeostasis can be disrupted by long-term therapy with certain antiepileptic drugs and the antimicrobial agent rifampin, resulting in drug-induced osteomalacia, which is attributed to vitamin D deficiency. We now report a novel CYP3A4-dependent pathway, the 4-hydroxylation of 25-hydroxyvitamin D3 (25OHD3), the induction of which may contribute to drug-induced vitamin D deficiency. The metabolism of 25OHD3 was fully characterized in vitro. CYP3A4 was the predominant source of 25OHD3 hydroxylation by human liver microsomes, with the formation of 4β,25-dihydroxyvitamin D3 [4β,25(OH)2D3] dominating (Vmax/Km = 0.85 ml · min−1 · nmol enzyme−1). 4β,25(OH)2D3 was found in human plasma at concentrations comparable to that of 1α,25-dihydroxyvitamin D3, and its formation rate in a panel of human liver microsomes was strongly correlated with CYP3A4 content and midazolam hydroxylation activity. Formation of 4β,25(OH)2D3 in primary human hepatocytes was induced by rifampin and inhibited by CYP3A4-specific inhibitors. Short-term treatment of healthy volunteers (n = 6) with rifampin selectively induced CYP3A4-dependent 4β,25(OH)2D3, but not CYP24A1-dependent 24R,25-dihydroxyvitamin D3 formation, and altered systemic mineral homeostasis. Our results suggest that CYP3A4-dependent 25OHD3 metabolism may play an important role in the regulation of vitamin D3 in vivo and in the etiology of drug-induced osteomalacia.
Introduction
Vitamin D3 is the major source of vitamin D in humans, which is essential for the maintenance of calcium and phosphate homeostasis (DeLuca, 1988; Plum and DeLuca, 2010). Insufficiency or deficiency of vitamin D is a risk factor for metabolic bone diseases such as rickets, osteoporosis, and osteomalacia (Holick, 2007; Zhang and Naughton, 2010; Rosen, 2011). Vitamin D3 is initially converted to 25-hydroxyvitamin D3 (25OHD3) in the liver; this is considered to be the first step in vitamin D activation (Ohyama and Yamasaki, 2004). 25OHD3 is the most abundant circulating form of vitamin D3, which, under normal conditions, is present at 20 to 50 ng/ml (Plum and DeLuca, 2010; Rosen, 2011). It is the substrate for a second hydroxylase, the mitochondrial cytochrome P450 27B1, which is found abundantly in the kidney, resulting in the production of the most biologically active form of vitamin D3, 1α,25-dihydroxyvitamin D3 [1α,25(OH)2D3] (Sutton and MacDonald, 2003; DeLuca, 2008). The biological activities of 1α,25(OH)2D3 are mediated primarily through the vitamin D receptor, a member of the nuclear superfamily of ligand-activated transcription factors (Pike, 1991). Binding of 1α,25(OH)2D3 to vitamin D receptor initiates transcriptional cascades, leading to the expression of, among others, genes involved in calcium metabolism, cellular proliferation, and immune responses (Bouillon et al., 2008; Pike and Meyer, 2010).
Metabolism of 25OHD3 at the alkyl side chain is considered to be the critical step in the hormone inactivation pathway. In particular, the carbon centers C-23, C-24, and C-26 are the most susceptible sites for oxidation. Mitochondrial CYP24A1 is recognized as a key enzyme for hydroxylation at either C-23 or C-24 of both 1α,25(OH)2D3 and 25OHD3, to give the terminal products of calcitroic acid or a cyclic lactone (Sakaki et al., 2000; Prosser and Jones, 2004). However, Gupta et al. (2004, 2005) reported that CYP3A4 is a microsomal 25-hydroxylase of vitamin D, exhibiting 24- and 25-hydroxylation activities for 1α-hydroxyvitamin D3, 1α-hydroxyvitamin D2, and vitamin D2, after screening 16 major human hepatic P450s expressed in baculovirus-infected insect cells. In contrast, similar experiments performed by Kamachi et al. (2001) revealed no 25-hydroxylation activity toward 1α-hydroxyvitamin D3 for 14 major P450s including CYP3A4 prepared in β-lymphoblastoid cells. We reported that CYP3A4 can catalyze the hydroxylation of 1α,25(OH)2D3 at C-23 and C-24 (Xu et al., 2006). In contrast to CYP24A1 that catalyzes C-23S and C-24R hydroxylation of 1α,25(OH)2D3, CYP3A4 exhibited an opposite product stereoselectivity, with preferential formation of C-23R- and C-24S-hydroxy products (Sakaki et al., 2000; Xu et al., 2006). Therefore, perturbation of 1α,25(OH)2D3 elimination through changes in gene expression of these catalytic enzymes can contribute to vitamin D deficiency and emergent disease, as it is thought to be the case for drug-induced osteomalacia (Pack et al., 2004; Pascussi et al., 2005; Xu et al., 2006; Zhou et al., 2006).
Long-term treatment (for at least 2 weeks) with the PXR agonist rifampin results in a marked reduction in 25OHD3 plasma levels, presumably by enhanced clearance of the hormone (Brodie et al., 1980, 1982; Shah et al., 1981). This in turn would limit the availability of substrate for the formation of the biologically active 1α,25(OH)2D3. It is well known that CYP3A4 is induced by certain drugs, such as rifampin via the PXR (Gonzalez, 2007; Zhou, 2008). Thus, induction of CYP3A4 gene expression by certain drugs may enhance 25OHD3 catabolism and hence modulate vitamin D3 effects in the body. Therefore, it was of interest to us to investigate whether CYP3A4 could catalyze the alkyl side chain oxidation of 25OHD3, as it does with 1α,25(OH)2D3, leading to vitamin D inactivation. We also examined whether activation of PXR, after treatment with rifampin, could alter the formation of CYP27B1-, CYP3A4- and CYP24A1-dependent metabolites of 25OHD3. The results presented here demonstrate that CYP3A4 is a multifunctional enzyme capable of hydroxylating the alkyl side chain and A-ring of 25OHD3 reactions, which might contribute to the regulation of vitamin D3 metabolism, and that formation of the CYP3A4-dependent, but not the CYP24A1-dependent, products is selectively induced by rifampin in healthy subjects.
Materials and Methods
Materials.
6′,7′-Dihydroxybergamottin (DHB), EDTA, ketoconazole (KTZ), n-butylboronic acid, NADPH, 25OHD3, 24R,25-dihydroxyvitamin D3 [24R,25(OH)2D3], 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD), sodium periodate, and N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% trimethylsilylchlorosilane were purchased from Sigma-Aldrich (St. Louis, MO). 1α,25(OH)2D3 was purchased from Calbiochem (San Diego, CA). Authentic standards of 23R,25-dihydroxyvitamin D3 [23R,25(OH)2D3], 24S,25-dihydroxyvitamin D3 [24S,25(OH)2D3], and 25,26-dihydroxyvitamin D3 [25,26(OH)2D3] were custom-synthesized by SAFC Pharma (Madison WI). 23S,25-Dihydroxyvitamin D3 [23S,25(OH)2D3] and 3-epi-1α,25-dihydroxyvitamin D3 [1α,25(OH)2-3-epi-D3] were gifts from Dr. T. Fujishima (Tokushima Bunri University, Fujishima, Japan) and Dr. T. Sakaki (Toyama Prefectural University, Toyama, Japan), respectively. Deuterated d6-25OHD3 and d6-1α,25(OH)2D3 (containing six deuterium atoms at C-26 and C-27) were purchased from Medical Isotopes Inc. (Pelham, NH). Supersomes containing cDNA-expressed human P450 enzymes and P450 reductase (some with cytochrome b5) were purchased from BD Gentest (Woburn, MA). Human liver microsomes (HLM), prepared previously, were generated from the University of Washington School of Pharmacy Human Tissue Bank (Seattle, WA). Basic demographic information and characterization of CYP3A4/5 expression in these samples have been published previously (Lin et al., 2002). Cryopreserved primary human hepatocytes from three different human liver donors were obtained from either BD Gentest or Invitrogen (Carlsbad, CA).
Characterization of 25OHD3 Metabolism.
Because of the light sensitivity of vitamin D, the following procedures were conducted under low-light conditions. All 25OHD3 metabolism experiments with recombinant P450 enzymes and HLM were carried out in 0.1 M potassium phosphate buffer containing 1 mM EDTA at 37°C, in a total volume of 0.5 ml. After preincubation of the enzyme with 25OHD3 for 5 min, the reaction was initiated by the addition of 1 mM NADPH. Incubations without NADPH served as negative controls. 25OHD3 was dissolved in methanol, and the total percentage of organic solvent in the incubation volume was 1% for all the incubations.
Reaction kinetic parameters (Vmax and Km) for the formation of two major products [4α,25(OH)2D3 and 4β,25(OH)2D3] were determined using 5 pmol of CYP3A4 (coincubated with P450 reductase and cytochrome b5) or 50 μg of HLM and 2.5 to 50 μM 25OHD3, incubated for 5 min. These conditions resulted in linear product formation rates at all substrate concentrations tested (data not shown). For screening of the various recombinant P450 enzymes for 4-hydroxylation activity, formation rates were determined using 10 pmol of enzyme incubated with 12.5 μM 25OHD3 for 30 min, except for CYP3A4 and CYP3A5 for which incubations were conducted for 10 min. After incubation, an aliquot (20 μl) was subjected to liquid-liquid extraction (LLE) and LC-MS/MS analysis. For inhibition studies, we used two CYP3A4 inhibitors, DHB and KTZ. DHB (50 μM) or vehicle was preincubated with CYP3A4 and NADPH for 30 min at 37°C before a 65-μl aliquot (with 10 pmol of enzyme) was then transferred to a second incubation tube containing 25OHD3. In separate experiments, KTZ (0.5 μM), a competitive inhibitor, or vehicle was added directly to the incubation mixture before addition of NADPH to initiate the reaction. The total volume was 0.5 ml, and the final concentrations were 20 nM CYP3A4, 1 mM NADPH, and 12.5 μM 25OHD3.
Reactions were terminated with the addition of 0.5 ml of ethyl acetate. The vitamin D3 products were then isolated by addition of 5 ml of ethyl acetate. After extraction, removal of solvent, and reconstitution in the mobile phase, the isolated products were analyzed by HPLC with a UV diode array detector. For some experiments, the incubation mixtures were spiked with 2 ng of d6-1α,25(OH)2D3 as an internal standard. After LLE and PTAD derivatization, the derivatized vitamin D3 metabolites were subjected to LC-MS/MS analysis.
HPLC and LC-MS/MS Analysis.
Method I.
Initial analysis of the in vitro 25OHD3 incubation products was performed using an 1100 series HPLC system (Agilent Technologies, Santa Clara, CA). Chromatographic separation of the major analytes was achieved on a Symmetry C8 (2.1 × 150 mm, 3.5 μm) column (Waters, Milford, WA). The mobile phase consisted of a linear gradient from 70% methanol at 0.25 ml/min as follows: 0 to 40 min, 70 to 80% methanol; 40.1 to 45 min, 90% methanol; and 45.1 to 60 min, 70% methanol. An aliquot (5 μl) of reconstituted residue was injected for HPLC-UV analysis, and seven authentic vitamin D3 standards were analyzed in parallel. All eight peaks isolated from incubation extracts that eluted between 26 and 36 min were collected and subjected to periodate cleavage, followed by LC-MS/MS analysis.
Method II.
LC-MS/MS was performed using an Agilent 1200 series high-performance liquid chromatograph and an Agilent 6410 triple quadrupole tandem mass spectrometer equipped with an electrospray ionization source, as described previously (Wang et al., 2011). Separation was achieved on a Hypersil Gold (2.1 × 100 mm, 1.9 μm) column (Thermo Scientific Fisher, Waltham, MA). Multiple reaction monitoring (MRM) of the transitions m/z 574 → 314, 574 → 298, 558 → 298, 564 → 298, and 580 → 314 was used to detect 1α,25(OH)2D3, 24R,25(OH)2D3, 25OHD3, d6-25OHD3, and d6-1α,25(OH)2D3, respectively.
Purification of Two Major Metabolites Using HPLC.
Vitamin D3 products from in vitro incubations were isolated using LLE, pooled, and concentrated under a nitrogen stream. The major vitamin D3 metabolites were purified by HPLC. The vitamin D3 metabolites were monitored using UV absorbance at 265 nm. Because multiple metabolites with similar structures were formed, the chromatographic conditions were selected to achieve complete separation of the two major metabolites. Chromatographic isolations were accomplished using an Eclipse XDB-C8 (4.6 × 150 mm, 5 μm) column (Agilent Technologies) at a flow rate of 1 ml/min under two different mobile phase conditions. An isocratic separation (85% methanol-15% water) was performed initially, and fractions, which contained the two major unknown metabolites, were collected between 4.3 and 5.0 min. These fractions were then concentrated under a nitrogen stream and subjected to a second separation using a gradient mobile phase. The proportion of methanol in the mobile phase was as follow: 0 min, 75%; 16 min, 79%; 16.2 to 19 min, 95%; and 19.2 to 25 min, 75%. Two major peaks were collected and designated as M1 (retention time 13.4 min) and M2 (14.2 min), which proved to be the isomers of 4,25(OH)2D3 as defined below.
Structure Identification by GC-MS and 1H NMR.
GC-MS analysis of the trimethylsilyl derivatives of M1 (∼0.5 μg) and M2 (∼0.8 μg) was performed using a QP2010 GC-MS system (Shimadzu, Columbia, MD), as described previously (Xu et al., 2006). NMR analysis was accomplished on a Varian Unity-Inova 500 MHz NMR spectrometer. The M1 (∼30 μg) and M2 (∼15 μg) metabolites were dissolved separately in 0.5 ml of deuteromethanol, and 1H NMR spectra were acquired at 25°C and referenced to residual solvent at 4.87 ppm.
Addition of the 4-Hydroxylated Metabolites into Plasma.
After we identified the structures of M1 and M2, the concentrations of these two major metabolites were estimated using their UV absorbance at 265 nm and the calibration curve of 25OHD3 plotted as concentration versus absorbance. An aliquot (50 or 100 pg) of purified M1 or M2 was spiked into 1 ml of plasma and then prepared for LC-MS/MS analysis after LLE and derivatization. In addition, we incubated these plasma extracts with 0.5% sodium periodate to determine the presence of vicinal hydroxyl groups in the vitamin D3 metabolites as indicated by the disappearance of mass spectral signals for M1 and M2 after periodate cleavage.
Isolation of the 4-Hydroxlated Metabolites from Human Plasma.
Isolation of the circulating 4-hydroxylated vitamin D3 metabolites from human plasma was performed as follows. Outdated plasma (∼350 ml) from the Puget Sound Blood Center (Seattle, WA) was divided into 20 portions. After protein precipitation by addition of 2 volumes of acetonitrile, the supernatants were concentrated by removing acetonitrile under a stream of nitrogen and extracted with an equal volume of methyl tert-butyl ether. The organic extracts were combined and concentrated under a nitrogen stream. Because an aqueous component was observed in the residue, 3 volumes of methyl tert-butyl ether were added for a second round of extraction. The organic extracts were then dried under a nitrogen stream, reconstituted in 100 μl of methanol, and subjected to HPLC-UV analysis. The UV signals of the 4-hydroxylated metabolites, however, were not observed because of interference from signals from other unknown compounds in the extracts. Thus, fractions were collected on the basis of the retention times at which the 4-hydroxylated metabolites were expected to elute. After repeated injections, all fractions putatively containing the metabolites were combined, dried, and then subjected to PTAD derivatization. The resulting derivatives were then subjected to LC-MS/MS analysis.
Interliver Differences in the Formation of the 4-Hydroxylated Metabolites.
To evaluate interliver differences in the formation of 4α,25(OH)2D3 and 4β,25(OH)2D3, we screened a panel of 42 HLM, which were prepared as described previously (Lin et al., 2002). The assay was performed as described above with a 10-min incubation time. The final protein concentration of HLM was 100 μg/ml in a total volume of 0.5 ml. After PTAD derivatization and LC-MS/MS analysis, the formation rates of the 4-hydroxylated metabolites were correlated with the previously reported CYP3A4 content (determined by Western blot analysis) or total CYP3A activity (sum of the rates of 1′- and 4-hydroxymidazolam formation from MDZ) (Lin et al., 2002).
Metabolism of 25OHD3 in Human Hepatocytes.
Cryopreserved hepatocytes were thawed at 37°C, centrifuged at 100g in cryopreserved hepatocyte recovery medium for 10 min, and resuspended in plating media (Williams' E plus plating supplements: 5% fetal bovine serum, 100 nM dexamethasone, 100 U/ml penicillin and streptomycin, 4 μg/ml insulin, 2 mM GlutaMAX, and 15 mM HEPES, pH 7.4). Viability and density were measured by trypan blue exclusion, and 52,000 cells/well were plated onto 96-well collagen I-coated plates. Hepatocytes were allowed to attach for 4 to 6 h, and then plating media was removed and replaced with maintenance media (Dulbecco's modified Eagle's medium plus maintenance supplements: 100 U/ml penicillin and streptomycin, 6.25 μg/ml insulin, 6.25 μg/ml transferrin, 6.25 ng/ml selenous acid, 1.25 mg/ml bovine serum albumin, 5.35 μg/ml linoleic acid, 2 mM GlutaMAX, and 15 mM HEPES, pH 7.4) containing 0.25 mg/ml Matrigel. On the following day, cells were treated with rifampin (10 μM, 100 μl/well) or vehicle alone (0.1% DMSO) to elicit induction of CYP3A4. After 48 h of treatment, the medium was removed, and the cells were washed twice with buffer solution and then preincubated with DHB (20 μM) or vehicle alone (0.2% ethanol) for 4 h. The culture medium was replaced with fresh medium containing 25OHD3 (5 μM) in the presence or absence of DHB (20 μM) for 24 h. At the end of treatment, culture medium (1 ml) was collected and pooled for the quantification of 4-hydroxylated metabolites by LC-MS/MS. Total RNA was isolated using the MagMax 96 RNA Isolation Kit as needed for quantitative reverse transcriptase-polymerase chain reaction.
Subject Recruitment.
A human pilot study was approved by the University of Washington Human Subjects Review Board and conducted at the University of Washington Medical Center. Potential participants (ages 18–40 years) were interviewed and provided written informed consent before undergoing study procedures. After a screening evaluation, subjects were excluded from further participation if they had a history or laboratory tests indicative of liver, kidney, gastrointestinal, or heart disease, diabetes, or bone problems; were allergic to rifampin; were taking prescription, over-the-counter medications, or herbal medications; used oral contraceptives or hormone patches; smoked; or were pregnant at the time of the study. Eligible subjects were asked to abstain from prescription and over-the-counter medications, herbal products, nutritional supplements, oral contraceptives, and grapefruit juice for at least 1 week before and during the study period. They were also requested to abstain from alcohol before the 1st day and during the study period and to fast for 8–12 h before all study visits.
Study Procedures.
Subjects were scheduled for a total of four visits for blood and spot urine collection, all taking place on different days at approximately 9 am. Two visits occurred before rifampin administration: day 0 and on the morning before the first rifampin dose, day 1. Subjects received 600 mg of rifampin at 9:00 PM daily with 8 ounces of water, beginning on day 1, for 7 consecutive days. The final two blood and urine collection visits occurred on day 7 and day 8. Blood was obtained by venipuncture, and serum or plasma was isolated by centrifugation. All biological samples were stored at −80°C before analysis.
Vitamin D and Biomarker Analysis.
The concentrations of 1α,25(OH)2D3, 24R,25(OH)2D3, 4β,25(OH)2D3, and 25OHD3 in plasma were measured as described previously (Wang et al., 2011). Free and total calcium and inorganic phosphate concentrations in serum, as well as calcium, inorganic phosphate, and creatinine concentrations in urine, were measured using standard clinical laboratory procedures in the Department of Laboratory Medicine at the University of Washington. For each analyte, the average of measurements for days 0 and 1 (prerifampin) and days 7 and 8 (postrifampin) is reported.
Statistical Analyses.
All kinetic and inhibition experiments with recombinantly expressed enzymes and HLM were performed in triplicate. Resulting data are expressed as the mean ± S.D. and were compared using one-way analysis of variance analysis with Dunnett or Tukey multiple comparison tests. An assessment of interliver differences in 4α,25(OH)2D3 and 4β,25(OH)2D3 formation were obtained from two separate experiments. We used GraphPad Prism (version 5; GraphPad Software Inc., La Jolla, CA) for the statistical analyses. Statistical comparison of the pre- and postrifampin treatment measurements were conducted with SAS. Generalized estimating equations, with treatment period as the independent variable, was used; p < 0.05 was considered statistically significant. For regression analysis, sample correlation coefficients and corresponding p values were calculated; the null hypothesis that the population correlation is 0 was tested with a t test, under the assumption that the population was bivariate normally distributed.
Results
Metabolism of 25OHD3 by CYP3A4 and HLM.
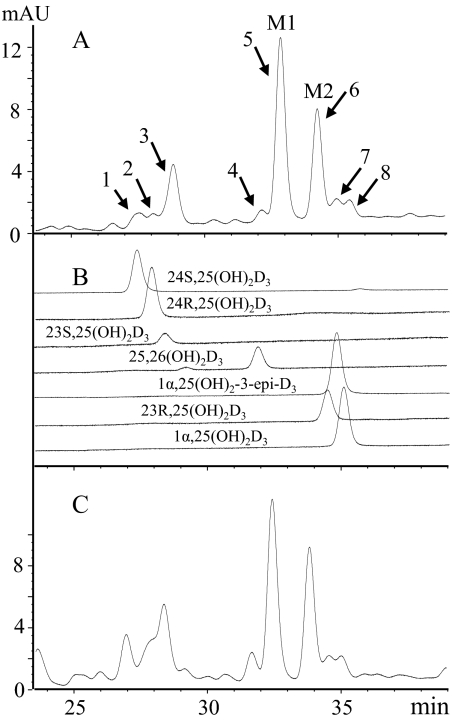
After incubation of 25OHD3 with CYP3A4 (coexpressed with P450 reductase and cytochrome b5) or HLM for 90 min, vitamin D3 products were extracted and subjected to HPLC-UV analysis. The HPLC profiles of 25OHD3 metabolites produced by CYP3A4 and HLM are depicted in Fig. 1, A and C, respectively. Eight major peaks were detected by UV absorbance, all of which demonstrated the typical vitamin D chromophore (λmax of 265 nm). To identify the metabolites by retention time, seven authentic standards with concentrations ranging from 1 to 4 μg/ml were analyzed under the same LC conditions (Fig. 1B). To further verify the identity of these metabolites, all eight peaks were collected individually and subjected to periodate cleavage and LC-MS/MS analysis. As summarized in Table 1, nine putative metabolites were identified, seven of which matched the authentic standards by comparison of LC retention times, mass spectra, and sensitivity to sodium periodate treatment. Peaks 1 through 4 were identified as 24S,25(OH)2D3, 24R,25(OH)2D3, 23S,25(OH)2D3, and 25,26(OH)2D3, respectively. Peaks 5 and 6, the two major metabolites designated as M1 and M2, were suspected to be unknown dihydroxyvitamin D3 metabolites with two vicinal hydroxyl groups on the A-ring. Peaks 7 and 8 were assigned as mixtures of 23R,25(OH)2D3, 1α,25(OH)2D3, and 1α,25(OH)2-3-epi-D3.
Fig. 1.
HPLC profiles of the vitamin D metabolites produced from 25OHD3 by incubation with recombinant CYP3A4 and HLM. A, extracts from 25OHD3 incubation with recombinant CYP3A4. B, seven authentic vitamin D3 metabolites. C, extracts from 25OHD3 incubation with HLM. M1 and M2 denote two major unknown metabolites. mAU, milliabsorbance units.
TABLE 1.
Identification of dihydroxyvitamin D3 metabolites by HPLC-UV, periodate cleavage, and LC-MS/MS analysis
After incubation of 25OHD3 with recombinant CYP3A4 or HLM, the vitamin D3 metabolites were extracted by LLE and reconstituted in mobile phase for HPLC-UV analysis. The fractions corresponding to eight peaks were collected, dried, and subjected to PTAD derivatization. The derivatives were then divided into two parts, one for LC-MS/MS analysis directly and the other for periodate cleavage followed by LC-MS/MS analysis. The results were compared with those for authentic standards, which were conducted under the same conditions. HPLC analysis was performed using method I and LC-MS/MS analysis was performed using method II. After reaction with NaIO4 or water as vehicle, metabolites were extracted by ethyl acetate; after evaporation, samples were reconstituted in acetonitrile (40%) for LC-MS/MS analysis. Experimental details are described under Materials and Methods.
| Vitamin D Metabolites | HPLC-UV RT | LC-MS/MS after PTAD Derivatizationa |
Periodate Cleavage (Vicinal–OH Groups?) | Assigned Structures | |
|---|---|---|---|---|---|
| MRM (m/z) | RT | ||||
| min | min | ||||
| Peak 1 | 27.36 | 574 → 298 | 8.61; 10.18 | Yes | 24S,25(OH)2D3 |
| Peak 2 | 27.81 | 574 → 298 | 8.56; 10.13 | Yes | 24R,25(OH)2D3 |
| Peak 3 | 28.61 | 574 → 298 | 8.59; 9.97 | No | 23S,25(OH)2D3 |
| Peak 4 | 31.95 | 574 → 298 | 8.78; 9.81 | Yes | 25,26(OH)2D3 |
| Peak 5 | 32.62 | 574 → 314 | 11.31; 12.15 | Yes | 4β,25(OH)2D3 (M1) |
| Peak 6 | 34.01 | 574 → 314 | 11.15; 11.91 | Yes | 4α,25(OH)2D3 (M2) |
| Peak 7 | 34.70 | 574 → 298 | 11.37 | No | 23R,25(OH)2D3 |
| 574 → 314 | 11.62 | No | 1α,25(OH)2D3 | ||
| 574 → 314 | 12.36 | No | 1α,25(OH)2-3-epi-D3 | ||
| Peak 8 | 35.21 | 574 → 314 | 12.37 | No | 1α,25(OH)2-3-epi-D3 |
| 574 → 314 | 11.62 | No | 1α,25(OH)2D3 | ||
RT, retention time.
Identification of the Two Major Metabolites.
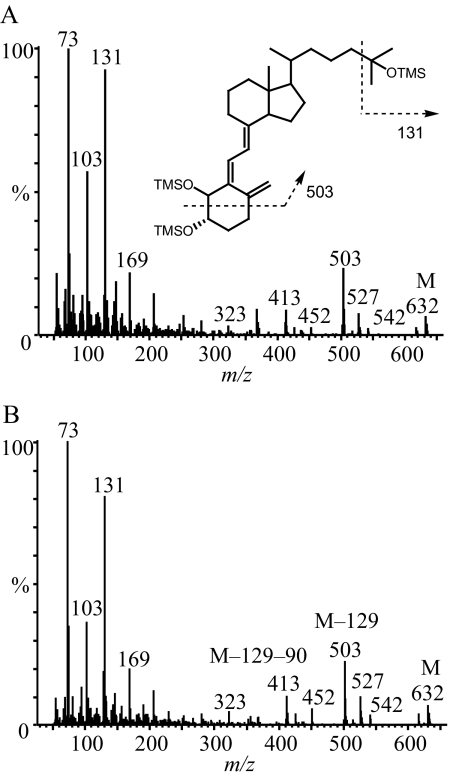
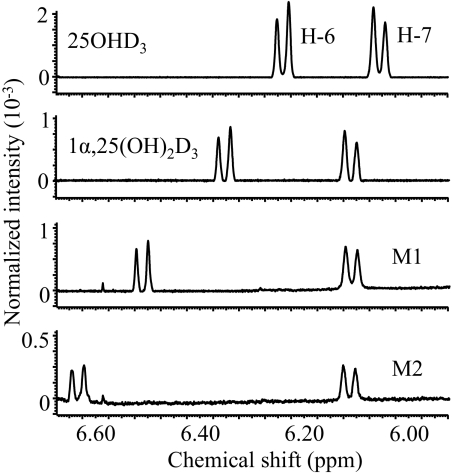
The two major metabolites, M1 and M2, were initially thought to be either 2,25-dihydroxyvitamin D3 or 4,25(OH)2D3 regioisomers based on the following criteria: 1) both contained two hydroxyl groups on the A-ring, based on a characteristic daughter ion of m/z 314 from the PTAD derivative [M − H2O + H]+ (m/z 574); 2) neither metabolite corresponded to 1α,25(OH)2D3 or 1α,25(OH)2-3-epi-D3 because of their different retention times (Supplemental Fig. 1); and 3) both were sensitive to periodate cleavage, although M1 was much more sensitive than M2. To fully define the structures, sufficient quantities (>10 μg) of these two metabolites were purified and characterized by three additional experiments. In the first experiment, the metabolites were converted to the trimethylsilyl ether derivatives and subjected to GC-MS analysis. As shown in Fig. 2, the molecular ions of both M1 and M2 were at m/z 632, confirming that they were vitamin D3 derivatives with three hydroxyl groups. The abundant ion at m/z 131 indicated the presence of an octadecyltrimethoxysilane group at C-25. Structurally informative fragment ions were found at m/z 503 (M − 129) and 413 (M − 129 − 90) for both M1 and M2. The loss of a 129-Da neutral mass from analogous structures has been attributed to the elimination of C-1, C-2, and C-3 together with an octadecyltrimethoxysilane group at C-3 (Dumaswala et al., 1989; Araya et al., 2003). Thus, M1 and M2 are likely to be 4,25(OH)2D3 isomers. This conclusion was further confirmed by results from a 1H NMR experiment. As shown in Fig. 3, the chemical shifts of the vinylic hydrogens at C-6 of M1 (6.49 ppm) and M2 (6.61 ppm) were shifted downfield, compared with the same hydrogens in the spectra of both 25OHD3 and 1α,25(OH)2D3, whereas the chemical shifts of the hydrogen at C-7 of M1 (6.09 ppm) and M2 (6.09 ppm) were similar compared with the same hydrogen in the standards, 25OHD3 (6.03 ppm) and 1α,25(OH)2D3 (6.09 ppm). The downfield shift is most likely the result of an inductive, deshielding field effect of the electronegative hydroxyl group at C-4 on the C-6 proton. Hydroxylation at C-2 would not be expected to have this effect because the hydroxyl group would be too distant from the C-6 proton (Araya et al., 2003). Thus, both M1 and M2 were assigned as configurational isomers of 4,25(OH)2D3. After a third set of experiments, M1 was identified as 4β,25(OH)2D3 and M2 as 4α,25(OH)2D3 because of their differential reactivities with sodium periodate and n-butylboronic acid treatment (Xu et al., 2006). Both reactions were predicted to be more facile for adjacent cis-dihydroxy groups than trans-dihydroxy groups in a cyclic ring system (Alexander et al., 1951). The rate of A-ring cleavage after periodate treatment was much greater for M1 than for M2, and only M1 reacted with n-butylboronic acid (leading to a loss of an [M + Na]+ ion at m/z 439 under LC-MS analysis) to generate a product with a [M + Na]+ ion at m/z 505. This result is consistent with the formation of a cyclic boronate, a known product of cis-diols but not of trans-diols. The larger downfield shift of ∼0.12 ppm for the proton at C-6 in the proposed 4α-hydroxy isomer (M2) compared with the 4β-hydroxy isomer (M1) also is consistent with the assignments, because the 4α-hydroxy group will primarily exist in a pseudo-equatorial orientation that is closer to the C-6 hydrogen in M2 than the 4β-hydroxy group in M1 that will primarily exist in a pseudo-axial orientation.
Fig. 2.
Electron impact mass spectra of the trimethylsilyl ether (TMSO) derivatives of the two unknown metabolites, M1 (A) and M2 (B). The metabolites were isolated and purified by HPLC before GC-MS analysis. OTMS, octadecyltrimethoxysilane.
Fig. 3.
Identification of M1 and M2 by 1H NMR as 4-hydroxylated 25OHD3 metabolites. The chemical shifts of protons at C-6 and C-7 for 25OHD3, 1α,25(OH)2D3, M1, and M2 were assigned according to literature values (Eguchi and Ikekawa, 1990; Kamao et al., 2004).
Assessment of 4-Hydroxylation Activity by Various Recombinant P450 Enzymes.
In addition to CYP3A4, 12 other recombinantly expressed human P450 enzymes were evaluated for their ability to catalyze the 4-hydroxylation of 25OHD3. We found that CYP3A5 (coincubated with P450 reductase and cytochrome b5) catalyzed the 4-hydroxylation reaction, whereas the other enzymes had no detectable activity (Table 2). However, the reaction rate for CYP3A5 was ∼10% that of CYP3A4 under the same incubation conditions. The metabolism of 25OHD3 by CYP24A1 was also studied (incubation performed in Japan by Dr. Toshiyuki Sakaki) and, as expected, 24R,25(OH)2D3 was the predominant product, and no detectable amount of 4,25(OH)2D3 was formed (Table 2).
TABLE 2.
25OHD3 4-hydroxylase activity for various recombinant P450 enzymes
Incubations were performed with 10 pmol of enzyme for 30 min. The metabolite formation rates for CYP3A4 and CYP3A5 were obtained from triplicate incubations for 10 min. The substrate 25OHD3 concentration was 12.5 μM.
| P450s | Metabolite Formation Rate |
||
|---|---|---|---|
| 24R,25(OH)2D3 | 4β,25(OH)2D3 | 4α,25(OH)2D3 | |
| pmol · min−1 · pmol−1 | |||
| CYP3A4 (b5 coexpressed) | BLQ | 2.27 ± 0.22 | 1.26 ± 0.27 |
| CYP3A5 (b5 coexpressed) | BLQ | 0.24 ± 0.03 | 0.17 ± 0.02 |
| CYP24A1a | 2.39 ± 0.63 | N.D. | N.D. |
| Other P450sb | N.D. | N.D. | N.D. |
BLQ, below limit of quantification; N.D., not detected.
Metabolism of 25OHD3 by CYP24A1 was conducted in Dr. T. Sakaki's laboratory (Toyama Prefectural University). Incubation conditions: CYP24A1 (20 nM), adrenodoxin (200 nM), adrenodoxin reductase (20 nM), 1 mM NADPH, and 12.5 μM 25OHD3 in 100 mM Tris-HCl (pH 7.4, 1 mM EDTA) at 37°C for 10 min. After incubation, the products were extracted and shipped to the University of Washington. LC-MS/MS analysis was performed as described under Materials and Methods.
Other P450s: CYP1A1, -1A2, -2A6, -2B6, -2C8, -2C9, -2C19, -2D6, -2E1, -2J2, and -3A7.
Kinetic Parameters for 4β,25(OH)2D3 and 4α,25(OH)2D3 Formation from 25OHD3.
The kinetics of 4β,25(OH)2D3 and 4α,25(OH)2D3 formation were determined using recombinant CYP3A4 and HLM over a range of substrate concentrations (2.5–50 μM). The Michaelis-Menten equation was fit to the resulting data, because no evidence for allosterism was observed. As summarized in Table 3, the Vmax for 4β,25(OH)2D3 formation was ∼2-fold higher than that for 4α,25(OH)2D3, although the Km values were comparable when determined using either recombinant CYP3A4 or HLM. The total intrinsic clearance (Vmax/Km, sum of 4-hydroxylated metabolites) for CYP3A4 was approximately 1.32 ml · min−1 · nmol−1.
TABLE 3.
Kinetic parameters for the 4-hydroxylation of 25OHD3 by recombinant CYP3A4 and HLM
Values represent means ± S.D. for triplicate incubations. Total intrinsic CL is the sum of Vmax/Km for these two major products [4β,25(OH)2D3 and 4α,25(OH)2D3].
| 4β,25(OH)2D3 |
4α,25(OH)2D3 |
Total Intrinsic CL: Vmax/Km | |||
|---|---|---|---|---|---|
| Vmax | Km | Vmax | Km | ||
| pmol · min−1 · pmol−1 | μM | pmol · min−1 · pmol−1 | μM | ml · min−1 · nmol−1 | |
| CYP3A4 (b5 coexpressed) | 6.4 ± 0.99 | 7.5 ± 1.35 | 3.0 ± 0.25 | 6.4 ± 0.86 | 1.32 |
| HLMa | 229 ± 62.7 | 18.8 ± 4.6 | 105 ± 13.8 | 13.3 ± 1.8 | 0.22b |
CL, clearance.
Vmax is expressed as picomoles per minute per milligram of protein.
Calculated on the basis of the CYP3A4 protein level as determined by Western blot (Lin et al., 2002).
Inhibition of 25OHD3 4-Hydroxylation by DHB and KTZ.
To further confirm that the 4-hydroxylation of 25OHD3 is catalyzed predominantly by CYP3A4, the effects of a mechanism-based inhibitor DHB and a competitive inhibitor KTZ were evaluated using recombinant CYP3A4 and HLM. The results showed that preincubation with 50 μM DHB for 30 min blocked more than 97% of the 4-hydroxylation activities of both CYP3A4 and HLM (Table 4). Likewise, coincubation of 0.5 μM KTZ with 25OHD3 resulted in >80% loss of 4-hydroxylation activity. In contrast, DHB did not inhibit the 24-hydroxylation of 25OHD3 by CYP24A1 under similar incubation conditions (data not shown). These results support the conclusion that CYP3A4 is the predominant microsomal enzyme catalyzing 4-hydroxylation of 25OHD3 in human liver.
TABLE 4.
Inhibition of 25OHD3 4-hydroxylation activity by CYP3A4 and HLM by DHB and KTZ
Experimental details are described under Materials and Methods.
| Recombinant CYP3A4 |
Human Liver Microsomes |
|||
|---|---|---|---|---|
| 4β,25(OH)2D3 | 4α,25(OH)2D3 | 4β,25(OH)2D3 | 4α,25(OH)2D3 | |
| % of control | ||||
| DHB | 0.34 ± 0.04 | 0.77 ± 0.19 | 2.61 ± 0.61 | 2.90 ± 0.04 |
| KTZ | 17.6 ± 3.38 | 18.5 ± 2.28 | 11.0 ± 1.82 | 18.5 ± 5.61 |
Metabolism of 25OHD3 in Human Hepatocytes.
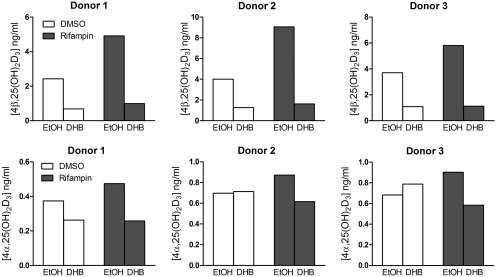
We also evaluated the contribution of CYP3A4 to the 4-hydroxylation of 25OHD3 in primary human hepatocytes incubated with 5 μM 25OHD3 for 24 h with and without pretreatment with rifampin (10 μM). The CYP3A4 mRNA level in hepatocytes was significantly increased above baseline by incubation with rifampin for 48 h (rifampin 43.6 ± 9.5 versus DMSO 1.02 ± 0.06). Both 4β,25(OH)2D3 and 4α,25(OH)2D3 metabolites were detected under control (vehicle-pretreated) and induced (rifampin-pretreated) conditions. On average, 4β,25(OH)2D3 formation compared with 4α,25(OH)2D3 formation was found to be approximately 6-fold higher under control conditions and 9-fold higher under induced conditions (Fig. 4). Cotreatment of control and rifampin-pretreated human hepatocytes with DHB resulted in 70 and 81% reductions of 4β,25(OH)2D3 formation, respectively. Of interest, DHB had no consistent effect on 4α,25(OH)2D3 formation in control cells but elicited a 30 to 56% reduction of metabolite formation in rifampin-pretreated cells (Fig. 4).
Fig. 4.
Metabolism of 25OHD3 in primary human hepatocytes. Cells were treated with rifampin (10 μM) or vehicle (0.1% DMSO) for 48 h and then preincubated with DHB (20 μM) or vehicle [0.1% ethanol (EtOH)] to block CYP3A4 activity. Experimental details are shown under Materials and Methods. The culture medium was collected and pooled for LC-MS/MS analysis.
4β,25(OH)2D3 Is a Newly Identified Circulating Metabolite in Human Plasma.
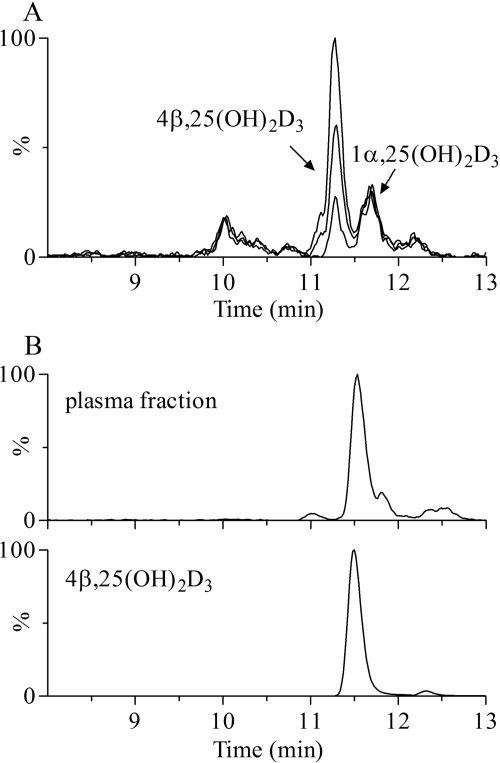
During the course of developing a sensitive LC-MS/MS assay for the quantification of 1α,25(OH)2D3, an endogenous interfering peak was observed during chromatography of human plasma extracts (Supplemental Fig. 2A). On the basis of its mass spectrum and sensitivity to periodate cleavage, this unknown appeared to be a metabolite of vitamin D3 that bore two hydroxyl groups on the A-ring (Wang et al., 2011). It was hypothesized, therefore, that the interfering peak corresponded to 2,25-dihydroxyvitamin D3 or 4,25(OH)2D3. Two additional experiments described here support the conclusion that this unknown is 4β,25(OH)2D3. As shown in Fig. 5A, purified 4β,25(OH)2D3 coeluted with the unknown, and the intensity of the mass spectrometric signal increased in proportion to the amount of 4β,25(OH)2D3 added to the plasma. In addition, we successfully isolated this compound from 350 ml of human plasma. After LLE and HPLC purification, the isolated fractions from repeated injections were subjected to LC-MS/MS analysis. The ion current chromatograms presented in Fig. 5B showed that the material isolated from plasma exhibited HPLC and MRM characteristics identical to those of 4β,25(OH)2D3 purified from the in vitro biosynthesis reaction.
Fig. 5.
Detection of 4β,25(OH)2D3 in human plasma. A, LC-MS/MS-based ion current chromatograms of native plasma and native plasma spiked with isolated 4β,25(OH)2D3 (50 and 100 pg). B, ion current chromatograms of HPLC-isolated fractions from native plasma (350 ml) and 4β,25(OH)2D3 standard. MRM was performed by monitoring the transition m/z 574 → 314.
With use of a novel LC-MS/MS assay, the plasma concentration of 4β,25(OH)2D3 was measured in 25 healthy adult volunteers. As reported previously (Wang et al., 2011), the average concentration of 4β,25(OH)2D3 was ∼40 pg/ml, which was comparable to the level of 1α,25(OH)2D3 (60 pg/ml) detected in the same subjects. Interestingly, the levels of 4β,25(OH)2D3 were highly variable among the study population, ranging from 2 to 128 pg/ml, but correlated well with the plasma levels of 25OHD3 (R2 = 0.73) (Supplemental Fig. 3). In agreement with its much lower rate of formation in human hepatocytes, 4α,25(OH)2D3 was detectable in some but not all of the study subjects (data not shown).
Interliver Differences in the Formation of 4β,25(OH)2D3 and 4α,25(OH)2D3 from 25OHD3.
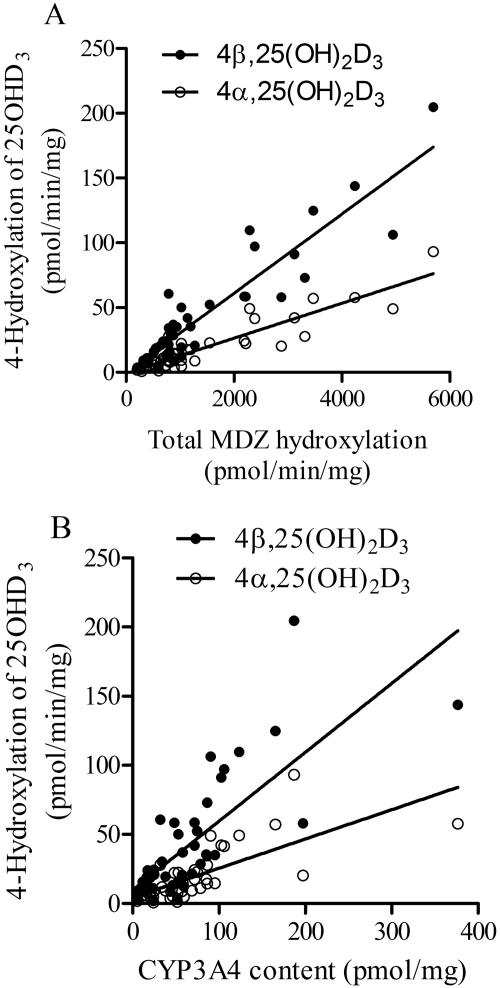
Because the formation of both 4β,25(OH)2D3 and 4α,25(OH)2D3 from 25OHD3 appears to be catalyzed predominantly by CYP3A4, we investigated whether the formation rates of these metabolites were correlated with CYP3A4 activity or content in livers from different donors. Preparations of HLM from 42 individual donors were used, in which the CYP3A4 activity was established using the total MDZ hydroxylation rate, and the enzyme content was determined by Western blot analysis (Lin et al., 2002). We incubated this panel of HLM with 25OHD3 to measure the rates of 4β,25(OH)2D3 and 4α,25(OH)2D3 formation. As shown in Fig. 6, the rates of 4β- and 4α-hydroxylation were highly variable, ranging from 1.9 to 204 and from 0.7 to 93 pmol · min−1 · mg protein−1, respectively. Both rates were correlated with the total rate of MDZ 1′- and 4-hydroxylation (4β,25(OH)2D3 r2 = 0.86; 4α,25(OH)2D3 r2 = 0.83), as well as CYP3A4 protein content (4β,25(OH)2D3 r2 = 0.59; 4α,25(OH)2D3 r2 = 0.53). As expected, the rates of 4β- and 4α-hydroxylation were highly correlated with each other (r2 = 0.99), indicating that the metabolites were being produced by the same enzyme in HLM.
Fig. 6.
Correlation between rates of 4-hydroxylation of 25OHD3 and CYP3A4 activity (A)/content (B) in HLM from different liver tissues (n = 42). CYP3A4 activity was expressed as the sum of 1′- and 4-hydroxylation of MDZ, and content was determined by Western blot (Lin et al., 2002).
Induction of 4β,25(OH)2D3 Formation after Oral Doses of Rifampin.
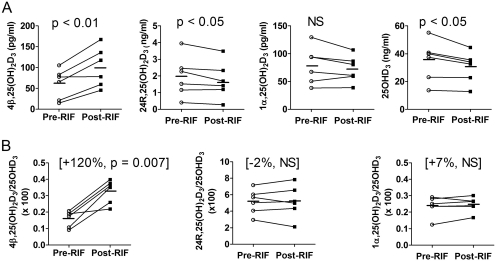
We analyzed plasma concentrations of 25OHD3 and three of its primary metabolites: 1α,25(OH)2D3, 24R,25(OH)2D3, and 4β,25(OH)2D3 (Supplemental Fig. 2B). Short-term treatment with rifampin for 7 days caused a slight reduction in the concentration of 25OHD3 (−11%, p < 0.05) and in the 24R,25(OH)2D3 level (−13%, p < 0.05). Although the average plasma concentration of 1α,25(OH)2D3 also declined with rifampin treatment, the change was not significant. In contrast, there was a significant, nearly 2-fold increase in the plasma concentration of 4β,25(OH)2D3 (Fig. 7A). To evaluate the effect of rifampin on the respective 25OHD3 metabolite formation clearances, we calculated the metabolite/parent concentration ratio for each pathway and compared the individual mean differences. As seen in Fig. 7B, an increase in the 4β,25(OH)2D3/25OHD3 ratio in all subjects was observed. The ratio change varied between 15 and 224%, with a mean increase of 120%, and the drug effect was highly significant (p < 0.001). In contrast, there was no significant change in the metabolite/parent concentration ratio for the 24R,25(OH)2D3 and 1α,25(OH)2D3 pathways. Assuming that rifampin does not cause a change in the clearance of the 25OHD3 metabolites, the data suggest that rifampin caused selective induction of the 4β,25(OH)2D3 formation clearance pathway, an effect mediated presumably by CYP3A4 induction.
Fig. 7.
Effect of short-term rifampin (RIF) treatment (600 mg. once a day, for 7 days) on the plasma concentrations of dihydroxyvitamin D metabolites (A) and the metabolite to 25OHD3 concentration ratios (B). Individual mean concentrations and ratios at baseline (average of day 0 and day 1, pre-RIF) and after rifampin treatment (average of day 7 and day 8, post-RIF) from six subjects are shown. Bars indicate the mean value for the group. The p value was calculated by paired t test. NS, not significant.
Biomarker Analysis of Mineral Homeostasis.
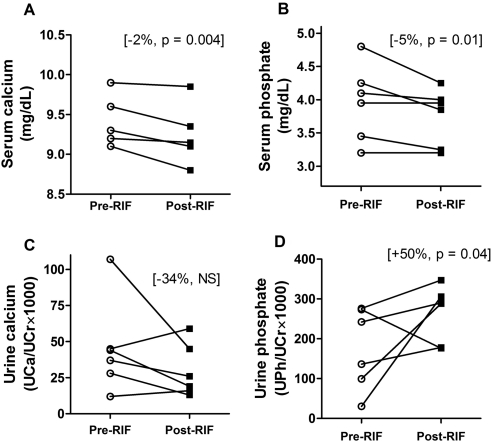
For the same six study subjects, we measured serum intact parathyroid hormone, ionized calcium, total calcium, and phosphate concentrations and urine calcium and phosphate concentrations (normalized for creatinine concentration) before and after rifampin treatment. The majority of these measurements were found within a normal physiological range, which was expected, given the health of the subjects and such a short-term treatment with rifampin. However, the results showed important changes after rifampin treatment: all six subjects had decreased serum calcium levels (Fig. 8A), four of them had reduced serum phosphate levels with no change in the other two subjects (Fig. 8B), four subjects had decreased creatinine-normalized urine calcium (Fig. 8C), and five of six subjects had increased creatinine-normalized urine phosphate (Fig. 8D). The magnitude of changes in the urine parameters was greater than that of serum parameters. Analyzing the individual prerifampin versus postrifampin treatment changes, we found that serum calcium levels were significantly decreased (total serum calcium declined by 1.9%; 9.27 versus 9.36 mg/dl, p = 0.004) and serum phosphate was reduced by 5% (3.75 versus 3.96 mg/dl, p = 0.01). In the urine, the only significant change was the urine creatinine-normalized phosphate concentration, which increased by 50% (0.18 versus 0.26, p = 0.04). The spot measurements of serum parathyroid hormone were unchanged by rifampin treatment.
Fig. 8.
Effect of short-term rifampin (RIF) treatment on individual serum and urine levels of calcium and phosphate. Serum calcium (A), serum phosphate (B), creatinine-normalized calcium (UCa/UCr × 1000) in spot urine (C), and creatinine-normalized phosphate (UPi/UCr × 1000) in spot urine (D) were measured in six subjects at baseline and after rifampin treatment. For each subject, the averages of two prerifampin measurements (day 0 and day 1) and two post-rifampin measurements (day 7 and day 8) are plotted. NS, not significant.
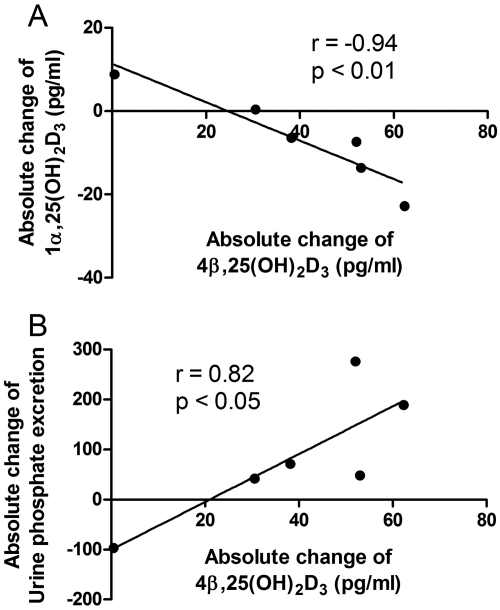
Although there were only six subjects in the study, we observed some interindividual differences in both vitamin D and mineral homeostasis data. Of importance, we noted that the subject with the smallest increase in 4β,25(OH)2D3 formation also stood out with regard to urinary phosphate excretion (decreased, instead of an increase for the other five subjects) (Fig. 8). Moreover, the same subject also had the lowest baseline plasma 1α,25(OH)2D3 level, and it increased 35% with rifampin treatment (Fig. 9A). Overall, there was a significant correlation between the absolute change in plasma 4β,25(OH)2D3 concentration after rifampin treatment and the absolute change in the plasma 1α,25(OH)2D3 concentration (r = −0.94, p < 0.01) and urinary phosphate excretion (r = 0.84, p < 0.05) (Fig. 9).
Fig. 9.
Correlation between absolute changes in plasma 4β,25(OH)2D3 concentration with absolute changes in the plasma 1α,25(OH)2D3 level (A) and urinary phosphate excretion (B) from six subjects. The absolute concentration change was calculated as the postrifampin minus prerifampin concentrations. Sample correlation coefficients and corresponding p values were calculated; the null hypothesis that the population correlation is zero was tested with a t test, under the assumption that the population was bivariate normally distributed.
Discussion
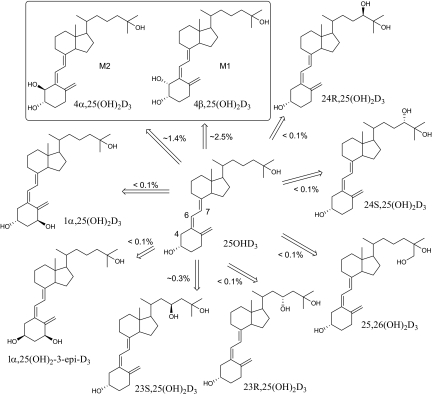
Metabolism of the major circulating vitamin D3 metabolite, 25OHD3 plays as a key role in vitamin D3 homeostasis. In general, the 1α-hydroxylation of 25OHD3 represents the final activation step, whereas side chain oxidations of 25OHD3 and 1α,25(OH)2D3 by CYP24A1 are the major inactivation pathways (Christakos et al., 2010). In a previous study, we reported that CYP3A4 also catalyzes 23-/24-hydroxylations of 1α,25(OH)2D3, which provides an alternative pathway for the inactivation of vitamin D3 (Xu et al., 2006). In the present study, we demonstrated that CYP3A4 also catalyzes 25OHD3 metabolism and produces nine monohydroxylated metabolites in vitro (Fig. 10). Seven minor metabolites were identified by comparison with authentic standards; these included the 23-, 24-, and 26-hydroxy products of the alkyl side chain. Production of these minor hydroxylated products of 25OHD3 [23R,25(OH)2D3, 23S,25(OH)2D3, 24R,25(OH)2D3, 24S,25(OH)2D3, and 25, 26(OH)2D3] by CYP3A4 is consistent with previous reports demonstrating that CYP3A4 can oxidize the vitamin D3 core structure at C-23, C-24, and C-26 positions (Xu et al., 2006). Although the relative efficiency of their formation was very low, formation of 1α,25(OH)2D3 identified in this study could possibly represent an alternative paracrine activation pathway because of the large abundance of CYP3A4 in liver and small intestine. Recent studies indicate that CYP27B1 can be expressed extrarenally in many normal tissues and under pathological situations and therefore could be responsible for the extrarenal formation of 1α,25(OH)2D3 (Hewison et al., 2000, 2007). However, the possibility that CYP3A4 might contribute to the formation of 1α,25(OH)2D3 as well cannot be excluded, particularly in tissues in which CYP3A4 is highly expressed.
Fig. 10.
Proposed metabolic profile of 25OHD3 by recombinant CYP3A4 in vitro. The relative percentage of metabolite/25OHD3 was estimated using HPLC-UV, assuming that each metabolite has the same UV absorption extinction coefficient at 265 nm. In vitro incubation conditions are described under Materials and Methods.
The two major metabolites of 25OHD3 produced by CYP3A4 were 4β,25(OH)2D3 and 4α,25(OH)2D3, and CYP3A4 is the dominant source of these A-ring hydroxylated products in human liver. This is supported by the fact that both 4β,25(OH)2D3 and 4α,25(OH)2D3 were generated by recombinant CYP3A4 and HLM. Moreover, the HLM reaction was inhibited substantially by addition of the selective CYP3A4 inhibitors DHB and KTZ. When other microsomal P450s were screened, only CYP3A5 catalyzed the 4-hydroxylation reactions, although the formation rates for CYP3A5 were much lower than those for CYP3A4. Using HLM isolated from 42 different liver tissues, we determined that the formation rates of 4β,25(OH)2D3 and 4α,25(OH)2D3 were well correlated with CYP3A4 MDZ hydroxylation and protein expression.
In contrast, we found that the mitochondrial enzyme, CYP24A1, lacked any detectable 4-hydroxylase activity. Incubation of primary human hepatocytes with DHB, a CYP3A4 selective, mechanism-based inhibitor, effectively inhibited the 4β-hydroxylation reaction, indicating the absence of any other appreciable source of 4β,25(OH)2D3 formation. However, there was a discrepancy between microsomal and hepatocyte 4α,25(OH)2D3 formation with regard to the inhibitory effect of DHB. The CYP3A4 inhibitor decreased 4α,25(OH)2D3 formation in HLM but was less effective in human hepatocytes under basal conditions, although the inhibitory effect was seen after rifampin induction. We do not have an explanation for the discrepancy, but note that the relative formation of the 4α-metabolite in hepatocytes was much less than that predicted by recombinant enzyme and microsomal activities. It is possible that most of the metabolite, once formed by CYP3A4, is rapidly cleared by another metabolic pathway (i.e., sequential oxidation or conjugation) and that the residual amounts detected in the control hepatocyte incubations do not derive from CYP3A4. Similar to human plasma, 4α,25(OH)2D3 was detectable in some but not all of the study subjects, and its level was relatively low.
4β,25(OH)2D3 was also identified as an endogenous circulating metabolite in human plasma. Because of its structural similarity to 1α,25(OH)2D3, a new LC-MS/MS method was developed to chromatographically separate and quantitate 4β,25(OH)2D3, 4α,25(OH)2D3, and 1α,25(OH)2D3 (Wang et al., 2011). In 25 healthy subjects, the mean levels of 4β,25(OH)2D3 (40 pg/ml) were comparable to that of 1α,25(OH)2D3 (60 pg/ml) but much less than those of 25OHD3 (25.6 ng/ml) and 24R,25(OH)2D3 (2.3 ng/ml). Although values of 4β,25(OH)2D3 and 25OHD3 correlated with each other, the ratio of 4β,25(OH)2D3 to 25OHD3 was still variable, possibly due to differing in vivo CYP3A4 activities in each subject. Indeed, we observed significant interindividual differences in 4β,25(OH)2D3 formation using HLM prepared from different liver donors. Thus, it is possible that 4β,25(OH)2D3 might be an endogenous biomarker for systemic CYP3A4 activity, which remains to be explored.
Although a novel finding, the production of A-ring hydroxylated metabolites from 25OHD3 by CYP3A4 is not surprising. Thierry-Palmer et al. (1988) reported that 25OHD3 was hydroxylated at the C-2 or C-4 position by rat renal microsomes in vitro, because the product was sensitive to periodate cleavage. Rao et al. (1999) isolated and identified a similar vitamin D2 metabolite, 4,25(OH)2D2, from serum in vitamin D2-intoxicated rats. In addition, Takeda et al. (2006) reported that 2α,25(OH)2D3 was produced from microbial hydroxylation of vitamin D3 by Pseudonocardia autotrophica. Finally, Araya et al. (2003) recently reported that 4β,25(OH)2D3 was produced in vitro by recombinant human CYP27A1, a hepatic mitochondrial vitamin D3 25-hydroxylase. However, the maximal rate of conversion of 25OHD3 to 4β,25(OH)2D3 by CYP27A1 (0.024 pmol · min−1 · mg protein−1) was much lower than what we observed for CYP3A4 in the present study (6.4 pmol · min−1 · mg protein−1). This does not necessarily equate to a difference in the formation clearance, but we found that DHB pretreatment of human hepatocytes greatly diminished the 4β-hydroxylation reaction, demonstrating the dominance of CYP3A4 in the metabolic process. It remains to be determined whether the CYP3A4 catalyzed 4-hydroxylation reaction contributes significantly to the clearance of 25OHD3 in vivo. However, the formation clearance of 4β,25(OH)2D3 by CYP3A4 is considerable and although circulating levels of 4β,25(OH)2D3 are far less than those of 24R,25(OH)2D3, this difference could be the result of much more rapid clearance of 4β,25(OH)2D3 than 24R,25(OH)2D3, rather than more efficient formation of 24R,25(OH)2D3.
Long-term treatment with certain drugs, e.g., rifamycins and barbiturates, has been associated with altered bone metabolism and decreased bone density (Shah et al., 1981; Pack et al., 2004). The adverse effect has been attributed to a dysregulation of vitamin D homeostasis and the transcription of some of the genes that it controls (e.g., intestinal TRPV6), principally through a drug-induced enhancement of 25OHD3 and/or 1α,25(OH)3D3 catabolic clearance (Pascussi et al., 2005; Xu et al., 2006; Zhou et al., 2006). Results from our pilot study showed that short-term rifampin treatment selectively induced CYP3A4-dependent 4β,25(OH)2D3 formation, while having no appreciable effect on the CYP24A1- and CYP27B1-catalyzed pathways of 25OHD3 elimination. Moreover, we observed a decline in the absolute plasma 25OHD3 level with short-term rifampin treatment, which is consistent with the more pronounced reduction in hormone level seen with a longer (2-week) duration of rifampin treatment (Brodie et al., 1980, 1982). In addition, the short duration of rifampin treatment (7 days) was associated with significant changes in the renal excretion of inorganic phosphate, as well as with plasma calcium and phosphate concentrations, suggesting the possibility of a significant reduction in the absorption of calcium as a result of enhanced intestinal vitamin D metabolism.
There are several caveats to our interpretation of the clinical study results beyond the relatively small subject number. First, the dietary intake of calcium and phosphate was uncontrolled, potentially introducing interday dietary noise to the biochemical measurements. In addition, we used creatinine-normalized calcium and phosphate measurements from spot urine samples as an approximation of the 24-h calcium and phosphate excretion rates. This introduces uncertainties about intraday variation (Gökçe et al., 1991; Topal et al., 2008). It is also possible that longer term treatment with rifampin could produce different effects, in particular, the 24R,25(OH)2D3 metabolite if it has a plasma half-life much longer than 7 days. These limitations can be easily addressed in a longer duration, prospective investigation, with a sample size powered on the basis of the mean effects and variation observed in the current pilot study.
We also note that the enhancement of vitamin D clearance by drugs such as rifampin and increased risk of drug-induced osteomalacia has been attributed to a PXR-mediated activation of CYP24A1 gene transcription (Pascussi et al., 2005). However, we have suggested previously that activation of CYP3A4 transcription represents an alternative or complementary mechanism behind the adverse effect (Xu et al., 2006; Zhou et al., 2006). Results from the current investigation support the hypothesis that CYP3A4 induction and enhanced 4β,25(OH)2D3 formation and not CYP24A1 induction and enhanced 24R,25(OH)2D3 formation contributes to the reduction in 25OHD3 levels after chronic treatment with PXR agonists and increases the risk of osteomalacia.
In summary, CYP3A4 is able to catalyze 25OHD3 hydroxylation at multiple positions. One of the major products, 4β,25(OH)2D3, a newly identified circulating metabolite, is produced predominantly by CYP3A4 in human liver and might be an endogenous biomarker for assessment of CYP3A4 activity in vivo. In addition, short-term treatment with the PXR agonist rifampin selectively induced CYP3A4-dependent 4β,25(OH)2D3 formation and altered systemic mineral homeostasis in vivo. These changes may underlie the increased risk of osteomalacia associated with chronic administration of rifampin and other PXR agonists. Whether the 4β/α-hydroxy metabolites possess biological activities, as do other A-ring-oxidized metabolites, remains to be explored.
Supplementary Material
Acknowledgments
We thank Dr. Toshie Fujishima for the gift of 23S,25(OH)2D3 and Dr. Toshiyuki Sakaki for the gift of 1α,25(OH)2-3-epi-D3 and incubation of 25OHD3 with CYP24A1. We also thank Michelle Doyle for assistance in the conduct of the clinical study.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM063666, P01-GM032165]; National Institutes of Health National Institute of Environmental Health Sciences [Grant P30-ES07033]; and National Institutes of Health National Institute of National Center for Research Resources [Grant GUL1-RR025014].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
- 25OHD3
- 25-hydroxyvitamin D3
- 1α,25(OH)2D3
- 1α,25-dihydroxyvitamin D3
- PXR
- pregnane X receptor
- DHB
- 6′,7′-dihydroxybergamottin
- KTZ
- ketoconazole
- 24R,25(OH)2D3
- 24R,25-dihydroxyvitamin D3
- PTAD
- 4-phenyl-1,2,4-triazoline-3,5-dione
- 23R,25(OH)2D3
- 23R,25-dihydroxyvitamin D3
- 24S,25(OH)2D3
- 24R,25-dihydroxyvitamin D3
- 25,26(OH)2D3
- 25,26-dihydroxyvitamin D3
- 23S,25(OH)2D3
- 23S,25-dihydroxyvitamin D3
- 1α,25(OH)2-3-epi-D3
- 1α,25-dihydroxy-3-epi-vitamin D3
- HLM
- human liver microsome(s)
- 4,25(OH2)D3
- 4,25-dihydroxyvitamin D3
- 4α,25(OH)2D3
- 4α,25-dihydroxyvitamin D3
- 4β,25(OH)2D3
- 4β,25-dihydroxyvitamin D3
- LLE
- liquid-liquid extraction
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- HPLC
- high-performance liquid chromatography
- MRM
- multiple reaction monitoring
- GC
- gas chromatography
- MS
- electron impact mass spectrometry
- MDZ
- midazolam
- DMSO
- dimethyl sulfoxide.
Authorship Contributions
Participated in research design: Wang, Lin, Zheng, Hebert, Davis, and Thummel.
Conducted experiments: Wang, Lin, Zheng, Senn, Hashizume, Scian, Davis, and Dickmann.
Contributed new reagents or analytic tools: Wang, Lin, and Senn.
Performed data analysis: Wang, Lin, Zheng, Senn, Hashizume, Scian, Dickmann, Nelson, Baillie, Blough, and Thummel.
Wrote or contributed to the writing of the manuscript: Wang, Lin, Nelson, Baillie, Hebert, Blough, and Thummel.
References
- Alexander BH, Dimler RJ, Mehltretter CL. (1951) d-Galactosan(1,4)α(1,6): its structure and resistance to periodate oxidation. J Am Chem Soc 73:4658–4659 [Google Scholar]
- Araya Z, Hosseinpour F, Bodin K, Wikvall K. (2003) Metabolism of 25-hydroxyvitamin D3 by microsomal and mitochondrial vitamin D3 25-hydroxylases (CYP2D25 and CYP27A1): a novel reaction by CYP27A1. Biochim Biophys Acta 1632:40–47 [DOI] [PubMed] [Google Scholar]
- Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, Lieben L, Mathieu C, Demay M. (2008) Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev 29:726–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie MJ, Boobis AR, Dollery CT, Hillyard CJ, Brown DJ, MacIntyre I, Park BK. (1980) Rifampicin and vitamin D metabolism. Clin Pharmacol Ther 27:810–814 [DOI] [PubMed] [Google Scholar]
- Brodie MJ, Boobis AR, Hillyard CJ, Abeyasekera G, Stevenson JC, MacIntyre I, Park BK. (1982) Effect of rifampicin and isoniazid on vitamin D metabolism. Clin Pharmacol Ther 32:525–530 [DOI] [PubMed] [Google Scholar]
- Christakos S, Ajibade DV, Dhawan P, Fechner AJ, Mady LJ. (2010) Vitamin D: metabolism. Endocrinol Metab Clin North Am 39:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca HF. (1988) The vitamin D story: a collaborative effort of basic science and clinical medicine. FASEB J 2:224–236 [PubMed] [Google Scholar]
- DeLuca HF. (2008) Evolution of our understanding of vitamin D. Nutr Rev 66 (10 Suppl 2):S73–S87 [DOI] [PubMed] [Google Scholar]
- Dumaswala R, Setchell KD, Zimmer-Nechemias L, Iida T, Goto J, Nambara T. (1989) Identification of 3α,4β,7α-trihydroxy-5β-cholanoic acid in human bile: reflection of a new pathway in bile acid metabolism in humans. J Lipid Res 30:847–856 [PubMed] [Google Scholar]
- Eguchi T, Ikekawa N. (1990) Conformational analysis of 1α,25-dihydroxyvitamin D3 by nuclear magnetic resonance. Bioorg Chem 18:19–29 [Google Scholar]
- Gökçe C, Gökçe O, Baydinç C, Ilhan N, Alaşehirli E, Ozküçük F, Taşçi M, Atilkeler MK, Celebi H, Arslan N. (1991) Use of random urine samples to estimate total urinary calcium and phosphate excretion. Arch Intern Med 151:1587–1588 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ. (2007) CYP3A4 and pregnane X receptor humanized mice. J Biochem Mol Toxicol 21:158–162 [DOI] [PubMed] [Google Scholar]
- Gupta RP, He YA, Patrick KS, Halpert JR, Bell NH. (2005) CYP3A4 is a vitamin D-24- and 25-hydroxylase: analysis of structure function by site-directed mutagenesis. J Clin Endocrinol Metab 90:1210–1219 [DOI] [PubMed] [Google Scholar]
- Gupta RP, Hollis BW, Patel SB, Patrick KS, Bell NH. (2004) CYP3A4 is a human microsomal vitamin D 25-hydroxylase. J Bone Miner Res 19:680–688 [DOI] [PubMed] [Google Scholar]
- Hewison M, Burke F, Evans KN, Lammas DA, Sansom DM, Liu P, Modlin RL, Adams JS. (2007) Extra-renal 25-hydroxyvitamin D3-1α-hydroxylase in human health and disease. J Steroid Biochem Mol Biol 103:316–321 [DOI] [PubMed] [Google Scholar]
- Hewison M, Zehnder D, Bland R, Stewart PM. (2000) 1α-Hydroxylase and the action of vitamin D. J Mol Endocrinol 25:141–148 [DOI] [PubMed] [Google Scholar]
- Holick MF. (2007) Vitamin D deficiency. N Engl J Med 357:266–281 [DOI] [PubMed] [Google Scholar]
- Kamachi S, Sugimoto K, Yamasaki T, Hirose N, Ide H, Ohyama Y. (2001) Metabolic activation of 1α-hydroxyvitamin D3 in human liver microsomes. Xenobiotica 31:701–712 [DOI] [PubMed] [Google Scholar]
- Kamao M, Tatematsu S, Hatakeyama S, Sakaki T, Sawada N, Inouye K, Ozono K, Kubodera N, Reddy GS, Okano T. (2004) C-3 epimerization of vitamin D3 metabolites and further metabolism of C-3 epimers: 25-hydroxyvitamin D3 is metabolized to 3-epi-25-hydroxyvitamin D3 and subsequently metabolized through C-1α or C-24 hydroxylation. J Biol Chem 279:15897–15907 [DOI] [PubMed] [Google Scholar]
- Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, Schuetz EG, Thummel KE. (2002) Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol 62:162–172 [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Yamasaki T. (2004) Eight cytochrome P450s catalyze vitamin D metabolism. Front Biosci 9:3007–3018 [DOI] [PubMed] [Google Scholar]
- Pack AM, Gidal B, Vazquez B. (2004) Bone disease associated with antiepileptic drugs. Cleve Clin J Med 71 (Suppl 2):S42–S48 [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Robert A, Nguyen M, Walrant-Debray O, Garabedian M, Martin P, Pineau T, Saric J, Navarro F, Maurel P, et al. (2005) Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J Clin Invest 115:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW. (1991) Vitamin D3 receptors: structure and function in transcription. Annu Rev Nutr 11:189–216 [DOI] [PubMed] [Google Scholar]
- Pike JW, Meyer MB. (2010) The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D3. Endocrinol Metab Clin North Am 39:255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum LA, DeLuca HF. (2010) Vitamin D, disease and therapeutic opportunities. Nat Rev Drug Discov 9:941–955 [DOI] [PubMed] [Google Scholar]
- Prosser DE, Jones G. (2004) Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci 29:664–673 [DOI] [PubMed] [Google Scholar]
- Rao DS, Dayal R, Siu-Caldera ML, Horst RL, Uskokovic MR, Tserng KY, Reddy GS. (1999) Isolation and identification of 4,25-dihydroxyvitamin D2: a novel A-ring hydroxylated metabolite of vitamin D2. J Steroid Biochem Mol Biol 71:63–70 [DOI] [PubMed] [Google Scholar]
- Rosen CJ. (2011) Clinical practice. Vitamin D insufficiency. N Engl J Med 364:248–254 [DOI] [PubMed] [Google Scholar]
- Sakaki T, Sawada N, Komai K, Shiozawa S, Yamada S, Yamamoto K, Ohyama Y, Inouye K. (2000) Dual metabolic pathway of 25-hydroxyvitamin D3 catalyzed by human CYP24. Eur J Biochem 267:6158–6165 [DOI] [PubMed] [Google Scholar]
- Shah SC, Sharma RK, Hemangini, Chitle AR. (1981) Rifampicin induced osteomalacia. Tubercle 62:207–209 [DOI] [PubMed] [Google Scholar]
- Sutton AL, MacDonald PN. (2003) Vitamin D: more than a “bone-a-fide” hormone. Mol Endocrinol 17:777–791 [DOI] [PubMed] [Google Scholar]
- Takeda K, Kominato K, Sugita A, Iwasaki Y, Shimazaki M, Shimizu M. (2006) Isolation and identification of 2α,25-dihydroxyvitamin D3, a new metabolite from Pseudonocardia autotrophica 100U-19 cells incubated with vitamin D3. Steroids 71:736–744 [DOI] [PubMed] [Google Scholar]
- Thierry-Palmer M, Gray TK, Napoli JL. (1988) Ring hydroxylation of 25-hydroxycholecalciferol by rat renal microsomes. J Steroid Biochem 29:623–628 [DOI] [PubMed] [Google Scholar]
- Topal C, Algun E, Sayarlioglu H, Erkoc R, Soyoral Y, Dogan E, Sekeroglu R, Cekici S. (2008) Diurnal rhythm of urinary calcium excretion in adults. Ren Fail 30:499–501 [DOI] [PubMed] [Google Scholar]
- Wang Z, Senn T, Kalhorn T, Zheng XE, Zheng S, Davis CL, Hebert MF, Lin YS, Thummel KE. (2011) Simultaneous measurement of plasma vitamin D3 metabolites, including 4β,25-dihydroxyvitamin D3, using liquid chromatography-tandem mass spectrometry. Anal Biochem 418:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hashizume T, Shuhart MC, Davis CL, Nelson WL, Sakaki T, Kalhorn TF, Watkins PB, Schuetz EG, Thummel KE. (2006) Intestinal and hepatic CYP3A4 catalyze hydroxylation of 1α,25-dihydroxyvitamin D3: implications for drug-induced osteomalacia. Mol Pharmacol 69:56–65 [DOI] [PubMed] [Google Scholar]
- Zhang R, Naughton DP. (2010) Vitamin D in health and disease: current perspectives. Nutr J 9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Assem M, Tay JC, Watkins PB, Blumberg B, Schuetz EG, Thummel KE. (2006) Steroid and xenobiotic receptor and vitamin D receptor crosstalk mediates CYP24 expression and drug-induced osteomalacia. J Clin Invest 116:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF. (2008) Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab 9:310–322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.