Abstract
Prior studies demonstrated that resistance to the ERBB1/2 inhibitor lapatinib could be overcome by the B cell CLL/lymphoma-2 (BCL-2) family antagonist obatoclax (GX15-070). Coadministration of lapatinib with obatoclax caused synergistic cell killing by eliciting autophagic cell death that was dependent upstream on mitochondrial reactive oxygen species generation and increased p62 levels and downstream on activation of p38 mitogen-activated protein kinase and inactivation of mammalian target of rapamycin. By immunohistochemical analysis, in drug combination-treated cells, microtubule-associated protein light chain 3 (LC3) associated with mitochondrial (cytochrome c oxidase), autophagosome (p62), and autolysosome (lysosomal associated membrane protein 2) proteins. Treatment of cells with 3-methyladenine or knockdown of beclin 1 was protective, whereas chloroquine treatment had no protective effect. Expression of myeloid cell leukemia-1 (MCL-1), compared with that of BCL-2 or BCL-2-related gene long isoform, protected against drug combination lethality. Lapatinib and obatoclax-initiated autophagy depended on NOXA-mediated displacement of the prosurvival BCL-2 family member, MCL-1, from beclin 1, which was essential for the initiation of autophagy. Taken together, our data argue that lapatinib and obatoclax-induced toxic autophagy is due to impaired autophagic degradation, and this disturbance of autophagic flux leads to an accumulation of toxic proteins and loss of mitochondrial function.
Introduction
Human epidermal growth factor receptor type 2 (HER2 or ErbB2) belongs to ErbB family of receptors, which consists of four members: ErbB1 (also known as epidermal growth factor receptor), ErbB2 (HER2), ErbB3 (HER3), and ErbB4 (HER4) (Olayioye et al., 2000; Yarden and Sliwkowski, 2001). As a transmembrane receptor tyrosine kinase, HER2 can homo- or heterodimerize with other ErbB receptors upon ligands binding to their extracellular domain, leading to autophosphorylation of specific tyrosine residues on the cytosolic domain of the receptors. HER family members are highly expressed in various tumors including glioblastoma and head and neck, lung, esophageal, colorectal, ovarian, and prostate cancers (Salomon et al., 1995).
Dysregulated ErbB receptor activities are associated with cancer development, progression, and resistance to antineoplastic treatment. Overexpression or constant activation of HER receptors results in the engagement and activation of prosurvival signal transduction events such as such as phosphatidylinositol 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) pathways, which contribute to uncontrolled cell proliferation, enhanced angiogenesis and metastasis, and increased resistance to apoptosis (Hynes and Lane, 2005). Amplification of ErbB2 occurs in approximately 15 to 30% of primary breast cancers, inflammatory breast cancer, and invasive breast cancers (Slamon et al., 1989; Hobday and Perez, 2005). HER2-positive breast cancer is more aggressive and is associated with a higher incidence of therapeutic failure, disease recurrence, and death (Slamon et al., 1987; Lin and Winer, 2007). Therefore, identification of HER2 expression has been adopted as a predictive and prognostic marker for cancer, and manipulation of ErbB activities and their downstream pathways is an appealing therapeutic target for antitumor strategies.
Lapatinib (Tykerb; GlaxoSmithKline, Collegeville, PA) is a dual tyrosine kinase inhibitor of ErbB1 and ErbB2. Lapatinib has been approved for patient use in more than 90 countries worldwide for treatment of ErbB2-positive breast cancer and off-label for other cancers that overexpress ErbB2. In particular, it was adopted as a therapeutic agent for the treatment of patients with HER2-positive refractory advanced or metastatic breast cancer, who had received previous failed treatments such as trastuzumab, anthracyclines, and taxanes (Rusnak et al., 2001; Wood et al., 2004). In vitro and in vivo studies demonstrated that lapatinib was able to inhibit proliferation of HER2 and epidermal growth factor receptor-overexpressing cancer cells and in a few cell lines caused profound cell killing as a single agent (Rusnak et al., 2001; Xia et al., 2002; Konecny et al., 2006). Although lapatinib provides a new treatment option for the management of ErbB2-positive cancer, lapatinib monotherapy more frequently demonstrated only modest activity in intermediate HER2-positive breast cancer cells (Burstein et al., 2008).
A number of mechanisms could account for lapatinib treatment failure. Resistance to lapatinib could be caused by genetic/epigenetic alterations in tumor cells. Mutations in ErbB receptor kinase domains lead to the ligand-independent constitutive activation of the receptor, which abrogates the ability of lapatinib to suppress the kinase activities (Pao et al., 2005; Sok et al., 2006). Amplification of PI3K signaling can be a resistance factor as a result of PI3K gene mutation-induced constitutively activated PI3K or loss of tumor suppressor phosphatase and tensin homolog expression (Nagata et al., 2004; Berns et al., 2007; Serra et al., 2008) or overexpression of receptor tyrosine kinase Axl (Liu et al., 2009), compensatory HER3 expression (Amin et al., 2010). In addition, antiapoptotic molecules belonging to the BCL-2 family have also been linked to lapatinib resistance (Raffo et al., 1995). As with all drugs, lapatinib is metabolized by several enzymes such as cytochrome P450 members, CYP3A4 and CYP3A5, and can be secreted from cell ATP-binding cassette drug transporter excretion by P-glycoprotein (Szakács et al., 2006).
Radiotherapy and a spectrum of clinically or experimentally available chemotherapeutic agents have been reported to induce autophagy in cell lines and animal models (Kondo et al., 2005; Amaravadi and Thompson, 2007). Whether autophagy induced by antineoplastic therapies functions as a direct death execution approach or represents a self-defense mechanism for resisting therapy-mediated killing remains controversial. Autophagy can be substantially elevated when cells are exposed to certain types of therapeutic agents as an alternative survival strategy to delay programmed cell death. However, if the stresses persist, eventually essential cellular organelles are degraded and beyond a certain threshold, cell damage exceeds the capacity for cells to survive. In those cases, cells digest themselves completely by autophagy and undergo programmed cell death. The ability of certain chemotherapies to cause cell death in cancer cell lines that display resistance to apoptosis may be dependent on autophagy (Kondo et al., 2005). Nevertheless, there is no unique paradigm addressing the role of autophagy in antineoplastic therapies and autophagy may promote survival or death, depending on the therapeutic agents and on the stages, or contexts, of tumorigenesis.
Studies from our group showed that lapatinib-resistant cells increased expression of the prosurvival B cell CLL/lymphoma-2 (BCL-2) family members MCL-1 and BCL-XL and decreased expression of proapoptosis BCL-2 family members BAX and BAK (Martin et al., 2008, 2009). As an alternative to lapatinib monotherapy, coadministration of lapatinib with the BCL-2/BCL-XL/MCL-1 antagonist obatoclax (GX-15-070) attenuated lapatinib resistance and produced synergistic cancer cell killing by eliciting autophagic cell death in a wide range of human breast and colon cancer cells (Martin et al., 2008, 2009; Mitchell et al., 2010). However, molecular mechanisms by which obatoclax and lapatinib interact to cause toxic autophagy have not been fully described or understood.
Herein we find that obatoclax and lapatinib treatment caused a toxic form of autophagy that depends on mammalian target of rapamycin (mTOR) inhibition and p38 MAPK activation. Early autophagy vesicles were associated with mitochondria, suggestive of mito-autophagy taking place, which was supported by the fact that Rho zero cells were resistant to drug combination lethality. Inhibition of autophagy through either pharmacological [3-methyladenine (3-MA)] or genetic means (knockdown of ATG5 or Beclin1) attenuated cell death. Obatoclax and lapatinib treatment increased the level of NOXA, which displaced the prosurvival Bcl-2 family member, Mcl-1, from beclin 1 and therefore allowed for autophagy initiation. Abrogation of NOXA expression alleviated the drug-induced autophagy and cell death.
Materials and Methods
Materials.
Breast cancer cell lines BT474, MCF7, HCC38, BT549, and SKBR3 cell lines were purchased from American Type Culture Collection (Manassas, VA). The phoenix-Ampho packaging cell line was from Allele Biotechnology (San Diego, CA). RPMI 1640 and Dulbecco's modified Eagle's medium, antibiotics-antimycotics (100 units/ml penicillin, 100 μg/ml streptomycin, and 250 μg/ml amphotericin B), and trypsin-EDTA were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from HyClone (Logan, UT). The microtubule-associated protein light chain 3 (GFP-LC3) retrovirus expression construct, was kindly given by Dr. Jayanta Debnath from the University of California. BCL-2, BCL-XL, NOXA, and MCL-1 expression plasmids were purchased from Addgene (Cambridge, MA). Validated ATG5, beclin 1, and ATM short hairpin RNA molecules were purchased QIAGEN (Valencia, CA). The commercially validated shRNA plasmids against BAD, BIM, BID, or NOXA and antibodies against p62/SQSTM1, ATG7, and LAMP-2 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) (Supplemental Figs. S1–S3). All other antibodies were purchased from Cell Signaling Technology (Danvers, MA). All the secondary antibodies and Immobilon-FL polyvinylidene difluoride membrane were purchased from LI-COR Biosciences (Lincoln, NE). Obatoclax was provided by Gemin X Pharmaceuticals (Malvern, PA). Lapatinib was provided by GlaxoSmithKline (Collegeville, PA). Chloroquine, N-acetylcysteine (NAC), and 3-MA were purchased from Sigma-Aldrich (St. Louis, MO). Lipofectamine 2000 transfection reagent was purchased from Invitrogen. LysoTracker Red DND-99, MitoTracker Deep Red FM, MitoTracker Green FM, reactive oxygen species (ROS) detection reagent (carboxy-H2DCFDA), and an Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit were purchased from Invitrogen. The Lab-Tek II Chamber Slide System was purchased from Nalge Nunc International (Rochester, NY). The siPORT NeoFX Transfection Agent was purchased from Applied Biosystems/Ambion (Austin, TX).
Cell Culture and Transfection.
Phoenix-Ampho cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% antibiotic-antimycotic in a humidified incubator under an atmosphere containing 5% CO2 at 37°C. BT474, MCF7, HCC38, BT549, and SKBR3 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% antibiotic-antimycotic. A retroviral vector (pBABE) was used to generate cell lines stably expressing GFP-LC3. For this viral infection, approximately 10 million Phoenix-Ampho cells were transfected with the plasmid by using Lipofectamine 2000 transfection reagent. Viral supernatants were collected 48 h after transfection, diluted 1:1 in fresh medium in the presence of 4 μg/ml hexadimethrine (Polybrene), and added to target cells seeded in six-well plates at 30% confluence. After 24 h at 37°C, the viral supernatant was replaced with a fresh aliquot. Three sequential rounds of infection were performed for each condition, after which more than 90% of the cells expressed the exogenous proteins.
Transfection with siRNA.
siRNA transfection was performed with siPORT NeoFX Transfection Agent following the manufacturer's procedures. In brief, a 10 nM concentration of prevalidated siRNA was diluted into 50 μl of serum-free media. On the basis of the manufacturer's instructions, an appropriate amount of siPORT NeoFX Transfection Agent was diluted into a separate vial containing serum-free media. The two solutions were incubated separately at room temperature for 15 min and mixed together by pipetting up and down several times, and the mixture was added dropwise to the target cells. Twenty-four hours after transfection, the transfection medium was replaced with complete medium, and 12 h later the cells were subjected to treatments.
Generation of Rho Zero Cells.
Rho zero cells lack mitochondrial function. To achieve this, BT474 and MCF7 cells were cultured in 50 μM uridine and 1 mM sodium pyruvate, and the growth medium was supplemented with 10 μg/l of ethidium bromide. Cells were cultured in this medium for 1 month before any further experimentation. Rho zero cells exhibited little or no expression of mitochondrial proteins.
Western Blotting.
The breast cancer cells subjected to drug treatments were collected with whole cell lysis buffer (0.5 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, and 0.02% bromphenol blue) in the presence of a protease inhibitor cocktail (Sigma-Aldrich). The collected samples were sonicated and boiled for 5 min. The boiled samples were loaded onto 10 to 14% SDS-polyacrylamide gel electrophoresis gels and were fractionated in a Bio-Rad Protean II system. After proteins were transferred to an Immobilon-FL polyvinylidene difluoride membrane, the membrane was blocked with Odyssey Blocking buffer from LI-COR Biosciences for 60 min at room temperature and incubated with the primary antibody at appropriate dilutions in Odyssey Blocking buffer at 4°C overnight. After overnight incubation with appropriate primary antibodies, the membrane was washed (three times) with Tris-buffered saline-Tween 20 for a total of 15 min, probed with fluorescently labeled secondary antibody (1:5000) for 80 min at room temperature and washed (three times) with Tris-buffered saline-Tween 20 for a total of 15 min. The immunoblots were visualized by an Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunocytochemistry and Confocal Microscopy.
The breast cancer cells were seeded onto a Lab-Tek II Chamber Slide to 70% confluence. Twenty-hour hours later, the cells were applied to the treatment. To terminate the treatment, slides were fixed with 3% paraformaldehyde in PBS for 15 min, followed by permeabilization with 0.1% Triton X-100 for 5 min. After blocking in 3% bovine serum albumin-1× PBS containing 3% rabbit serum for 40 min, the slides were incubated with primary antibodies with appropriate dilution at 4°C overnight. After overnight incubation, the slides were washed three times with PBS and then incubated with Alexa Fluor 568 goat anti-mouse IgG (H+L) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) secondary antibody for 1.5 h (1:200 dilution). The slides were then washed three times with PBS, counterstained in Prolong Gold antifade reagent with 4,6-diamidino-2-phenylindole and visualized with a Zeiss LSM 510 META laser scanning confocal microscope.
Flow Cytometry for Mitochondrial Membrane Potential and ROS Detection.
The mitochondrial membrane potential (Δψ) of the treated BT474/MCF7 cells was measured using MitoTracker Deep Red FM staining. In brief, the cell culture medium was aspirated after treatment, and the cells were incubated with MitoTracker Deep Red FM (100 nM in RPMI 1640 without FBS) for 15 min at 37°C. The fluorescence emission was either detected under a microscope or analyzed by flow cytometry using a FACSVantage system. Mitochondrial mass was measured by MitoTracker Green FM staining. Mitochondrion staining by MitoTracker Green FM follows the same protocol as that for MitoTracker Deep Red FM staining. The fluorescence emission was detected by flow cytometry (excitation wavelength 488 nm, emission filter 530/30 nm). For ROS detection, the treated cells were incubated with carboxy-H2DCFDA (25 μM in RPMI 1640 without FBS) for 30 min at 37°C, protected from light. Dichlorofluorescin fluorescence was measured with a FACSCalibur flow cytometry system (excitation wavelength 488 nm, emission filter 530/30 nm).
Colony Formation Assay.
BT474 and MCF7 cells were plated in 12-well plates (∼400–1200 cells/well) and subjected to the indicated treatments, with drug being removed to terminate the treatment. Two weeks later, plates were washed in PBS, fixed with 100% methanol, and stained with a filtered solution of crystal violet (5% w/v). After washing with tap water, the colonies were counted both manually (by eye) and digitally using a ColCount plate reader (Oxford Optronix, Oxford, UK).
Cell Viability and Apoptotic Cell Death Measurement.
Cell viability was measured with Vi-CELL Series Cell Viability Analyzers (Beckman Coulter, Fullerton, CA), a procedure that is based on the traditional cell viability method of trypan blue exclusion. In addition, an Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit was used to determine cell viability by annexin V and propidium iodide (PI) staining-based flow cytometry. Alexa Fluor 488 annexin V and PI fluorescence were measured with a FACSCalibur flow cytometry system.
NOXA Promoter Activity Measurement.
Two NOXA promoter reporter constructs were purchased from Addgene. The promoter construct was cotransfected with a Rellina luciferase construct into target breast cancer cells. The promoter activities were measured using a Dual-Luciferase Reporter (DLR) Assay System (Promega).
Statistics.
The probability of statistically significant differences between two experimental groups was determined by Student's t test. p < 0.05 was considered statistically significant in all calculations.
Results
Lapatinib and Obatoclax Combination Treatment Causes Autophagy in ERBB2-Negative or Positive Breast Cancer Cells.
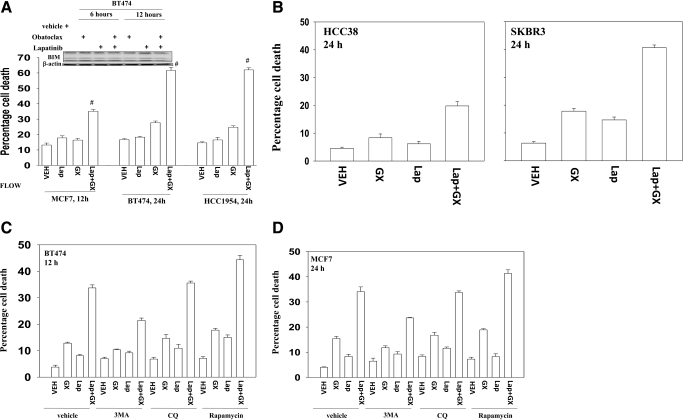
Lapatinib is a dual tyrosine kinase inhibitor of HER1 (ErbB1) and HER2 (ErbB2). We noted in breast cancer cells with either relatively low (MCF7and HCC38), or high (BT474, HCC1954, and SKBR3) expression of ErbB1/ErbB2 that the BCL-2 family inhibitor obatoclax enhanced lapatinib toxicity (Fig. 1, A and B) (Martin et al., 2009). Lapatinib and obatoclax exposure did not alter the expression of the proapoptotic protein BIM in BT474 cells (Fig. 1A, upper inset). Obatoclax enhanced toxicity in a synergistic fashion in long-term viability assays (Table 1). Our previous studies showed that the polycaspase inhibitor benzyloxycarbonyl-Val-Ala-Asp(OMe) fluoromethylketone failed to abrogate lapatinib and obatoclax toxicity but that inhibition of autophagy suppressed killing. In agreement with killing proceeding via a toxic form of autophagy, use of the small molecule inhibitor of the class III PI3K Vps34, 3MA suppressed drug combination lethality (Fig. 1, C and D). Rapamycin, which stimulates autophagy, promoted drug combination toxicity. However, and in contrast to 3MA, chloroquine, which acts to block autophagosome fusion with lysosomes (Amaravadi et al., 2007), had no effect on drug combination killing.
Fig. 1.
Lapatinib (Lap) interacts with obatoclax (GX) in a greater than additive fashion to kill ErbB2-negative or -positive breast cancer cells. A, MCF7, BT474, and HCC1954 cells were treated with vehicle (VEH, DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for the indicated time. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). Top inset, BT474 cells were subjected to the indicated treatments for 6 or 12 h. Floating and attached cells were collected, and the cell lysates were immunoblotted against BIM. B, HCC38 and SKBR3 cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 24 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). BT474 (C) and MCF7 (D) cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax in the presence or absence of 3-MA (2 mM), chloroquine (CQ, 5 μM), or rapamycin (50 nM) for 12 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3).
TABLE 1.
Lapatinib and obatoclax synergize to kill tumor cells
MCF7 and BT474 cells were plated as single cells (250–1500 cells/well) in sextuplicate, and 24 h after this plating, the cells were treated with vehicle (VEH, DMSO) or with lapatinib (0.5–1.5 μM) or obatoclax (33–99 or 15–45 nM as indicated), or with both drugs combined as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. After drug exposure (12 h for BT474 cells and 24 h for MCF7 cells), the medium was changed, and cells were cultured in drug-free medium for an additional 10 to 14 days. Cells were fixed and stained with crystal violet, and colonies of >50 cells/colony were counted. Colony formation data were entered into the CalcuSyn program, and CI values were determined. A CI value of less than 1.00 indicates synergy.
| Cell Line | GX | Lap | VEH | GX | Lap | GX + Lap | CI |
|---|---|---|---|---|---|---|---|
| nM | μM | ||||||
| BT474 | 33.0 | 0.5 | 100 | 88 | 97 | 66 | 0.55 |
| 66.0 | 1.0 | 100 | 64 | 96 | 33 | 0.60 | |
| 99.0 | 1.5 | 100 | 37 | 88 | 7 | 0.39 | |
| MCF7 | 33.0 | 0.5 | 100 | 91 | 96 | 65 | 0.45 |
| 66.0 | 1.0 | 100 | 73 | 95 | 27 | 0.50 | |
| 99.0 | 1.5 | 100 | 45 | 85 | 4 | 0.27 |
VEH, vehicle; Lap, lapatinib; GX, obatoclax (GX-15-070); CI, combination index.
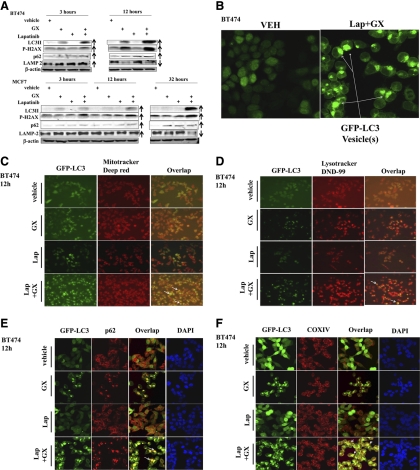
Mature autolysosomes are subject to autophagic proteolysis, leading to a reduced level of autophagic substrates as well as autophagosome and autolysosome components such as p62/SQSTM1 and LAMP-2. In BT474 and MCF7 cells, within 12 h of drug exposure LC3-II expression had increased (Fig. 2A). This change correlated with both enhanced DNA damage (P-H2AX) and p62 levels. The levels of LAMP2 also initially increased, arguing for less lysosomal degradation of this protein, and were followed by a decrease in LAMP2 levels at later times. This finding argues that early stages of autophagy are stimulated by drug exposure, but for several hours autophagy does not apparently progress beyond autophagosome formation, suggesting that dissociation between the early and late autophagy events may be occurring.
Fig. 2.
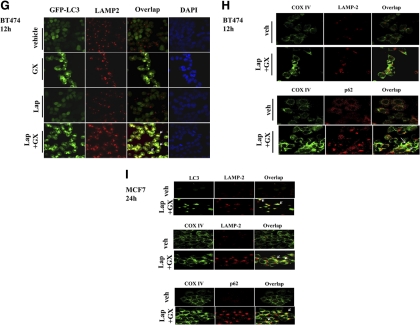
Lapatinib (Lap) + obatoclax (GX)-caused cell death is associated with enhanced autophagic responses. A, BT474 and MCF7 cells were treated with vehicle (VEH, DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for the indicated time. Floating and attached cells were collected, and the cell lysates were immunoblotted against the indicated autophagy and DNA damage markers. Arrows indicate whether expression of specific proteins appeared to increase or decline (n = 3). B, BT474 cells expressing GFP-LC3 were treated with vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Representative images of cells were taken with a Zeiss LSM 510 META laser scanning confocal microscope using a 100 × oil immersion objective lens. Arrows indicate formation of large GFP-LC3 vesicles. C and D, BT474 cells expressing GFP-LC3 were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 12 h. Cells subjected to the indicated treatment were stained with MitoTracker Deep Red FM (C) and LysoTracker Red DND-99 (D). E, F, and G, BT474 cells expressing GFP-LC3 were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 12 h. Treated cells were stained against p62 (E), COX IV (F), or LAMP-2 (G). H, BT474 cells were treated with vehicle (veh, DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h and were stained for COX IV/LAMP-2 (top) or COX IV/p62 (bottom). I, MCF7 cells were treated with vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h were stained for LC3/LAMP-2 (top left), COX IV/LAMP-2 (top right), or COX IV/p62 (bottom). DAPI, 4,6-diamidino-2-phenylindole.
We next examined the colocalization of autophagy regulatory proteins after drug exposure. In BT474 cells, obatoclax and to a greater extent lapatinib and obatoclax exposure triggered the formation of characteristic punctate GFP-LC3 vesicles, suggestive of autophagosome formation (Fig. 2, B and C). Of note was the drug treatment-stimulated formation of a few very large autophagosomes in contrast to the much smaller autophagic puncta often observed by other investigators (Fig. 2B). GFP-LC3 vesicles strongly colocalized with mitochondria as judged using MitoTracker Deep Red FM staining. The formation of acidic vesicular organelles was monitored by LysoTracker Red DND-99 staining, and, upon drug exposure, lysosome and GFP-LC3 staining also coincided, although the costaining effect was apparently weaker than that observed for GFP-LC3 with mitochondria (Fig. 2, cf. C and D).
The protein p62 is another biochemical indicator of autophagy, and this protein also exhibited punctate distribution and strongly colocalized with GFP-LC3 vesicles, suggesting that p62 was targeted onto the autophagosomes (Fig. 2E). The protein COX IV, is a large transmembrane protein located in the mitochondrion. In agreement with our data in Fig. 2C, GFP-LC3 strongly associated with COX IV after drug stimulation (Fig. 2F).
The protein LAMP2 is a constituent of the lysosomal membrane, displaying punctate structures after drug exposure, and colocalized with GFP-LC3 vesicles, indicating that some form of autolysosome development had occurred (Fig. 2G). Finally, we examined the colocalization of mitochondrial COX IV with other autophagy regulatory proteins. In BT474 cells, staining for COX IV and LAMP2 showed a weak coassociation whereas in MCF7 cells COX IV and LAMP2 strongly colocalized (Fig. 2, H and I). In both BT474 and MCF7 cells, COX IV strongly localized with p62, further supporting the role of mitochondria in the autophagy response.
Lapatinib and Obatoclax-Induced Killing Depends in Part on the Inhibition of the mTOR Signaling Pathway and Activation of the p38 MAPK Pathway.
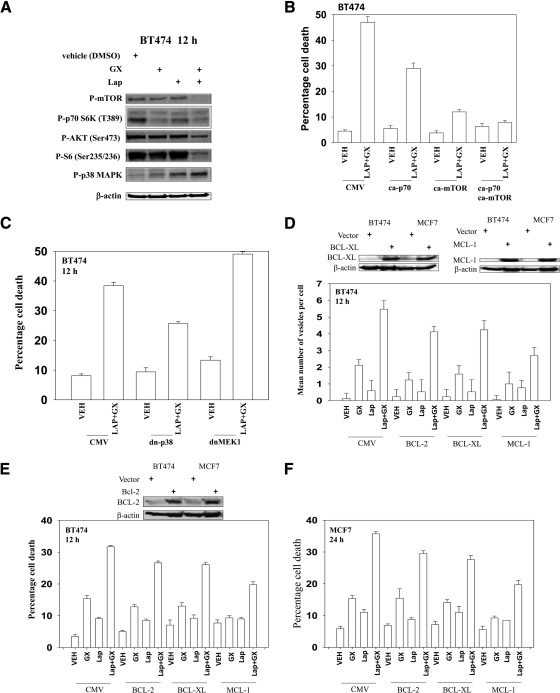
It has been well established that mTOR negatively regulates autophagy (Mathew et al., 2007). Lapatinib and obatoclax treatment reduced AKT, p70 S6K, and mTOR activity and increased p38 MAPK activity in BT474 and MCF7 cells (Fig. 3A; data not shown). Transfection of cells with constitutively active mTOR attenuated lapatinib and obatoclax-induced autophagic cell death to a greater extent than a plasmid to express a constitutively active p70S6K (Fig. 3B; Supplemental Fig. S1). Expression of dominant-negative p38 MAPK blocked drug-induced killing, whereas expression of dominant-negative mitogen-activated protein kinase kinase 1 facilitated drug lethality (Fig. 3C; Supplemental Figs. S1–S3).
Fig. 3.
Lapatinib (Lap) and obatoclax (Gx)-induced autophagy depends on the inhibition of mTOR signaling pathway. A, BT474 cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for the indicated time. Cell lysates were immunoblotted against the phosphorylated (P) form of p38, mTOR, p70S6K, and AKT (n = 3). B, BT474 cells were transfected with constitutively active (ca) p70S6K or mTOR. Twenty-four hours after transfection, cells were treated with vehicle (VEH, DMSO), or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). C, BT474 cells were infected with recombinant adenoviruses to express dominant-negative (dn) p38 or mitogen-activated protein kinase kinase (MEK1). Twenty-four hours after the infection, cells were treated with vehicle(DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). D, BT474 cells expressing GFP-LC3 were transfected with BCL-2-, BCL-XL-, or MCL-1-expressing constructs. The overexpression of each protein was confirmed by Western blot (top blotting panels in D and E). Twenty-four hours after transfection, cells were treated with vehicle (DMSO) or lapatinib (2 μM) + obatoclax (GX, 50 nM) for 12 h. Cells were examined using a fluorescence microscope for the formation of GFP-LC3 vesicles (±S.E.M., n = 3). E and F, BT474 and MCF7 cells were transfected with BCL-2-, BCL-XL-, or MCL-1-expressing constructs. Twenty-four hours after transfection, the cells were treated with vehicle (DMSO), lapatinib (2 μM) + obatoclax (50 nM) for 24 h. Cell viability was measured at the indicated times by trypan blue exclusion (±S.E.M., n = 3). CMV, cytomegalovirus.
In addition to changes in the activity of signal transduction pathways, protective BCL-2 family members can regulate both autophagy and pore formation in the mitochondrial outer membrane. Overexpression of MCL-1, and to a lesser extent of BCL-2 and BCL-XL, suppressed the formation of autophagic vesicles (Fig. 3D), and, in agreement with the data in Fig. 3D, expression of MCL-1, which preferentially interacts with NOXA (Chen et al., 2005), more effectively suppressed lapatinib and obatoclax toxicity than did BCL-2 and BCL-XL (Fig. 3, E and F).
Increased GFP-LC3 puncta formation and LC3-II levels may be interpreted as evidence for enhanced autophagosome synthesis; however, such an observation may also instead indicate that delayed trafficking of autophagosomes to the lysosomes or impaired lysosomal proteolytic activity has occurred. To further verify the cytotoxic role of autophagy in the lapatinib and obatoclax combination, autophagy was suppressed via knockdown of beclin 1, which is required for the autophagy initiation, or ATG5, which controls autophagosome formation. Knockdown of beclin 1 or ATG5 expression in BT474 and MCF7 cells markedly attenuated lapatinib and obatoclax-induced LC3-II formation, P-H2AX levels, and the late decrease in LAMP2 and increase in ATG4 levels (Tables 2 and 3; Supplemental Figs. S1–S3). Thus, the apparent DNA damage response we have observed is dependent on autophagy. The ROS quenching agent N-acetylcysteine blocked the drug-induced changes in protein expression, including LC3-II processing. Knockdown of either beclin 1 or ATG5 prevented drug combination killing (data not shown).
TABLE 2.
Autophagy-dependent changes in protein levels and protein phosphorylation in BT474 cells
BT474 cells were transfected with either siSCR, siATG5, or sibeclin 1. Thirty-six hours after transfection, the cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM),or lapatinib + obatoclax in the presence or absence of NAC (1 mM) for 12 h. Western blotting was performed to detect changes in phosphorylation/expression of proteins. The protein levels were quantified by densitometry analysis and are relative vehicle protein level (defined as 1.00) (±S.E.M., n = 3).
| BT474 | LC3II | P-H2AX | LAMP 2 | ATG4A |
|---|---|---|---|---|
| siSCR | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 1.57 | 1.00 | 0.82 | 1.43 |
| 2 μM Lap | 0.91 | 1.12 | 1.06 | 0.96 |
| 50 nM GX + 2 μM Lap | 4.44* | 4.10* | 0.72† | 3.14* |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.53 | 1.06 | 0.81 | 1.56 |
| 1 mM Nac | 1.29 | 1.00 | 0.92 | 1.78 |
| siATG5 | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 0.84 | 1.01 | 0.96 | 0.87 |
| 2 μM Lap | 0.89 | 1.05 | 1.02 | 0.89 |
| 50 nM GX + 2 μM Lap | 1.45*‡ | 1.74*‡ | 0.92 | 0.86 |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.38 | 1.44 | 0.95 | 0.95 |
| 1 mM Nac | 1.42 | 1.38 | 1.03 | 0.96 |
| siSCR | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 1.66 | 1.05 | 0.79 | 1.18 |
| 2 μM Lap | 0.98 | 1.08 | 0.98 | 0.84 |
| 50 nM GX + 2 μM Lap | 4.68* | 3.92* | 0.64† | 2.78* |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.69 | 1.05 | 0.71 | 1.08 |
| 1 mM Nac | 1.13 | 0.89 | 1.05 | |
| sibeclin 1 | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 1.47 | 1.00 | 1.18 | 1.09 |
| 2 μM Lap | 1.02 | 0.98 | 0.95 | 1.01 |
| 50 nM GX + 2 μM Lap | 2.13*‡ | 1.83*‡ | 1.07 | 0.93 |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.85 | 1.24 | 1.01 | 0.88 |
| 1 mM Nac | 1.08 | 1.09 | 1.09 |
VEH, vehicle; Lap, lapatinib; GX, obatoclax (GX-15-070).
p < 0.05 greater than the corresponding vehicle control.
p < 0.05 less than the corresponding vehicle control.
p < 0.05 less than the corresponding value in siSCR.
TABLE 3.
Autophagy-dependent changes in protein levels and protein phosphorylation in MCF7 cells
MCF7 cells were transfected with either siSCR, siATG5, or sibeclin 1. Thirty-six hours after transfection, the cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax in the presence or absence of NAC (1 mM) for 12 h. Western blotting was performed to detect changes in phosphorylation/expression of proteins. The protein levels were quantified by densitometry analysis and are relative vehicle protein level (defined as 1.00) (±S.E.M., n = 3).
| MCF7 | LC3II | P-H2AX | LAMP 2 | ATG4A |
|---|---|---|---|---|
| siSCR | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 1.28 | 1.01 | 0.88 | 0.95 |
| 2 μM Lap | 1.03 | 1.00 | 0.99 | 1.01 |
| 50 nM GX + 2 μM Lap | 5.21* | 5.31* | 0.81† | 2.16* |
| 50 nM GX + 2 μM Lap+ 1 mM Nac | 2.12 | 1.12 | 0.78 | 0.97 |
| 1 mM Nac | 1.51 | 1.22 | 0.85 | 1.23 |
| siATG5 | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 1.09 | 1.03 | 1.13 | 0.93 |
| 2 μM Lap | 1.03 | 1.06 | 1.02 | 0.98 |
| 50 nM GX + 2 μM Lap | 1.55*‡ | 2.23*‡ | 0.96 | 0.86 |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.17 | 1.02 | 1.10 | 1.07 |
| 1 mM Nac | 1.04 | 1.08 | 0.96 | 0.80 |
| siSCR | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 1.11 | 1.37 | 0.92 | 1.00 |
| 2 μM Lap | 1.08 | 0.86 | 1.05 | 0.99 |
| 50 nM GX + 2 μM Lap | 5.35* | 5.64* | 0.85† | 2.21* |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.98 | 1.71 | 0.82 | 1.06 |
| 1 mM Nac | 1.50 | 1.00 | 1.16 | |
| sibeclin 1 | ||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 |
| 50 nM GX | 0.99 | 1.18 | 0.99 | 0.95 |
| 2 μM Lap | 1.02 | 0.99 | 1.11 | 1.03 |
| 50 nM GX + 2 μM Lap | 2.69*‡ | 2.03*‡ | 0.91 | 1.01 |
| 50 nM GX + 2 μM Lap + 1 mM Nac | 1.89 | 1.24 | 1.01 | 0.98 |
| 1 mM Nac | 1.45 | 0.99 | 0.81 |
VEH, vehicle; Lap, lapatinib; GX, obatoclax (GX-15-070).
p < 0.05 greater than the corresponding vehicle control.
p < 0.05 less than the corresponding vehicle control.
p < 0.05 less than the corresponding value in siSCR.
Lapatinib and Obatoclax-Induced Autophagy Is Associated with Mitochondria Defects, Elevated Reactive Oxygen Species Generation and Increased DNA Damage.
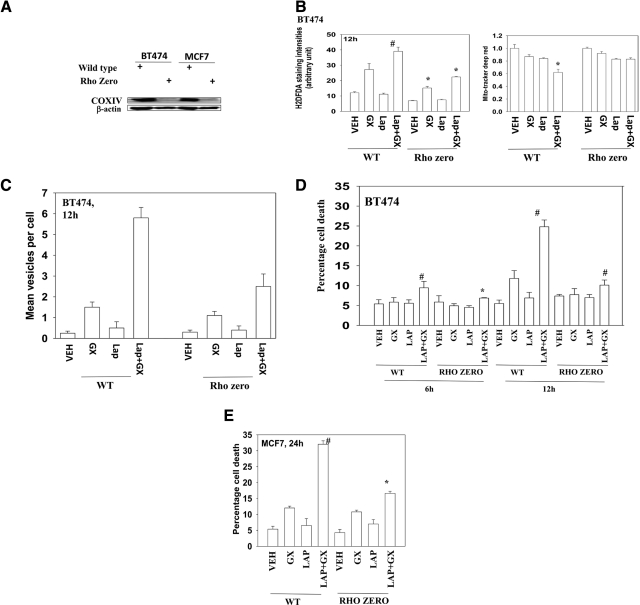
The actions of multiple chemotherapeutic agents often require the generation of ROS. Rho zero cells lack mitochondrial function because of prolonged exposure to ethidium bromide that specifically depletes mitochondrial DNA but not genomic DNA. We and others have previously observed that Rho zero cells have reduced ROS generating capacity. In wild-type cells, but not Rho zero cells, lapatinib and obatoclax interacted to generate ROS (Fig. 4, A and B). This event also correlated with reduced MitoTracker staining in wild-type cells compared with Rho zero cells. The drug combination was significantly less capable of stimulating autophagy in Rho zero cells (Fig. 4C).
Fig. 4.
Lapatinib (Lap) + obatoclax (GX)-caused toxic autophagy is associated with compromised mitochondrial membrane integrity. A, mitochondria were depleted from BT474 and MCF7 cells (“Rho zero”). The depletion of mitochondria was confirmed by Western blotting against COX IV. B, BT474 cells with depleted mitochondria were treated with vehicle (VEH, DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 12 h. The cells were stained with carboxy-H2DCFDA and MitoTracker Deep Red. The fluorescence intensities were analyzed by flow cytometry (±S.E.M., n = 3). C and D, BT474 and BT474 Rho cells expressing GFP-LC3 were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 12 h. Cells were examined in a fluorescent microscope for the formation of GFP-LC3 vesicles. Cell viability was measured by annexin V/PI staining. E, MCF7 and MCF7 Rho cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 24 h. Viability was determined by trypan blue exclusion (±S.E.M., n = 3). WT, wild-type.
We next performed time course studies examining the expression of autophagy marker proteins. In BT474 and MCF7 cells, drug combination exposure initially caused an increase in LC3-II, LAMP2, and p62 levels, as well as in P-H2AX, a marker for DNA damage, which were all reduced or abolished in Rho zero cells (Tables 4 and 5). At later times (12–32 h) after drug treatment, LAMP2 levels declined in wild-type but not in Rho zero cells. We found that the expression level of ATG4, which is essential for reactive oxygen species-triggered autophagy, was significantly increased after drug exposure in wild-type but not in Rho zero cells (Tables 4 and 5; data not shown). In agreement with a lack of change in autophagy markers in Rho zero cells, Rho zero cells were resistant to the toxic effects of lapatinib and obatoclax treatment (Fig. 4, D and E).
TABLE 4.
Loss of mitochondrial function suppresses lapatinib and obatoclax-induced changes in protein expression/protein phosphorylation in BT474 cells
BT474 cells and BT474 Rho zero cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 6 to 12 h. Western blotting was performed to detect changes in phosphorylation/expression of proteins. The protein levels were quantified by densitometry analysis and is relative vehicle protein level (defined as 1.00) (±S.E.M., n = 3).
| LC3II | P-H2AX | p62 | LAMP 2 | ATG4A | ATG7 | |
|---|---|---|---|---|---|---|
| BT474 | ||||||
| 12 h | ||||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 h | ||||||
| 50 nM GX | 1.71 | 1.24 | 0.99 | 1.02 | 1.89 | 1.20 |
| 2 μM Lap | 0.96 | 1.08 | 0.97 | 0.99 | 1.01 | |
| 50 nM GX + 2 μM Lap | 2.17* | 3.96* | 1.78* | 1.35* | 2.57* | 1.14 |
| 12 h | ||||||
| 50 nM GX | 1.98 | 1.29 | 1.08 | 0.84‡ | 1.77 | 1.15 |
| 2 μM Lap | 0.98 | 1.04 | 1.06 | 0.95 | 1.74 | 1.17 |
| 50 nM GX + 2 μM Lap | 5.71* | 4.55* | 1.49* | 0.49‡ | 3.01* | 1.18 |
| Rho zero | ||||||
| 12 h | ||||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 h | ||||||
| 50 nM GX | 1.26 | 1.03 | 0.83 | 0.98 | 0.99 | 0.96 |
| 2 μM Lap | 1.01 | 1.02 | 0.84 | 1.05 | 1.18 | 1.02 |
| 50 nM GX + 2 μM Lap | 1.49*† | 1.14 | 1.01 | 1.03 | 1.12 | 1.12 |
| 12 h | ||||||
| 50 nM GX | 1.95 | 1.13 | 1.02 | 0.92 | 1.17 | 0.92 |
| 2 μM Lap | 0.99 | 0.99 | 0.86 | 1.04 | 1.08 | 1.12 |
| 50 nM GX + 2 μM Lap | 3.17*† | 1.39*† | 1.20* | 0.97 | 1.08 | 1.19 |
VEH, vehicle; Lap, lapatinib; GX, obatoclax (GX-15-070).
p < 0.05 greater than the corresponding vehicle control.
p < 0.05 less than the corresponding value in siSCR cells.
p less than the corresponding vehicle control.
TABLE 5.
Loss of mitochondrial function suppresses lapatinib and obatoclax-induced changes in protein expression/protein phosphorylation in MCF7 cells
MCF7 cells and MCF7 Rho zero cells were treated with vehicle (DMSO), lapatinib (2 μM), obatoclax (50 nM), or lapatinib + obatoclax for 12 to 24 h. Western blotting was performed to detect changes in phosphorylation/expression of proteins. The protein levels were quantified by densitometry analysis and are relative vehicle protein level (defined as 1.00) (±S.E.M., n = 3).
| LC3II | P-H2AX | p62 | LAMP 2 | ATG4A | ATG7 | |
|---|---|---|---|---|---|---|
| BT474 | ||||||
| 12 h | ||||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 h | ||||||
| 50 nM GX | 1.69 | 1.55 | 0.96 | 0.89 | 1.23 | 0.89 |
| 2 μM Lap | 1.02 | 1.39 | 0.95 | 0.93 | 1.56 | 1.12 |
| 50 nM GX + 2 μM Lap | 2.32 | 2.80* | 1.68* | 1.39* | 1.91* | 1.13 |
| 12 h | ||||||
| 50 nM GX | 1.63 | 1.14 | 1.23 | 0.59 | 1.02 | 1.13 |
| 2 μM Lap | 1.00 | 1.14 | 1.25 | 1.02 | 2.29 | 1.04 |
| 50 nM GX + 2 μM Lap | 5.75 | 6.37* | 1.41* | 0.32‡ | 2.42* | 0.89 |
| Rho zero | ||||||
| 12 h | ||||||
| Vehicle | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 h | ||||||
| 50 nM GX | 1.37 | 1.64 | 0.96 | 0.90 | 0.99 | 0.96 |
| 2 μM Lap | 1.00 | 1.33 | 0.95 | 0.95 | 0.98 | 1.12 |
| 50 nM GX + 2 μM Lap | 1.52*† | 2.14*† | 1.03 | 0.93 | 0.98 | 1.11 |
| 12 h | ||||||
| 50 nM GX | 1.51 | 2.08 | 1.02 | 0.90 | 0.99 | 1.17 |
| 2 μM Lap | 1.02 | 0.95 | 1.03 | 0.91 | 1.02 | 1.05 |
| 50 nM GX + 2 μM Lap | 2.74*† | 2.51*† | 1.20*† | 0.90 | 1.12 | 1.12 |
VEH, vehicle; Lap, lapatinib; GX, obatoclax (GX-15-070).
p < 0.05 greater than the corresponding vehicle control.
p < 0.05 less than the corresponding value in siSCR cells.
p less than the corresponding vehicle control.
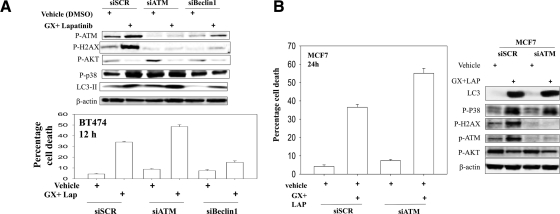
ATM and p38 MAPK Signaling Downstream of Autophagy.
As noted in other figures, lapatinib and obatoclax treatment increased the phosphorylation of histone H2AX, a marker indicative of DNA damage (Fig. 5, A and B). Knockdown of ATM, the kinase that phosphorylates H2AX, blocked H2AX phosphorylation but did not alter drug-induced inactivation of mTOR and AKT (Fig. 5, A and B, blots). Knockdown of ATM enhanced basal levels and drug-induced activation of p38 MAPK and promoted the drug combination-induced processing of LC3-II. In agreement with increased LC3-II processing, knockdown of ATM enhanced drug combination toxicity (Fig. 5, A and B, graphs).
Fig. 5.
Loss of ATM function increases LC3-II conversion and enhances drug lethality. A, BT474 cells were transfected with siSCR, siATM, or sibeclin1. Thirty-six hours after transfection, the cells were treated with either vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Bottom graph, floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). Top blot, in parallel sets of samples, cells were collected with SDS sample buffer and the phosphorylation and expression of the indicated proteins were detected by Western blot. B, MCF7 cells were transfected with siSCR or siATM. Thirty-six hours after the infection, the cells were treated with either vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Graph, floating and attached cells were isolated after drug exposure and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). Blot, in parallel sets of samples, cells were collected with SDS sample buffer and the phosphorylation and expression of the indicated proteins were detected by Western blot.
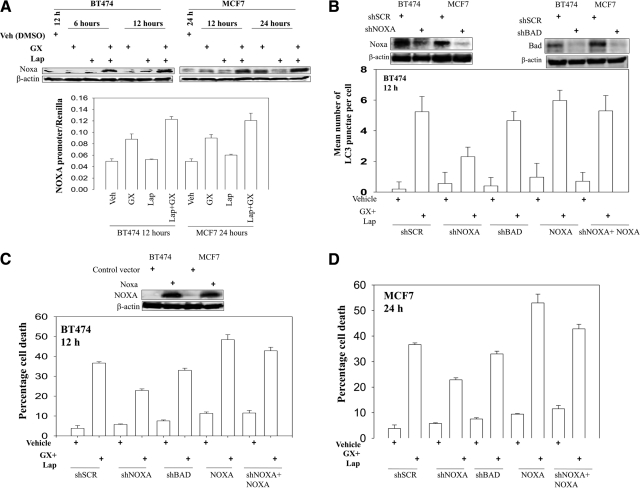
NOXA Plays a Central Role in Lapatinib and Obatoclax-Induced Autophagic Cell Death.
Expression of the BH3-only member of the BCL-2 family NOXA was increased after drug exposure, which was accompanied by elevated NOXA promoter activity (Fig. 6A). Considering that NOXA might interact with BCL-2 prosurvival proteins and neutralize their autophagy suppressive activities, we wondered whether an increase in NOXA expression is required or essential for drug-induced autophagy and subsequent cell death. Knockdown of NOXA expression significantly attenuated GFP-LC3 puncta formation upon lapatinib and obatoclax treatment (Fig. 6B). Lapatinib and obatoclax-induced killing was also reduced upon knockdown of NOXA expression (Fig. 6, C and D).
Fig. 6.
NOXA plays an important role in the regulation of drug-induced autophagy. A, bottom graph, BT474 and MCF7 cells were transfected with a plasmid containing the NOXA promoter linked to expression of luciferase. Thirty-six hours after transfection, cells were treated with either vehicle (Veh, DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 and 24 h, as indicated. Luciferase activity was determined (±S.E.M., n = 3). Upper blot, BT474 and MCF7 cells were treated with vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 6 to 24 h, as indicated. The expression of NOXA was determined (±S.E.M., n = 3). B and C, BT474 cells were transfected with scrambled shRNA (shSCR) to either knock down NOXA expression (shNOXA), knock down the levels of BAD (shBAD), or overexpress NOXA in a cell with NOXA knockdown (shNOXA+NOXA). Thirty-six hours after transfection, cells were treated with either vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. B, cells were examined in a fluorescent microscope for the formation of GFP-LC3 vesicles. C, cell viability was measured by trypan blue exclusion-based cell staining (± S.E.M., n = 3). D, MCF7 cells were transfected with scrambled shRNA to either knock down NOXA expression, knock down the levels of BAD, or overexpress NOXA in a cell with NOXA knockdown. Thirty-six hours after transfection, cells were treated with either vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 24 h, and cell viability was measured by trypan blue exclusion-based cell staining (±S.E.M., n = 3).
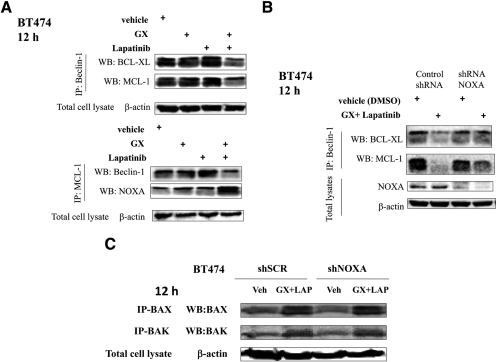
Lapatinib and Obatoclax-Induced Autophagic Cell Death Relies on NOXA-Mediated Displacement of Beclin 1 from MCL-1.
BCL-2 family members such as BCL-2 and BCL-XL suppress autophagy initiation by binding to beclin 1 (Liang et al., 1999), which compromises one portion of the beclin 1-class III PI3K complex (Noble et al., 2008). Recent studies revealed that BH3-only proteins could promote beclin 1-dependent autophagy through disruption of the interaction between beclin 1 and the prosurvival BCL-2 family members (Daido et al., 2004; Kanzawa et al., 2005; Pattingre et al., 2005; Erlich et al., 2007; Maiuri et al., 2007). When beclin 1 was immunoprecipitated after lapatinib and obatoclax treatment, the amount of coimmunoprecipitating MCL-1 and BCL-XL was significantly reduced (Fig. 7A); vice versa, when MCL-1 was immunoprecipitated, the amount of beclin 1 bound to MCL-1 was diminished compared with that for cells treated with vehicle (Fig. 7A). Of note, the amount of NOXA that coimmunoprecipitated with MCL-1 increased and the amount of beclin 1 decreased upon lapatinib and obatoclax treatment. Finally, we determined whether NOXA expression was required for disassembly of NOXA-MCL-1 complexes. Beclin 1 association with BCL-XL and MCL-1 was lost upon obatoclax and lapatinib treatment (Fig. 7B). Knockdown of NOXA abolished the reduction in NOXA-BCL-XL association and reduced the loss in association with MCL-1. Finally we determined whether activation of BAX and BAK, shown in our prior studies, was dependent on increased NOXA expression. Knockdown of NOXA did not alter drug-induced BAX or BAK activation (Fig. 7C).
Fig. 7.
NOXA regulates beclin 1 association with MCL-1. A, BT474 cells were treated with vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM). Cells were lysed, and portions of lysates immunoprecipitated using an anti-beclin 1 antibody and other portions using an anti-MCL-1 antibody. Immunoprecipitates were blotted for BCL-XL, MCL-1, NOXA, and beclin 1 as indicated (n = 3). B, BT474 cells were transfected with scrambled shRNA (shSCR) or to knockdown NOXA expression (shNOXA). Thirty-six hours after transfection, cells were treated with either vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Cells were lysed, and portions of lysates were immunoprecipitated using an anti-beclin 1 antibody. Immunoprecipitates were blotted for BCL-XL and MCL-1 as indicated (n = 3). C, BT474 cells were transfected with scrambled shRNA or to knockdown NOXA expression. Thirty-six hours after transfection, cells were treated with either vehicle (DMSO) or lapatinib (2 μM) + obatoclax (50 nM) for 12 h. Cells were lysed and portions of lysates immunoprecipitated using an anti-active BAX or anti-active BAK antibodies. Immunoprecipitates were probed for BAX and BAK as indicated (n = 3).
Discussion
Our prior studies demonstrated that obatoclax potentiated the cytotoxicity of lapatinib in human colon and breast cancer cells by inducing autophagic cell death (Martin et al., 2008, 2009). In the present article, we have endeavored to explore the mechanism by which obatoclax and lapatinib interacted to cause toxic autophagy. The drug combination compromised the integrity of mitochondria as evidenced by an increase in ROS generation and loss of mitochondria membrane potential, events that were lacking in Rho zero cells. Inhibition of ROS by NAC or depletion of mitochondria diminished obatoclax and lapatinib-induced autophagy and cytotoxicity. Based on protein-protein interaction studies, proteins associated with early autophagosome formation strongly coassociated with mitochondria. Defective autophagic degradation was reflected by accumulation of undigested large autophagosomes and toxic p62 proteins and unliquidated damaged mitochondria that collectively may account for obatoclax and lapatinib-induced autophagic cell death.
In mammalian cells, autophagy is initiated by ULK1 (mammalian homolog of Atg1), which forms a complex with Atg13 and FIP200 and is regulated by autophagy-related genes (Atg), which are implicated in four major steps: initiation, nucleation, cycling, and expansion/closure (Klionsky and Emr, 2000). The formation of beclin 1, class III (Vps34) PI3Ks, and UVRAG (UV radiation resistance-associated gene protein) complexes is indispensable for autophagy initiation (Klionsky and Emr, 2000). BCL-2 and its homologs (BCL-XL and MCL-1) inhibit the initial steps of autophagy by a direct interaction with beclin 1 (Pattingre et al., 2005). The competitive binding of BCL-2 family proteins to beclin 1 compromises beclin 1/Vps34/UVRAG complex formation and suppresses autophagy initiation (Erlich et al., 2007). As an alternative, increased expression or stabilization of BH3-only proteins may promote autophagy (Maiuri et al., 2007; Rashmi et al., 2008). Cetuximab, a therapeutic antibody that blocks ErbB1 function, is able to induce autophagy in cancer cells through the down-regulation of BCL-2 antiapoptotic proteins and activation of the beclin 1-hVps34 complex (Li and Fan, 2010). Consistent with this observation, BH3-only proteins BAD and BNIP3 or BH3 mimetic ABT737 disrupt beclin 1-BCL2/BCL-XL complexes and promote autophagy (Pattingre et al., 2005). Thus, the interaction between prosurvival BCL-2 family proteins and beclin 1 represents a key event in our cell system that determines the “on-and-off” of autophagy.
The clinically relevant small molecule obatoclax is a BH3 domain inhibitor that binds to all protective BCL-2 family proteins. The drug triggers the dissociation of a protective BCL-2 protein from a toxic BH3 domain protein, which leads to an enhanced level of free BH3 domain proteins. Prior studies have indicated that BAX and BAK play a role in lapatinib and obatoclax-stimulated autophagy (Martin et al., 2009). In the present study, lapatinib and obatoclax treatment increased the levels of NOXA. NOXA competed away the prosurvival BCL-2 family member, MCL-1, from beclin 1. Similar data were also seen with competition away from BCL-XL. Ectopic expression of MCL-1 or knockdown of NOXA diminished the potentiation of lapatinib lethality by obatoclax, and this result probably highlights a central role of the NOXA and MCL-1 interaction in autophagy initiation inhibition, together with BAX and BAK, whose activation was separate from that of NOXA.
BCL-2 inhibitors/antagonists [(R-(−)-gossypol acetic acid (AT-101), 4-[4-[[2-(4-chlorophenyl)-5,5-dimethylcyclohexen-1-yl]methyl]piperazin-1-yl]-N-[4-[[(2R)-4-morpholin-4-yl-1-phenylsulfanylbutan-2-yl]amino]-3-(trifluoromethylsulfonyl)phenyl]sulfonylbenzamide (ABT-263), and obatoclax] have been reported to cause compromised mitochondrial function due to permeabilization of the outer mitochondrial membrane (Kang and Reynolds, 2009; McCoy et al., 2010). In the present study, lapatinib and obatoclax led to ROS generation, which was concomitant with loss of mitochondrial membrane potential. Autophagy maintains cellular homeostasis by recognizing and liquidating damaged organelles and protein aggregates (Komatsu et al., 2006). In particular, the form of autophagy observed in our studies seems to specifically target depolarized mitochondria for degradation in a process often termed mitophagy, through which the defective mitochondria-originated ROS generation is attenuated and the release of proapoptotic factors is prevented (Zhang et al., 2008b; Tang et al., 2011). As shown, obatoclax or lapatinib treatment alone resulted in very limited colocalization between punctate GFP-LC3 and mitochondria, whereas lapatinib and obatoclax treatment induced strong colocalization between GFP-LC3 puncta and mitochondria. We also observed widespread colocalization between mitochondria with p62 and to a lesser extent with LAMP-2 in tumor cells treated with lapatinib and obatoclax. All of these observations strongly suggested that lapatinib and obatoclax caused malfunction of mitochondria and that because of this malfunction they were targeted by autophagy.
The increasing percentage of punctate GFP-LC3-positive cells together with the enhanced number of GFP-LC3 dots and intensity of fluorescent puncta per cell reflects the progression of autophagic flux (Kadowaki and Karim, 2009; Barth et al., 2010). In our prior autophagy studies and those conducted by other groups, drug exposure caused GFP-LC3 vesicle formation with the development of numerous (∼20–60) small intensely staining GFP-LC3 dots per cell (Park et al., 2008a,b; Zhang et al., 2008a; Kadowaki and Karim, 2009; Barth et al., 2010; Mizushima et al., 2010). In contrast, obatoclax and lapatinib and obatoclax treatment rapidly caused the formation of four to six very large, intensely staining GFP-LC3-positive vesicles per cell. We also found that lapatinib and obatoclax combination treatment resulted in the accumulation of LC3-II and p62 proteins. One explanation for the accumulation of gigantic GFP-LC3 vesicles and abolished p62 degradation is that lapatinib and obatoclax-induced autophagy is associated with impaired or retarded autophagic degradation. GFP-LC3 is sensitive to acidic pH and ceases to fluoresce once autophagosomes fuse with lysosomes, leading to failure of monitoring of the end-stage autophagy (Kimura et al., 2007; Kuma et al., 2007). We did observe widespread overlapping between punctate GFP-LC3 and lysosomal-associated membrane protein 2, which suggests that autophagosomes fused with lysosomes to generate autolysosomes whose clearance might be terminated because of impaired lysosomal acidification. In support of this hypothesis, 3-MA attenuated the cytotoxicity of lapatinib and obatoclax. In contrast, chloroquine exerted a weak effect on enhancing the accumulation of LC3-II and failed to further enforce lapatinib and obatoclax-induced cell death, which indicates that defective autophagic degradation occurred during lapatinib andobatoclax treatment. The protein p62 is a selective substrate of autophagy, and its level is often used as an indicator of autophagic activity (Bjørkøy et al., 2005; Pankiv et al., 2007; Ichimura et al., 2008; Shvets et al., 2008; Mizushima et al., 2010). As a consequence, impaired autophagic degradation leads to accumulation of p62 and a defect in the turnover of toxic polyubiquitinated protein aggregates. Accumulation of p62 then triggers a positive amplifying loop for ROS generation, oxidative stress, aggravated metabolic stress, and enhanced genomic instability (Scherz-Shouval and Elazar, 2007; Scherz-Shouval et al., 2007). In addition, given the importance of mitochondrial clearance in the regulation of cell homeostasis, impaired autophagic degradation disturbed the proper autophagy flux, leading to the accumulation of sequestered but undigested defective mitochondria and precipitating cell death. The precise molecular reasons for lapatinib and obatoclax causing defective autophagic degradation await further investigation.
Supplementary Material
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the Department of Defense [Breast Cancer Idea Award W81XWH-10-1-0009]; National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK52825]; National Institutes of Health National Cancer Institute [Grants CA141703, CA150218, F32-CA85159] (the last of these to Y.T. by way of the Massey Cancer Center).
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- HER
- human epidermal growth factor receptor
- PI3K
- phosphatidylinositol 3-kinase
- MAPK
- mitogen-activated protein kinase
- BCL-2/Bcl-2
- B-cell CLL/lymphoma-2
- MCL-1
- myeloid cell leukemia-1
- BCL-XL
- BCL-2-related gene long isoform
- BAX
- BCL-2-associated x protein
- BAK
- BCL-2 antagonist killer
- mTOR
- mammalian target of rapamycin
- 3-MA
- 3-methyladenine
- FBS
- fetal bovine serum
- GFP
- green fluorescent protein
- LC3
- microtubule-associated protein light chain 3
- ATG
- autophagy related
- ATM
- ataxia telangiectasia mutated
- shRNA
- short hairpin RNA
- BAD
- BCL-2 antagonist of cell death
- BIM
- BCL-2-interacting mediator of cell death
- BID
- BCL-2-interacting domain death agonist
- LAMP-2
- lysosomal associated membrane protein 2
- NAC
- N-acetylcysteine
- ROS
- reactive oxygen species
- DCFDA
- dichlorofluorescein diacetate
- si
- small interfering
- PBS
- phosphate-buffered saline
- PI
- propidium iodide
- DMSO
- dimethyl sulfoxide
- COX IV
- cytochrome c oxidase
- AT-101
- (R-(−)-gossypol acetic acid
- ABT-263
- 4-[4-[[2-(4-chlorophenyl)-5,5-dimethylcyclohexen-1-yl]methyl]piperazin-1-yl]-N-[4-[[(2R)-4-morpholin-4-yl-1-phenylsulfanylbutan-2-yl]amino]-3-(trifluoromethylsulfonyl)phenyl]sulfonylbenzamide
- Δψ
- mitochondrial membrane potential
- AVOs
- acidic vesicular organelles
- MOMP
- mitochondrial outer membrane permeabilization
- ER
- Estrogen receptor
- GX
- obatoclax (GX15–070).
Authorship Contributions
Participated in research design: Fisher, Grant, and Dent.
Conducted experiments: Tang, Hamed, and Cruickshanks.
Performed data analysis: Tang and Dent.
Wrote or contributed to the writing of the manuscript: Tang and Dent.
References
- Amaravadi RK, Thompson CB. (2007) The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res 13:7271–7279 [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. (2007) Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117:326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin DN, Sergina N, Ahuja D, McMahon M, Blair JA, Wang D, Hann B, Koch KM, Shokat KM, Moasser MM. (2010) Resiliency and vulnerability in the HER2-HER3 tumorigenic driver. Sci Transl Med 2:16ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Glick D, Macleod KF. (2010) Autophagy: assays and artifacts. J Pathol 221:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. (2007) A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12:395–402 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein HJ, Storniolo AM, Franco S, Forster J, Stein S, Rubin S, Salazar VM, Blackwell KL. (2008) A phase II study of lapatinib monotherapy in chemotherapy-refractory HER2-positive and HER2-negative advanced or metastatic breast cancer. Ann Oncol 19:1068–1074 [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. (2005) Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 17:393–403 [DOI] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. (2004) Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res 64:4286–4293 [DOI] [PubMed] [Google Scholar]
- Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S, Hirsch JA, Stein R, Pinkas-Kramarski R. (2007) Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy 3:561–568 [DOI] [PubMed] [Google Scholar]
- Hobday TJ, Perez EA. (2005) Molecularly targeted therapies for breast cancer. Cancer Control 12:73–81 [DOI] [PubMed] [Google Scholar]
- Hynes NE, Lane HA. (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 5:341–354 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kominami E, Tanaka K, Komatsu M. (2008) Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 4:1063–1066 [DOI] [PubMed] [Google Scholar]
- Kadowaki M, Karim MR. (2009) Cytosolic LC3 ratio as a quantitative index of macroautophagy. Methods Enzymol 452:199–213 [DOI] [PubMed] [Google Scholar]
- Kang MH, Reynolds CP. (2009) Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res 15:1126–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. (2005) Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene 24:980–991 [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. (2005) The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 5:726–734 [DOI] [PubMed] [Google Scholar]
- Konecny GE, Pegram MD, Venkatesan N, Finn R, Yang G, Rahmeh M, Untch M, Rusnak DW, Spehar G, Mullin RJ, et al. (2006) Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res 66:1630–1639 [DOI] [PubMed] [Google Scholar]
- Kuma A, Matsui M, Mizushima N. (2007) LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3:323–328 [DOI] [PubMed] [Google Scholar]
- Li X, Fan Z. (2010) The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1α and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res 70:5942–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676 [DOI] [PubMed] [Google Scholar]
- Lin NU, Winer EP. (2007) Brain metastases: the HER2 paradigm. Clin Cancer Res 13:1648–1655 [DOI] [PubMed] [Google Scholar]
- Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, Halsey W, Sathe GM, Martin AM, Gilmer TM. (2009) Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res 69:6871–6878 [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. (2007) BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 3:374–376 [DOI] [PubMed] [Google Scholar]
- Martin AP, Miller A, Emad L, Rahmani M, Walker T, Mitchell C, Hagan MP, Park MA, Yacoub A, Fisher PB, et al. (2008) Lapatinib resistance in HCT116 cells is mediated by elevated MCL-1 expression and decreased BAK activation and not by ERBB receptor kinase mutation. Mol Pharmacol 74:807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Mitchell C, Rahmani M, Nephew KP, Grant S, Dent P. (2009) Inhibition of MCL-1 enhances lapatinib toxicity and overcomes lapatinib resistance via BAK-dependent autophagy. Cancer Biol Ther 8:2084–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. (2007) Role of autophagy in cancer. Nat Rev Cancer 7:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy F, Hurwitz J, McTavish N, Paul I, Barnes C, O'Hagan B, Odrzywol K, Murray J, Longley D, McKerr G, et al. (2010) Obatoclax induces Atg7-dependent autophagy independent of beclin-1 and BAX/BAK. Cell Death Dis 1:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Yacoub A, Hossein H, Martin AP, Bareford MD, Eulitt P, Yang C, Nephew KP, Dent P. (2010) Inhibition of MCL-1 in breast cancer cells promotes cell death in vitro and in vivo. Cancer Biol Ther 10:903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. (2010) Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al. (2004) PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 6:117–127 [DOI] [PubMed] [Google Scholar]
- Noble CG, Dong JM, Manser E, Song H. (2008) Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. J Biol Chem 283:26274–26282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye MA, Neve RM, Lane HA, Hynes NE. (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 19:3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145 [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Yacoub A, Rahmani M, Zhang G, Hart L, Hagan MP, Calderwood SK, Sherman MY, Koumenis C, Spiegel S, et al. (2008a) OSU-03012 stimulates PKR-like endoplasmic reticulum-dependent increases in 70-kDa heat shock protein expression, attenuating its lethal actions in transformed cells. Mol Pharmacol 73:1168–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Yacoub A, Sarkar D, Emdad L, Rahmani M, Spiegel S, Koumenis C, Graf M, Curiel DT, Grant S, et al. (2008b) PERK-dependent regulation of MDA-7/IL-24-induced autophagy in primary human glioma cells. Autophagy 4:513–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939 [DOI] [PubMed] [Google Scholar]
- Raffo AJ, Perlman H, Chen MW, Day ML, Streitman JS, Buttyan R. (1995) Overexpression of bcl-2 protects prostate cancer cells from apoptosis in vitro and confers resistance to androgen depletion in vivo. Cancer Res 55:4438–4445 [PubMed] [Google Scholar]
- Rashmi R, Pillai SG, Vijayalingam S, Ryerse J, Chinnadurai G. (2008) BH3-only protein BIK induces caspase-independent cell death with autophagic features in Bcl-2 null cells. Oncogene 27:1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, Keith BR, Murray DM, Knight WB, Mullin RJ, et al. (2001) The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther 1:85–94 [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N. (1995) Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 19:183–232 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Elazar Z. (2007) ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 17:422–427 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26:1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al. (2008) NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res 68:8022–8030 [DOI] [PubMed] [Google Scholar]
- Shvets E, Fass E, Scherz-Shouval R, Elazar Z. (2008) The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci 121:2685–2695 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177–182 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244:707–712 [DOI] [PubMed] [Google Scholar]
- Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, Freilino ML, Graner MW, Wikstrand CJ, Bigner DD, et al. (2006) Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res 12:5064–5073 [DOI] [PubMed] [Google Scholar]
- Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. (2006) Targeting multidrug resistance in cancer. Nat Rev Drug Discov 5:219–234 [DOI] [PubMed] [Google Scholar]
- Tang Y, Chen Y, Jiang H, Nie D. (2011) Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differ 18:602–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. (2004) A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res 64:6652–6659 [DOI] [PubMed] [Google Scholar]
- Xia W, Mullin RJ, Keith BR, Liu LH, Ma H, Rusnak DW, Owens G, Alligood KJ, Spector NL. (2002) Anti-tumor activity of GW572016: a dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene 21:6255–6263 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2:127–137 [DOI] [PubMed] [Google Scholar]
- Zhang G, Park MA, Mitchell C, Walker T, Hamed H, Studer E, Graf M, Rahmani M, Gupta S, Hylemon PB, et al. (2008a) Multiple cyclin kinase inhibitors promote bile acid-induced apoptosis and autophagy in primary hepatocytes via p53-CD95-dependent signaling. J Biol Chem 283:24343–24358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. (2008b) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283:10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.