Abstract
Hepatic nuclear factor 4α (HNF4A) is a nuclear transcription factor that regulates the expression of many genes involved in drug disposition. To identify additional molecular mechanisms that regulate HNF4A, we identified microRNAs (miRNAs) that target HNF4A expression. In silico analyses suggested that HNF4A is targeted by many miRNAs. We conducted in vitro studies to validate several of these predictions. With use of an HNF4A 3′-untranslated region (UTR) luciferase reporter assay, five of six miRNAs tested significantly down-regulated (∼20–40%) the luciferase activity. In HepG2 cells, miR-34a and miR-449a also down-regulated the expression of both the HNF4A protein and an HNF4A target gene, PXR (∼30–40%). This regulation appeared without reduction in HNF4A mRNA expression, suggesting that they must be blocking HNF4A translation. Using additional bioinformatic algorithms, we identified polymorphisms that are predicted to alter the miRNA targeting of HNF4A. Luciferase assays indicated that miR-34a and miR-449a were less effective in regulating a variant (rs11574744) than the wild-type HNF4A 3′-UTR. In vivo, subjects with the variant HNF4A had lower CYP2D6 enzyme activity, although this result was not statistically significant (p = 0.16). In conclusion, our findings demonstrate strong evidence for a role of miRNAs in the regulation of HNF4A.
Introduction
Hepatic nuclear factor 4α (HNF4A) is an important transcriptional “master regulator” that regulates the expression of many genes involved in drug disposition. They include phase I enzymes, phase II enzymes, transporters, and additional transcriptional factors that also regulate these genes (Kamiyama et al., 2007). Hepatic HNF4A expression appears to be regulated by a complex set of positive and negative transcription factors in the HNF4A promoter (Hatzis and Talianidis, 2001). Because there remains substantial unexplained variability in the expression of HNF4A and its downstream targets (e.g., cytochrome P450 genes) (Wortham et al., 2007), we sought to identify additional mechanisms that regulate HNF4A and may ultimately contribute to the variable drug disposition. In these studies, we focused on identifying microRNAs (miRNAs) that regulate HNF4A expression.
miRNAs are small (18–25 nucleotides), noncoding RNAs that regulate gene expression post-transcriptionally. In animals, miRNAs typically bind to the 3′-untranslated region (3′-UTR) of mRNAs and negatively regulate gene expression by one of two mechanisms: by blocking protein translation or by degrading the mRNA (Olsen and Ambros, 1999; Ambros et al., 2003). As more miRNAs are identified and studied, new target sites and functions are being recognized. For example, it has now been shown that miRNAs can also bind to coding regions and repress gene expression (Duursma et al., 2008); this mechanism may explain some of the differential expression of mRNA splice variants. miRNAs also appear to be involved in the induction of gene expression; this induction occurs through binding to complementary regions in the promoter (Place et al., 2008) and the 5′-UTR (Ørom et al., 2008).
In the human genome, 1424 mature miRNAs have been reported so far (miRBase Registry version 17.0) (Griffiths-Jones et al., 2006). The bioinformatic algorithms predict that miRNAs can regulate 20 to 90% of the human transcripts (Lewis et al., 2005; Xie et al., 2005; Miranda et al., 2006). Each miRNA can regulate multiple genes, and each gene can be regulated by multiple miRNAs; therefore, these miRNAs form a broad and complex regulatory network.
miRNAs are involved in a wide range of biological activities including cell differentiation, cell death, cancer, and noncancerous human diseases (John et al., 2004). Emerging evidence indicates that these miRNAs also regulate genes involved in drug metabolism and disposition. At least 12 of the CYP genes are predicted by bioinformatic algorithms to be targets of miRNAs (Ramamoorthy and Skaar, 2011). In vitro studies have validated several of these predictions and have shown that several other drug disposition genes are also targets of miRNAs. Those studies have focused on genes such the ATP-binding cassette xenobiotic transporter ABCG2 (To et al., 2008), pregnane X receptor (Takagi et al., 2008), CYP1B1 (Tsuchiya et al., 2006), and CYP3A4 (Pan et al., 2009). Others have speculated that interindividual variability in cytochrome P450 expression, and drug response may be due to the action of miRNAs (Ingelman-Sundberg et al., 2007).
The activity of miRNAs can be affected by single nucleotide polymorphisms (SNPs) that occur either in the miRNA or in the miRNA target site on the mRNA. Such SNPs are called miRSNPs (Mishra et al., 2007). These miRSNPs can alter miRNA gene processing and/or the normal mRNA-miRNA interactions. Thus, these SNPs can create new miRNA target sites or destroy old target sites. Hence, these miRSNPs may also contribute to the interindividual variability in the enzyme expression and activity.
In this study, we hypothesized that endogenous miRNAs regulate the expression of HNF4A. To test this hypothesis, we first performed a comprehensive bioinformatic analyses to predict miRNAs that target HNF4A. Based on the bioinformatic analyses, we selected miRNAs that target HNF4A to perform further in vitro functional validation studies. Last, we identified polymorphisms in the miRNA target sites on the HNF4A 3′-UTR, which are predicted to alter the mRNA-miRNA interactions. Taken together, these results suggest that miRNAs are likely to play an important role in the regulation of drug metabolism.
Materials and Methods
In Vitro Studies.
Cell culture.
HeLa and HepG2 cells that were used in our in vitro transfection assays were obtained from American Type Culture Collection (Manassas, VA). All tissue culture reagents were purchased from Invitrogen (Carlsbad, CA). HeLa and HepG2 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Generation of luciferase reporter gene constructs.
The pIS-0 vector (plasmid 12178; Addgene, Cambridge, MA) (Yekta et al., 2004) was used to study 3′-UTR function. The 3′-UTR of HNF4A (1448 bp of the full-length 1724 bp; nucleotides 1558–3005 of NM_000457) was amplified using genomic DNA from the Coriell panel using primers (Integrated DNA Technologies, Coralville, IA) with NheI and SacI restriction sites (forward primer 5′-GGTGTTGAGCTCCTAAGAGAGCACCTGGTGA-3′ and reverse primer 5′-GGGTTTGCTAGCGGAGACCTGGGTTCAAG-3′; the restriction sites are italicized and the HNF4A sequence is underlined). The PCR product was cloned into the TOPO TA vector (Invitrogen), and the insert sequence was verified by DNA sequencing. The sequencing data revealed that the clone had the “variant” rs322210 (C allele); because the reported frequency of this allele is 55% [National Center for Biotechnology Information SNP database (dbSNP), http://www.ncbi.nlm.nih.gov/projects/SNP/], it seems to be the more common allele. Therefore, we used this clone. The insert was then subcloned into pIS-0 vector using the NheI and SacI restriction sites and transformed into DH5α competent cells. Colonies with inserts were identified by restriction digestion, and the sequence was verified by direct DNA sequencing. Plasmids were purified using the Plasmid Maxi Kit (QIAGEN, Valencia, CA). The DNA concentrations were determined using a Quant-iT DNA Broad Range kit (Invitrogen). The control vector is referred to as pIS-0, and the vector with HNF4A 3′-UTR sequence inserted is referred to as pIS-HNF4A. A plasmid with the variant HNF4A 3′-UTR (rs11574744) was created by site-directed mutagenesis (GenScript USA Inc., Piscataway, NJ). The presence of this mutation was confirmed by resequencing the plasmid. This mutant plasmid is referred to as pIS-HNF4A_SNP.
Transfection.
For luciferase assays, 0.9 × 105 HeLa cells were seeded into each well of a 24-well plate. The cells were transfected with 200 ng of either pIS-0 or pIS-HNF4A plasmid. Renilla luciferase reporter plasmid pGL4.74 was used as a transfection control; the pIS/pGL4.74 ratio was 50:1. Transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions using Opti-MEM (Invitrogen) and culture media without any antibiotics. At 24 h after transfection, the cells were harvested, and dual luciferase assays were performed as per manufacturer's instructions (Promega, Madison, WI). Transfections for the site-directed mutagenesis experiments were performed using the same protocol using 200 ng of pIS-0 or pIS-HNF4A or pIS-HNF4A_SNP plasmids.
For luciferase and miRNA cotransfection experiments, 1.5 × 106 HeLa cells were seeded in a T-25 flask. At 24 h, the cells were then transfected with 4 μg of pIS-0 or pIS-HNF4A plasmid, along with 80 ng of Renilla luciferase reporter plasmid as a transfection control. At 24 h after transfection, the cells were trypsinized, and 1 × 105 cells were seeded into each well of a 24-well plate. The cells were reverse-transfected (i.e., plated onto the well containing the transfection mix) with either a 30 nM concentration of the miRNA or combinations of miRNAs or negative control (miRIDIAN Mimics; Dharmacon, Lafayette, CO) using Lipofectamine 2000. At 24-h after miRNA transfection, the cells were harvested, and dual luciferase assays were performed. Cotransfections for the site-directed mutagenesis experiment were performed with the same protocol using the pIS-0, pIS-HNF4A, or pIS-HNF4A_SNP plasmids and the specific miRNAs. All transfections and luciferase assays were performed in triplicate on 3 different days.
To verify that the transfected synthetic miRIDIAN Mimics were efficiently taken up by the cells, we transfected HeLa cells (2 × 105 cells/well in 96-well plates) with a 30 nM concentration of either an hsa-miR-34a Mimic or a negative control Mimic (cel-miR-67). RNA isolation and reverse transcription were performed using a microRNA Cell-to-CT kit (Applied Biosystems, Foster City, CA) and specific TaqMan MicroRNA Assays (Applied Biosystems) for hsa-miR-34a, U6 small nuclear RNA (snRNA) (endogenous control), and hsa-miR-449a (nonspecific control) following the manufacturer's instructions. Specific details on the PCR are found under Quantification of miRNAs. The relative quantities of miRNA were calculated using the ΔΔCt method with U6 snRNA and negative control (cel-miR-67) transfections as controls. The relative miRNA expressions are reported as 2−(ΔΔCt) (Kreuzer et al., 1999). The miRNA transfections were replicated on 3 separate days, and the RT-PCR was performed in triplicate for each of the transfections. In an additional control experiment, we tested the effect of the transfection of the luciferase plasmids on miRNA expression. The transfections did not alter the expression of either hsa-miR-449a or hsa-miR-34a (data not shown).
RNA isolation.
Fresh human hepatocytes were isolated by Vitacyte LLC (Indianapolis, IN) from liver specimens that were collected after Indiana University's institutional review board approval. These hepatocytes were flash-frozen until RNA isolation. Total RNA, including small RNAs, was isolated using an miRNeasy kit (QIAGEN) following the manufacturer's instructions with the exception that phase separation was performed using MaXtract tubes (QIAGEN). The on-column DNase treatment step was included in the RNA isolation procedure and was done using a DNase set (QIAGEN). The RNA yield was determined using a Quant-iT RNA Broad Range kit (Invitrogen). RNA quality and integrity were assessed with an RNA 6000 Nano Labchip and BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA).
Quantification of miRNAs.
The miRNA expression levels were determined using specific TaqMan MicroRNA Assays following the manufacturer's instructions in a StepOne Plus real-time PCR instrument (Applied Biosystems). The PCR included a 10-min polymerase enzyme activation step at 95°C, 50 cycles each of denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 min. Relative quantities of miRNAs were calculated as 2−(ΔCt) using U6 snRNA as the endogenous control and was multiplied by 103 to simplify data presentation.
Quantification of HNF4A protein and mRNA.
In a six-well plate, 1 × 106 HepG2 cells/ml were reverse-transfected with 100 nM synthetic miRNA (hsa-miR-34a, hsa-miR-449a, hsa-miR-493*, and control cel-miR-67; miRIDIAN microRNA Mimics) using siPORT NeoFX transfection reagent (Ambion, Austin, TX) following the manufacturer's specifications. We also used a genome-wide siRNA for human HNF4A (Hs_HNF4A_9) and an AllStars Negative Control siRNA (QIAGEN) as described by Iwazaki et al. (2008) as process controls. At ∼72 h after transfection, the cells were harvested for HNF4A protein and RNA analyses. The transfections were repeated on 3 separate days.
For protein analyses, nuclear protein extract was isolated using the NucBuster protein extraction kit (Novagen, Madison, WI). Protein concentrations were determined with a bicinchoninic acid reagent protein assay kit (Thermo Fisher Scientific, Waltham, MA). The nuclear protein lysate (20 μg) was electrophoresed using a 4 to 20% Tris-glycine gel (Invitrogen) and transferred to a polyvinylidene difluoride transfer membrane (Millipore Corporation, Billerica, MA) using a Novex semidry blotter (Invitrogen). The membranes were blocked with 3% milk and then incubated overnight at 4°C with mouse anti-human HNF4A mouse monoclonal antibody (Perseus Proteomics, Tokyo, Japan) at a dilution of 1:5000, followed by horseradish peroxidase-conjugated ImmunoPure goat anti-mouse secondary antibody (Thermo Fisher Scientific) at a dilution of 1:10,000 for 1 h. As described previously for their HNF4A study by Iwazaki et al. (2008), β-actin was used as the internal control; a horseradish peroxidase-conjugated β-actin antibody (Abcam, Cambridge, MA) was used at a dilution of 1:5000. All three antibodies were diluted in Starting Block (T20) blocking buffer (Thermo Fisher Scientific). Protein bands were developed using a SuperSignal enhanced chemiluminescence kit (Thermo Fisher Scientific). The protein bands were visualized on a LAS-1000 plus system (Fujifilm, Tokyo, Japan).
For RNA analyses, total RNA, including small RNAs, was isolated and quantified as described above. cDNA was generated from 1 μg of total RNA with the reverse transcription system (Promega) following the manufacturer's instructions. The expression of HNF4A, PXR, and GAPDH (endogenous control) mRNAs was analyzed using specific TaqMan Gene Expression Assays (Applied Biosystems) following the manufacturer's instructions in a StepOne Plus real-time PCR instrument. The relative quantity of HNF4A mRNA was calculated using the ΔΔCt method using GAPDH and negative controls (cel-miR-67 and negative control siRNA) as reference controls. The RT-PCR was performed in triplicate for each of the three transfections.
Bioinformatics Studies.
Bioinformatic analysis to predict miRNAs.
To predict miRNAs that target the HNF4A gene, we used six different programs: miRanda (John et al., 2004), miRBase Targets (Griffiths-Jones et al., 2006), TargetScan (Lewis et al., 2003), PicTar (Krek et al., 2005), RNA22 (Miranda et al., 2006), and PITA (Kertesz et al., 2007). The parameter settings were either the default settings or those used in the publications describing the programs. For RNA22, which is a downloadable program with user defined mRNA and miRNA sequences, we used the UCSC Genome Browser (http://genome.ucsc.edu) to identify the HNF4A 3′-UTR sequence. The mature miRNA sequences (version 10.0) were downloaded from the miRBase Sequence database (Griffiths-Jones et al., 2006).
Identification and bioinformatic analysis of SNPs located in the HNF4A 3′-UTR.
SNPs from the HNF4A 3′-UTR were obtained from the dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and the UCSC Genome Browser database (http://genome.ucsc.edu/). The minor allele frequencies were obtained from dbSNP and SeattleSNPs database (http://pga.gs.washington.edu). We used two programs, PolymiRTS Database (Bao et al., 2007) and Patrocles (Georges et al., 2006) to predict the effect of miRSNPs in the HNF4A 3′-UTR on the mRNA-miRNA interaction.
In Vivo Human Studies.
Genotyping of HNF4A 3′-UTR SNP.
A custom TaqMan SNP genotyping assay (Applied Biosystems) was developed for HNF4A 3′-UTR SNP (rs11574744). The primer and probe sequences were CCCGAGAACATGGCCTAAGG as forward primer, CCAGAGCAGGGCGTCAA as reverse primer, VIC-ATCCCACAGCCACCC as probe 1, and FAM-ATCCCACTGCCACCC as probe 2 (variant sequence is underlined). Genotyping was performed in a StepOne Plus real-time PCR system using the recommended genotyping PCR conditions, that is, an initial denaturation for 10 min at 95°C, followed by 40 cycles of denaturation for 15 s at 92°C and annealing and extension for 1 min at 60°C. Because this was a custom assay, we confirmed the genotyping results by resequencing 96 DNA samples from the Coriell biorepository (48 white and 48 African-American samples). DNA resequencing was performed by Polymorphic DNA Technologies (Alameda, CA).
In addition, 151 white and 243 African-American subjects previously phenotyped with the CYP2D6 probe drug dextromethorphan and genotyped for CYP2D6 (Gaedigk et al., 2008) were screened for the presence of the SNP. To determine the effect of the HNF4A SNP on CYP2D6 enzyme activity, the urinary dextromethorphan metabolic ratio was compared after taking into account the CYP2D6 activity score. The CYP2D6 gene score was assigned on the basis of the expected enzyme activity (Gaedigk et al., 2008; Borges et al., 2010). The fully functional CYP2D6 alleles (*1 and *2) were assigned a score of 1, alleles associated with reduced enzyme activity (*10 and *41) were assigned a score of 0.5, and the nonfunctional alleles (*3–*6) were assigned a score of 0. The CYP2D6 activity score was the summation of the two values assigned to the individual alleles.
Statistical Analysis.
Statistical analyses were performed as described in the figure legends, using SAS 9.1 software. The comparisons of relative luciferase activities and ΔCt among different treatment groups were analyzed with linear mixed models, in which between-day and within-day variances are treated as random effects. p < 0.05 was considered statistically significant.
Results
HNF4A 3′-UTR Has Repressive Luciferase Activity In Vitro.
In our first functional study, we cloned the 3′-UTR of the HNF4A gene into the 3′-UTR of the luciferase gene of the pIS-0 luciferase reporter construct, pIS-HNF4A (Supplemental Fig. 1A). The pIS-0 reporter system has been used by others to study the miRNA regulation of several other target genes (Yekta et al., 2004; Adams et al., 2007). The pIS-HNF4A plasmid had significantly lower luciferase activity (∼60%; p < 0.001) compared with the pIS-0 (control) plasmid (Supplemental Fig. 1B). The effect of HNF4A 3′-UTRs was consistent across two other concentrations of transfected plasmids that we tested (100 and 400 ng; p < 0.001; data not shown). This result indicated that the 3′-UTR of the HNF4A gene has repressive activities, possibly through miRNA targeting.
Bioinformatic Predictions to Identify miRNAs Predicted to Target HNF4A.
We used six bioinformatic algorithms to identify miRNAs that are predicted to target HNF4A. The HNF4A gene has a 1724-bp-long 3′-UTR and was predicted to be targeted by 350 different miRNAs (Supplemental Tables 1 and 2). Candidate miRNAs for functional testing were selected on the basis of two criteria: 1) the miRNA predicted by two or more of the bioinformatic algorithms; and 2) the presence of a predicted favorable energy of binding (ΔG ≤ −25 kcal/mol) between the miRNA and the target sequence on the mRNA. Among those that fit these criteria, we initially selected hsa-miR-34c-5p, hsa-miR-449a, and hsa-miR-766, on the basis of the number of predicting algorithms and their ranking energy of binding. As a negative control, we chose hsa-miR-493* (formerly referred to as hsa-miR-493-5p), which was not predicted to target HNF4A.
miRNAs Regulate HNF4A 3′-UTR Luciferase Activity In Vitro.
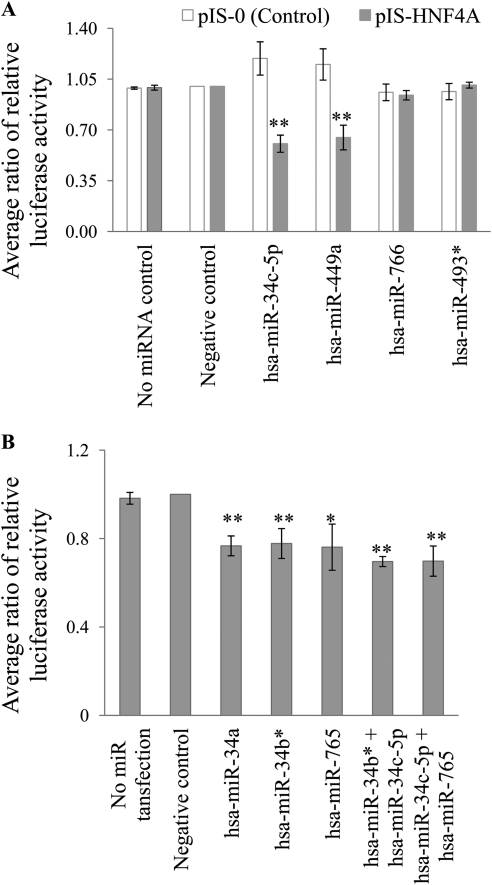
The next set of studies focused on testing the effect of individual miRNAs on the pIS-HNF4A plasmid. The selected miRNAs were cotransfected with either pIS-0 or pIS-HNF4A. Relative to the negative control miRNA (cel-miR-67), hsa-miR-34c-5p and hsa-miR-449a reduced the luciferase activity of the pIS-HNF4 plasmid by 40% (p < 0.001) and 35% (p < 0.001), respectively (Fig. 1A). In contrast, hsa-miR-766 did not have an impact on the expression of pIS-HNF4A and neither did the nonspecific control, hsa-miR-493*.
Fig. 1.
Regulation of pIS-HNF4A by miRNAs. HeLa cells were cotransfected with the following. A, pIS-0 or pIS-HNF4A luciferase plasmids, along with the Renilla reporter plasmid for normalization. These cells were also transfected with miRNA Mimics. The data are expressed as the pIS-HNF4A luciferase activity corrected for Renilla luciferase and normalized to the negative control (cel-miR-67) within each experiment (mean ± S.E.M.; n = 3 independent experiments). B, pIS-HNF4A luciferase plasmid along with the Renilla reporter plasmid for normalization. These cells were also transfected with miRNA Mimics individually (cel-miR-67, hsa-miR-34a, hsa-miR-34b*, or hsa-miR-765; 30 nM) or together (hsa-miR-34b* + hsa-miR-34c-5p or hsa-miR-34c-5p + hsa-miR-765; 15 nM each). All data are expressed as pIS-HNF4A luciferase activity corrected for Renilla luciferase and normalized to the negative control (cel-miR-67) within each experiment (mean ± S.E.M.; n = 3 independent experiments). *, p < 0.05; **, indicates p < 0.001, compared with the negative control.
In another set of transfections, we tested three additional miRNAs for activity against the pIS-HNF4A including hsa-miR-34a (same family as hsa-miR-34c-5p), hsa-miR-34b* (which is a part of a single large precursor miRNA transcript that makes hsa-miR-34c-5p), and hsa-miR-765. These miRNAs were selected because they were predicted by more than two bioinformatic algorithms and the predicted energy of binding was favorable. Compared with the negative control miRNA (cel-miR-67), hsa-miR-34a, hsa-miR-34b*, and hsa-miR-765 reduced pIS-HNF4A luciferase activity by 23% (p < 0.001), 22% (p < 0.001), and 24% (p < 0.001), respectively (Fig. 1B). Because multiple miRNAs can simultaneously interact with a target mRNA (Krek et al., 2005), we cotransfected hsa-miR-34c-5p with hsa-miR-34b* and hsa-miR-34c-5p with hsa-miR-765. These combinations of miRNAs significantly down-regulated (p < 0.001) pIS-HNF4A luciferase activity. However, we did not observe any additive or synergistic effects of the miRNAs when they were transfected together. In the case of hsa-miR-34c-5p cotransfection with hsa-miR-34b*, the lack of an additional or synergistic effect may be because of competition, as they are predicted to share one target site in common.
Expression of miRNAs in Human Hepatocytes.
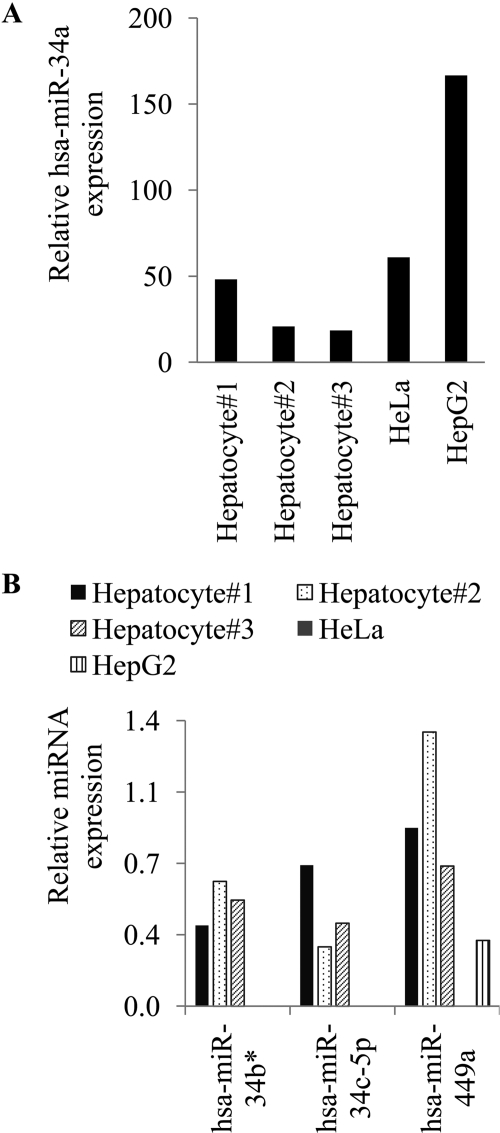
To determine whether the miRNAs that appear to target HNF4A are expressed in hepatocytes, we measured their expression in primary hepatocytes isolated from three individual subjects. In addition, we also measured their expression in HeLa and HepG2 cells. We performed quantitative real-time PCR using specific TaqMan miRNA assays. U6 snRNA was used as endogenous control. hsa-miR-34a was easily detectable in all three hepatocyte preparations, as well as in HeLa and HepG2 cells (Fig. 2A). The other three hsa-miRNAs (hsa-miR-34b*, hsa-34c-5p, and hsa-miR-449a) were also expressed in hepatocytes, although at a lower level (Fig. 2B). Neither hsa-miR-34b* nor hsa-miR-34c-5p was detectable in HeLa or HepG2 cells. hsa-miR-449a was expressed in HepG2 but was not detectable in HeLa cells. On the basis of their expression, hsa-miR-34a and hsa-miR-449a were selected for further in vitro studies.
Fig. 2.
Mature miRNA expression in human cell lines and hepatocytes. Total RNA, including the small RNAs, was isolated from three different human hepatocytes preparations and from the HeLa and HepG2 cell lines. To show the variability between hepatocyte preparations, the results from each hepatocyte preparation are graphed separately. RT-PCR assays were performed with U6 snRNA as the internal control. The relative miRNA expression was calculated as 2−(ΔCt). Values were multiplied by 103 to simplify data presentation. The values shown in the graph were obtained from triplicate assays in a single PCR experiment. Error bars are not shown because true biological replicates (plated on different days) cannot be obtained from human hepatocyte isolations. Therefore, separate bars are shown for each hepatocyte preparation. A, expression of hsa-miR-34a. B, expression of hsa-miR-34b*, hsa-miR-34c-5p, and hsa-miR-449a.
Effect of miRNAs on HNF4A mRNA and Protein Expression.
To investigate whether these miRNAs regulate HNF4A mRNA and protein expression, we transfected HepG2 cells with hsa-miR-34a, hsa-miR-449a, hsa-miR-493*, and the negative control cel-miR-67. Control experiments demonstrating successful miRNAs transfection into the cells are shown in Supplemental Fig. 2. Both hsa-miR-34a and hsa-miR-449a were predicted to target HNF4A at two positions, and they both target the same two locations: 1) positions 164 to 171 and 2) positions 254 to 260 of HNF4A 3′-UTR corresponding to the miRNA seed sequence. hsa-miR-493* is not predicted to target HNF4A and hence was used as a negative control. We also used HNF4A siRNA and a negative control siRNA as described by Iwazaki et al. (2008) as process controls.
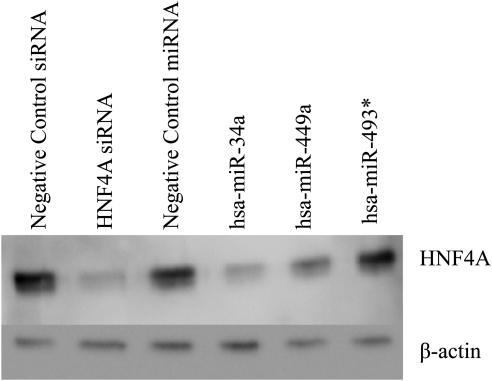
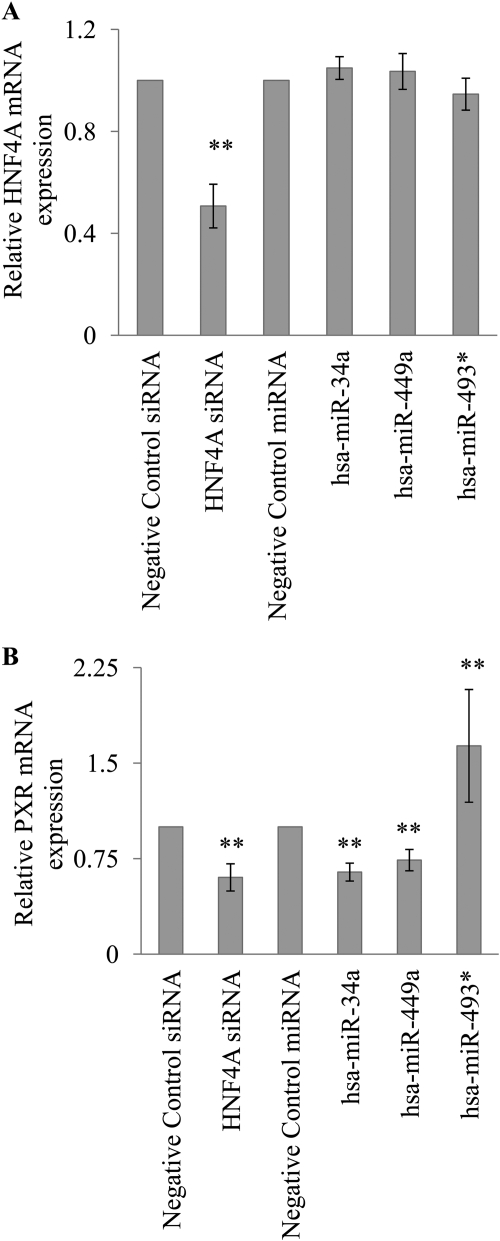
HNF4A protein expression was down-regulated by hsa-miR-449a, hsa-miR-34a, and the positive control HNF4A siRNA (Fig. 3). The HNF4A mRNA expression was down-regulated by the HNF4A siRNA but not by the miRNAs (Fig. 4A), indicating that these miRNAs must be blocking HNF4A translation and not causing degradation of the HNF4A mRNA. To determine whether the down-regulation of the HNF4A gene resulted in altered expression of downstream HNF4A target genes, we determined the effect of the miRNAs on PXR mRNA expression. PXR is a target of HNF4A that has been used as a marker of HNF4A activity (Iwazaki et al., 2008). The expression of PXR mRNA was significantly down-regulated (p < 0.001) by ∼30% by hsa-miR-449a, ∼40% by hsa-miR-34a, and ∼40% by HNF4A siRNA (Fig. 4B). In contrast, hsa-miR-493* did not alter HNF4A mRNA or HNF4A protein expression. We were surprised to find that hsa-miR-493* appeared to up-regulate PXR mRNA expression (p < 0.001); however, the mechanism is unclear.
Fig. 3.
Regulation of HNF4A protein by miRNAs. Representative Western blot using anti-HNF4A and anti-β-actin (internal control) antibodies. HepG2 cells were transfected with miRNAs (hsa-miR-34a, hsa-miR-449a, hsa-miR-493*, or cel-miR-67; 100 nM) or siRNAs (HNF4A siRNA, or negative control siRNA; 100 nM). At 72 h after transfection, nuclear protein was isolated and Western blot assays were performed.
Fig. 4.
Regulation of HNF4A and PXR mRNA by miRNAs. HepG2 cells were transfected with miRNAs (hsa-miR-34a, hsa-miR-449a, hsa-miR-493*, or cel-miR-67; 100 nM) or siRNAs (HNF4A siRNA or negative control siRNA; 100 nM). At 72 h after transfection, RNA was isolated, and RT-PCR assays were performed. GAPDH was used as the internal control. The miRNA transfections were normalized to cel-miR-67, and the siRNA transfections were normalized to negative control siRNA. The relative mRNA expression was calculated as 2−(ΔΔCt) (mean ± S.E.M.; n = 3 independent experiments performed in triplicate). A, expression of HNF4A mRNA. B, expression of PXR mRNA. **, p < 0.001.
SNPs in the miRNA Target Sites.
Because SNPs in the miRNA target sites of mRNA 3′-UTRs can alter normal miRNA-mRNA interactions, we searched for SNPs in the 3′-UTR of HNF4A that may disrupt or create miRNA-mRNA interactions. The dbSNP database indicated that many SNPs are present in the 3′-UTRs of HNF4A. We used two programs: PolymiRTS Database (Bao et al., 2007) and Patrocles (Georges et al., 2006) to determine whether any of these 3′-UTR SNPs that exists in miRNA target sites are predicted to affect the base-pairing between HNF4A mRNA and the target miRNAs. There were five SNPs in the HNF4A gene that are predicted to destroy six miRNA target sites and create two new miRNA target sites (Supplemental Table 3).
In Vitro Validation of SNP Predictions.
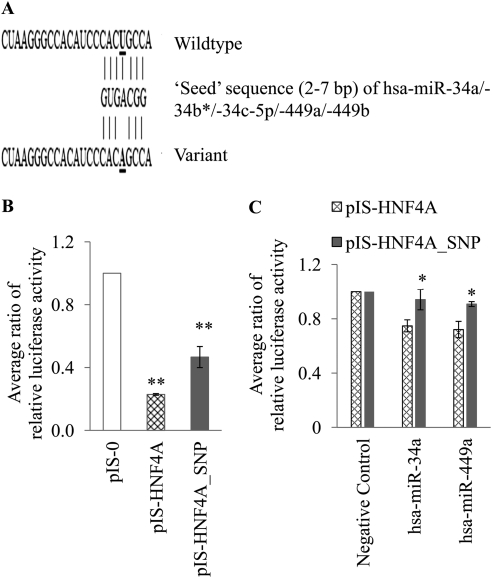
To test the hypothesis that germline variations can alter mRNA-miRNA interactions, we selected a SNP (rs11574744; T>A) in the HNF4A 3′-UTR for further in vitro validation (Fig. 5A). This SNP was predicted by both PolymiRTS Database and Patrocles programs to destroy a miRNA binding site (Supplemental Table 3). The expected result of this would be increased luciferase activity because of reduced negative regulation by the miRNA. In our first functional study, we compared the activity of pIS-0, pIS-HNF4A, and pIS-HNF4A_SNP plasmids. The luciferase activity from the plasmid with the variant HNF4A was 2-fold higher (p < 0.001) than the wild type pIS-HNF4A plasmid (Fig. 5B).
Fig. 5.
In vitro validation of SNP predictions. A, predicted miRNA interaction with wild-type and variant (rs11574744) HNF4A 3′-UTR. B, transfection of HeLa cell line with 200 ng of control pIS-0 plasmid or pIS-HNF4A or pIS-HNF4A_SNP constructs with the Renilla luciferase plasmid as internal control. Dual luciferase assays were performed at 24 h. Data are expressed as the pIS-HNF4A and pIS-HNF4A_SNP luciferase activity corrected for Renilla luciferase and normalized to pIS-0 within each experiment (mean ± S.E.M.). C, HeLa cells were cotransfected with 4 μg of pIS-HNF4A or pIS-HNF4A_SNP luciferase constructs, along with Renilla reporter plasmid for normalization. The cells were also transfected with miRNA Mimics (hsa-miR-34a or hsa-miR449a; 30 nM). All data are expressed as the pIS-HNF4A or pIS-HNF4A_SNP luciferase activity corrected for Renilla luciferase and normalized to the negative control (cel-miR-67) within each experiment (mean ± S.E.M.; n = 3 independent experiments). The assays were done in triplicate on three different days. *, p < 0.05.
In our second functional study, we cotransfected the plasmids (pIS-0 or pIS-HNF4A or pIS-HNF4A_SNP) with hsa-miR-34a, hsa-miR-449a, and the negative control (cel-miR-67). Compared with the negative control miRNA (cel-miR-67), hsa-miR-34a and hsa-miR-449a reduced the pIS-HNF4A luciferase activity by 25 and 28%. respectively, whereas these two miRNAs reduced the pIS-HNF4A_SNP luciferase activity by only 6 and 9%, respectively (Fig. 5C). The luciferase activity was significantly higher (p < 0.05) in the pIS-HNF4A_SNP plasmid transfected cells compared with the pIS-HNF4A transfected cells, when transfected with either the miR-34a or the miR-449a miRNAs (Fig. 5C).
We used RNAfold (Gruber et al., 2008) to determine whether this SNP changed the predicted mRNA secondary structure. We included 70-bp nucleotide flank on either side of the SNP (Kertesz et al., 2007) to assess the minimum folding energy and secondary structure. The minimum folding energy was the same for both the wild-type and SNP sequences (Supplemental Fig. 3). Likewise, no differences were observed when we extended the flanking sequence to 200 bp on either side of the SNP.
To determine the genotype frequency of rs11574744 in different ethnicities, we designed a custom TaqMan assay (Applied Biosystems) to genotype the Coriell human diversity panel comprising 94 whites, 89 African-American, and 87 Asian DNA samples. The results of our genotyping suggested that the SNP is present only in African Americans. The minor allele frequency was 3.4%.
To determine whether this HNF4A SNP affects CYP2D6 enzyme activity, we genotyped the rs11574744 SNP in 151 whites and 243 African Americans that were previously phenotyped for CYP2D6 with dextromethorphan (Gaedigk et al., 2008). The hypothesis was that the SNP destroys a mRNA-miRNA interaction, thereby increasing HNF4A mRNA protein expression and resulting in increased expression of downstream target mRNA (e.g., CYP2D6). The SNP was present only in the African-American cohort (minor allele frequency 4.6%). We compared the urinary dextromethorphan/dextrorphan (DM/DX) ratio in subjects with wild-type versus rs11574744 carrier (both homozygous and heterozygous) genotypes. Although not statistically significant (p = 0.10), the DM/DX ratio was numerically lower in subjects who were carriers of the variant, which is consistent with our in vitro mechanistic studies. When the analysis was focused on only subjects who were genotypically extensive metabolizers (functional CYP2D6 activity scores of 1.5 or 2.0), there was also a trend in the same direction (p = 0.16; means ± S.E.M. of log DM/DX for the wild type and carriers were −1.84 ± 0.05 and −2.07 ± 0.14, respectively).
Discussion
Interindividual variability in drug metabolism remains a significant contributor to differences in drug efficacy and toxicity. Some of this variability is due to known genetic variations and environmental factors that inhibit the enzymes or alter their expression levels. However, the mechanisms that underlie much of the variability are as yet unknown. The studies presented here provide evidence for a role of miRNAs in the regulation of drug disposition through transcriptional factors that regulate drug-metabolizing enzymes.
Our bioinformatic analysis predicted that HNF4A is targeted by many miRNAs. HNF4A is an important transcriptional “master regulator” of phase I and II enzymes, transporters, and transcriptional factors (Kamiyama et al., 2007). Thus, regulation of HNF4A gene expression by miRNAs would probably affect many genes involved in drug metabolism and disposition. HNF4A is also expressed in kidney, intestine, and pancreas; in those tissues, it controls lipid (Hayhurst et al., 2001) and glucose metabolism (Stoffel and Duncan, 1997). Therefore, targeting of HNF4A by miRNAs may also affect other functions. Nine different isoforms of HNF4A have been reported (Harries et al., 2008). Because the predominant isoform of HNF4A expressed in the liver is isoform 2 (Ihara et al., 2005), which contains the full-length 3′-UTR, it is likely to be regulated by miRNAs.
As with many bioinformatic predictions, there was substantial interalgorithm variability in the miRNAs that were predicted to target each gene. Part of this variability may be due to the different miBase database versions that are used by each algorithm; they ranged from versions 9 to 11. The variability may also be due to differences in the algorithms; these include differences in parameters such as degree of complementarity and differences in 3′-UTR annotations and species conservation. The total number of miRNAs predicted by the algorithms will continue to change as more miRNAs are being discovered and as the prediction algorithms are fine-tuned. These predicted miRNAs provided a starting point for subsequent laboratory analyses.
After our bioinformatic predictions, our in vitro studies also supported a role of miRNAs in the regulation of HNF4A. Five of the miRNAs regulated the pIS-HNF4A luciferase plasmid, and at least two of those also regulated HNF4A protein expression. Because the miRNAs did not significantly reduce the HNF4A mRNA levels, we presume that they are regulating HNF4A expression by blocking the translation of HNF4A mRNA into protein, which is a common mechanism of regulation by miRNAs (Ambros et al., 2003; Bartel, 2004). This presumption is supported by our results showing that the HNF4A-targeting miRNAs also suppressed the expression of a downstream HNF4A target, PXR. Similar to the actions of many miRNAs reported so far, the miRNAs only partially blocked HNF4A expression and activity. This result is consistent with the role of miRNAs as a fine-tuning mechanism in the regulation of the target genes. However, it could also be that when multiple miRNAs together target a gene, there are actually very large effects. Because HNF4A mRNA is predicted to be targeted by many miRNAs, additional testing will be required to determine which other miRNAs may target HNF4A and their collective impact on HNF4A gene expression.
There are a wide variety of HNF4A-regulated genes that we could study, however, we chose to focus on PXR first, because it is regulated by HNF4A in HepG2 cells (Iwazaki et al., 2008). Two of the miRNAs did in fact alter PXR expression. Although the bioinformatic analysis suggested that PXR itself may also be a weak target of these miRNAs, they are less likely to cause the observed reduction in PXR mRNA expression because the PXR target sequence contained several mismatches. Similar to PXR, it is likely that several of the other drug-metabolizing enzymes that are downstream targets of HNF4A are also indirectly regulated by miRNAs that target HNF4A. Testing those downstream targets will be best done in future studies using primary hepatocytes, because the mRNA and protein expression of most of the drug-metabolizing enzymes are either low or absent in the immortalized cell lines available. It will also be of interest to determine whether any of these miRNAs are regulated by xenobiotics and drugs. One of the HNF4A-regulating miRNAs that we identified in this study, hsa-miR-34a, was also recently shown to regulate HNF4A protein, but not mRNA levels, in a different model system, which further support our findings (Takagi et al., 2010).
miRNA functions can be altered by genetic variants that affect the miRNA binding to the target mRNA; these variants are called miRSNPs (Mishra et al., 2007). They occur either in the miRNA or in the mRNA. miRSNPs that create new functional miRNA binding sites would cause additional down-regulation of the target gene. In contrast, miRSNPs that destroy miRNA target sites would result in a loss of targeting and elevated expression of the target gene. miRSNPs have been shown to alter mRNA-miRNA interactions in genes such as dihydrofolate reductase (Mishra et al., 2007) and estrogen receptor α (Adams et al., 2007). Genetic variants in the 3′-UTRs of genes have not typically been given high priority in functional studies; however, on the basis of our analyses, they may contribute to interindividual variability in the expression of the drug-metabolizing enzymes. Some studies have reported on the SNPs in the 3′-UTRs of CYP genes that may be associated with altered phenotypes; these include CYP19A1 (Dunning et al., 2004) and CYP2A6 (Wang et al., 2006). It is conceivable that these SNPs may be a target of miRNAs. We have identified SNPs in the HNF4A 3′-UTR that are predicted to alter the miRNA interactions (Supplemental Table 3). Our in vitro luciferase assay revealed that at least one of these genetic variants can alter mRNA-miRNA interactions (Fig. 5). Furthermore, in our clinical study, we observed a trend in the expected direction toward lower CYP2D6 activity in subjects with the variant HNF4A. Because of the low minor allele frequency of this SNP, larger African-American population studies will be required to confirm these findings.
Both the algorithms, PolymiRTS Database and Patrocles, only predicted whether the SNPs in the “seed” region of the miRNA target sites affect the base-pairing between mRNA and miRNA. However, SNPs in “nonseed” regions can also affect miRNAs that bind either upstream or downstream of the SNP (Mishra et al., 2007). Neither of these algorithms predict such loss or gain of these mRNA-miRNA interactions. Likewise, SNPs in the mature miRNAs and premiRNA may also affect the mRNA-miRNA interaction. Because very few of the miRNA genes have been resequenced in depth, the genetic variants in those genes are not well characterized. Therefore, we have not included them in this analysis.
Our findings provide evidence for the role of miRNAs in the regulation of the transcriptional factor HNF4A. Recent studies have shown that other proteins involved in drug metabolism (Tsuchiya et al., 2006; Takagi et al., 2008; To et al., 2008; Pan et al., 2009) are also subject to miRNA regulation. Our results, taken together with those findings, suggest a complex regulatory mechanism for cytochromes P450 by miRNAs. The identification of the endogenous hepatic miRNAs that regulate CYP genes directly or indirectly should help us to better understand the variability in therapeutic efficacy and toxicity for patients to numerous commonly used drugs. Furthermore, identifying polymorphisms that alter the drug-metabolizing mRNA-miRNA interactions would probably be a clinically important biomarker for guiding the use of P450-metabolized drugs. Ultimately, we expect that these new biomarkers would help improve the efficacy and reduce the side effects of the commonly prescribed drugs.
Supplementary Material
Acknowledgments
We are grateful to Dr. Robert McCarthy (Vitacyte, Indianapolis, IN) for providing us with isolated human hepatocytes.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant 1R01-GM088076] (to T.C.S., A.G., L.L.); the National Institutes of Health Pharmacogenomics Research Network [Grant 5U01-GM061373-07] (to D.A.F.); the National Institutes of Health Agency for Healthcare Research and Quality [Grant R01-HS019818CM]; the Indiana University Cancer Center; and a Department of Defense predoctoral fellowship [Grant BC083078] (to A.R.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
This work was previously presented in part as follows: Ramamoorthy A (2010) Mechanisms of Variability in CYP2D6 Metabolism: The Contributions of Polymorphisms, Copy Number Variations and MicroRNA. Ph.D. thesis, Indiana University, Indianapolis; Ramamoorthy A, Flockhart DA, and Skaar TC (2008) Identification of microRNAs that target HNF4A. Proceedings of the Pharmacogenomics Meeting; 2008 Nov 19–22; Cold Spring Harbor, NY. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. Ramamoorthy A, Flockhart DA, and Skaar TC (2009) Identification of microRNAs that target HNF4A. Proceedings of the ASCPT Annual Meeting; 2009 Mar 18–21; Washington, DC. American Society for Clinical Pharmacology and Therapeutics, Alexandria, VA; Ramamoorthy A, Li L, Flockhart DA, and Skaar TC (2010) Identification of microRNAs that target HNF4A. Proceedings of the Workshop on Genetic Polymorphisms in Drug Disposition; 2010 Apr 13; Indianapolis, IN. International Society for the Study of Xenobiotics, Washington, DC.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- HNF4A
- hepatic nuclear factor 4α
- miRNA
- microRNA
- 3′-UTR
- 3′-untranslated region
- SNP
- single nucleotide polymorphism
- miRSNP
- microRNA SNP
- bp
- base pair
- PCR
- polymerase chain reaction
- snRNA
- small nuclear RNA
- cel-miR
- Caenorhabditis elegans microRNA
- hsa-miR
- Homo sapiens microRNA
- RT
- reverse transcriptase
- siRNA
- small interfering RNA
- PXR
- pregnane X receptor
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DM/DX
- dextromethorphan/dextrorphan.
Authorship Contributions
Participated in research design: Ramamoorthy, Benson, Flockhart, and Skaar.
Conducted experiments: Ramamoorthy, Gaedigk, Bradford, and Benson.
Performed data analysis: Ramamoorthy and Li.
Wrote or contributed to the writing of the manuscript: Ramamoorthy, Li, Gaedigk, Bradford, Benson, Flockhart, and Skaar.
References
- Adams BD, Furneaux H, White BA. (2007) The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21:1132–1147 [DOI] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. (2003) A uniform system for microRNA annotation. RNA 9:277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, Cui Y. (2007) PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res 35:D51–D54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- Borges S, Desta Z, Jin Y, Faouzi A, Robarge JD, Philips S, Philip S, Nguyen A, Stearns V, Hayes D, et al. (2010) Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 50:450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Dowsett M, Healey CS, Tee L, Luben RN, Folkerd E, Novik KL, Kelemen L, Ogata S, Pharoah PD, et al. (2004) Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst 96:936–945 [DOI] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. (2008) miR-148 targets human DNMT3b protein coding region. RNA 14:872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. (2008) The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 83:234–242 [DOI] [PubMed] [Google Scholar]
- Georges M, Clop A, Marcq F, Takeda H, Pirottin D, Hiard S, Tordoir X, Caiment F, Meish F, Bibé B, et al. (2006) Polymorphic microRNA-target interactions: a novel source of phenotypic variation. Cold Spring Harb Symp Quant Biol 71:343–350 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. (2008) The Vienna RNA websuite. Nucleic Acids Res 36:W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries LW, Locke JM, Shields B, Hanley NA, Hanley KP, Steele A, Njølstad PR, Ellard S, Hattersley AT. (2008) The diabetic phenotype in HNF4A mutation carriers is moderated by the expression of HNF4A isoforms from the P1 promoter during fetal development. Diabetes 57:1745–1752 [DOI] [PubMed] [Google Scholar]
- Hatzis P, Talianidis I. (2001) Regulatory mechanisms controlling human hepatocyte nuclear factor 4α gene expression. Mol Cell Biol 21:7320–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. (2001) Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara A, Yamagata K, Nammo T, Miura A, Yuan M, Tanaka T, Sladek FM, Matsuzawa Y, Miyagawa J, Shimomura I. (2005) Functional characterization of the HNF4α isoform (HNF4α8) expressed in pancreatic β-cells. Biochem Biophys Res Commun 329:984–990 [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116:496–526 [DOI] [PubMed] [Google Scholar]
- Iwazaki N, Kobayashi K, Morimoto K, Hirano M, Kawashima S, Furihata T, Chiba K. (2008) Involvement of hepatocyte nuclear factor 4α in transcriptional regulation of the human pregnane X receptor gene in the human liver. Drug Metab Pharmacokinet 23:59–66 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. (2004) Human microRNA targets. PLoS Biol 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. (2007) Role of human hepatocyte nuclear factor 4α in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab Pharmacokinet 22:287–298 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284 [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500 [DOI] [PubMed] [Google Scholar]
- Kreuzer KA, Lass U, Bohn A, Landt O, Schmidt CA. (1999) LightCycler technology for the quantitation of bcr/abl fusion transcripts. Cancer Res 59:3171–3174 [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. (2003) Prediction of mammalian microRNA targets. Cell 115:787–798 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217 [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. (2007) A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA 104:13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. (1999) The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol 216:671–680 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH. (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30:460–471 [DOI] [PubMed] [Google Scholar]
- Pan YZ, Gao W, Yu AM. (2009) MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos 37:2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA 105:1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy A, Skaar TC. (2011) In silico identification of microRNAs predicted to regulate the drug metabolizing cytochrome P450 genes. Drug Metabolism Letters 5:126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel M, Duncan SA. (1997) The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA 94:13209–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. (2010) MicroRNAs regulate human hepatocyte nuclear factor 4α, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 285:4415–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Mohri T, Yokoi T. (2008) Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem 283:9674–9680 [DOI] [PubMed] [Google Scholar]
- To KK, Zhan Z, Litman T, Bates SE. (2008) Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol 28:5147–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. (2006) MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66:9090–9098 [DOI] [PubMed] [Google Scholar]
- Wang J, Pitarque M, Ingelman-Sundberg M. (2006) 3′-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem Biophys Res Commun 340:491–497 [DOI] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. (2007) Expression of constitutive androstane receptor, hepatic nuclear factor 4α, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos 35:1700–1710 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594–596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.