Abstract
NADPH-cytochrome P450 oxidoreductase (CYPOR) variants have been described in patients with perturbed steroidogenesis and sexual differentiation, related to Antley-Bixler syndrome (ABS). It is important to determine the effect of these variants on CYP3A4, the major drug-metabolizing cytochrome P450 (P450) in humans. In this study, 12 CYPOR_ABS variants were separately coexpressed with CYP3A4 in a robust in vitro system to evaluate the effects of these variants on CYP3A4 activity in a milieu that recapitulates the stoichiometry of the mammalian systems. Full-length CYPOR variants were coexpressed with CYP3A4, resulting in relative expression levels comparable to those found in hepatic tissue. Dibenzylfluorescein (DBF), a CYP3A-specific reporter substrate (Biopharm Drug Dispos 24:375–384, 2003), was used to compare the variants and wild-type (WT) CYPOR activities with that of human liver microsomes. CYP3A4, combined with WT CYPOR, demonstrated kinetic parameters (kcat and Km) equal to those for pooled human liver microsomes. CYPOR variants Y181D, Y459H, V492E, L565P, and R616X all demonstrated maximal loss of CYP3A4 catalytic efficiency, whereas R457H and G539R retained ∼10 and 30% activities, respectively. Conversely, variants P228L, M263V, A287P, and G413S each showed WT-like capacity (kcat/Km), with the A287P variant being formerly reported to exhibit substantially lower catalytic efficiency. In addition, Q153R exhibited 60% of WT CYPOR capacity to support the DBF O-debenzylation reaction, contradicting increased catalytic efficiency (kcat/Km) relative to that for the WT, reported previously. Our data indicate the importance of use of simulated, validated in vitro systems, employing full-length proteins with appropriate stoichiometric incorporation of protein partners, when pharmacogenetic predictions are to be made for P450-mediated biotransformation.
Introduction
Cytochromes P450 (P450s) are members of a large superfamily of heme proteins, playing an important role in xenobiotic biotransformation, as well as in the endobiotic biosynthesis and catabolism of steroid hormones, bile acids, lipid-soluble vitamins, and fatty acids (Guengerich, 2005). There are more than four dozen human microsomal P450s (Guengerich, 2005) (http://www.cypalleles.ki.se/), activities of which are supported by electron transfer from NADPH-cytochrome P450 oxidoreductase (CYPOR). This reductase, encoded by the POR gene, is a ∼78-kDa microsomal flavoprotein, containing three distinct domains for binding its cofactors NADPH, flavin adenine dinucleotide (FAD), and flavin mononucleotide (FMN) (Porter and Kasper, 1986; Porter et al., 1990; Wang et al., 1997). The fourth CYPOR domain is the connecting domain, containing a flexible hinge region, which is involved in the recently established transition between its closed and open conformations (Aigrain et al., 2009; Ellis et al., 2009; Hamdane et al., 2009). Moreover, CYPOR has a hydrophobic N-terminal transmembrane segment (25–30 amino acids), responsible for its orientation in the endoplasmic reticulum and for the interaction of CYPOR with P450s, because its deletion leads to loss of its P450 reductase activity (Black and Coon, 1982).
More than 50 different haplotypes of the human POR gene have been described so far (http://www.cypalleles.ki.se/por.htm). Considerable numbers of these haplotypes have been found in patients with Antley-Bixler syndrome (ABS) (OMIM 201750), a disorder characterized by severe developmental abnormalities (Antley and Bixler, 1975). The majority of these ABS-related POR mutations cause a reduction in CYPOR activity, such as Y181D, involved in FMN binding (Arlt et al., 2004; Huang et al., 2005; Marohnic et al., 2010), Y459H and V492E, involved in FAD binding (Marohnic et al., 2006; Kranendonk et al., 2008), and G539R, positioned in the NADPH binding domain (Huang et al., 2005). Of interest, some mutant proteins such as variant Q153R were reported to support increased P450 activity, compared with the wild-type enzyme (Agrawal et al., 2008, 2010; Flück et al., 2010; Sandee et al., 2010). Variant A287P has been described as having differential behavior with different P450s (Fukami et al., 2005; Dhir et al., 2007). Many of these studies were performed with in vitro (reconstituted) systems, using N-terminal end-deleted CYPOR and extreme CYPOR/P450 ratios in favor of CYPOR (>1), the opposite of those found in vivo, estimated to be 1:3 to 10 (CYPOR/P450) (Paine et al., 1997; Venkatakrishnan et al., 2000) in favor of P450. These in vivo CYPOR/P450 ratios implicate a highly dynamic process in which P450 (as well as other redox partners; see above, this section) must compete for CYPOR.
Although the majority of studies on these CYPOR variants were initially focused on steroidogenesis, studies of our group (Kranendonk et al., 2008; Marohnic et al., 2010) and of others (Agrawal et al., 2008; Hart et al., 2008; Gomes et al., 2009) have attempted to evaluate the effects of these CYPOR variants on drug-metabolizing P450s. Two recent studies (Agrawal et al., 2010; Flück et al., 2010) described the effect of ABS-related CYPOR variants on the major drug-metabolizing enzyme in humans, CYP3A4, which together with CYP3A5 is estimated to be involved in the metabolism of more than 50% of currently prescribed drugs (Guengerich, 2005).
In this current report, we have studied 12 different CYPOR_ABS variants in a robust in vitro Escherichia coli expression system designed to evaluate the effect of these CYPOR variants on CYP3A4 activity. To draw conclusions regarding the effect of CYPOR mutants on drug metabolism, it is important to maintain the physiological ratios of CYPOR to P450 as practically as possible. We have used the same system as used in our former studies of ABS-related CYPOR variants in combination with human P450, namely the bacterial cell model BTC_CYP (Kranendonk et al., 2008; Marohnic et al., 2010). This E. coli model contains a biplasmid expression system, allowing the coexpression of full-length CYPOR variants with human P450s, at physiologically relevant stoichiometries of the P450 and CYPOR proteins (Duarte et al., 2005). We combined the different CYPOR variants separately with human CYP3A4 in this BTC_CYP strain. Dibenzylfluorescein (DBF) was used for kinetic measurements of CYP3A4 activities, which is a highly specific substrate for CYP3A (Ghosal et al., 2003), permitting a rigorous validation of our system against human liver microsomes at saturating concentrations not achievable with other fluorescent substrates. Our data demonstrate substantial differences compared with the outcomes of recently published reports on ABS-related CYPOR variants and their effect on CYP3A4.
Materials and Methods
Reagents.
l-Arginine, δ-aminolevulinic acid, ampicillin, kanamycin sulfate, chloramphenicol, tetracycline HCl, cytochrome c, isopropyl β-d-thiogalactoside (dioxane-free), thiamine, glucose 6-phosphate, NADPH, and NADP+ were obtained from Sigma-Aldrich (St. Louis, MO). Bacto agar, Bacto peptone, Bacto tryptone, and Bacto yeast extract were obtained from Difco (Detroit, MI). Pooled (n = 200) human liver microsomes (XTreme 200) were obtained from XenoTech, LLC (lot 0910251; Lenexa, KS; with the following characteristics: total protein, 20 mg/ml; P450 content, 391 pmol/mg; cytochrome b5, 346 pmol/mg; and activity in NADPH-cytochrome c reduction, 153 ± 9 nmol · mg protein−1 · min−1). Dibenzylfluorescein was obtained from BD Biosciences (San Jose, CA) and fluorescein was obtained from Invitrogen (Carlsbad, CA). The Vivid CYP450 substrates BOMCC and DBOMF were purchased from Invitrogen. A polyclonal antibody purified by Protein A-Sepharose from rabbit serum raised against recombinant rat CYPOR was used for CYPOR immunodetection (Kranendonk et al., 2008).
Expression Plasmids.
The expression of human CYPOR variants in strain BTC-CYP (Duarte et al., 2005) was accomplished with plasmid pLCM_PORwt (Kranendonk et al., 2008), containing human WT POR cDNA, based on National Center for Biotechnology Information sequence NM_000941, encoding the CYPOR consensus protein sequence NP_000932. The POR cDNA of pLCM_ PORwt was adapted to encode the different CYPOR_ABS variants. The POR cDNA of all pLCM_POR plasmids was confirmed by extensive sequencing of the POR cDNA. Plasmid pLCM (Kranendonk et al., 1998) was used as the POR mock plasmid. The expression plasmid for human CYP3A4 (pCWh3A4) was described previously (Kranendonk et al., 1999). Each of the pLCM_POR plasmids was cotransformed with pCWh3A4 to strain PD301 [mother strain of BTC_CYP (Duarte et al., 2005)] by electroporation.
Cultures and Membrane Preparations and Their CYP3A4/CYPOR Content.
Cultures and membrane fractions of the different strains were prepared as described previously (Kranendonk et al., 2008). Membranes were characterized for their protein content, according to the method described by Bradford (1976), following the manufacturer's protocol (Bio-Rad Laboratories, Hercules, CA), using bovine serum albumin as standard. The CYP3A4 content of membranes was determined using the CO-difference spectral technique as reported previously (Kranendonk et al., 2008). CYPOR content of membrane fractions was quantified by immunoblotting against a standard curve of purified human, full-length WT CYPOR, using polyclonal rabbit anti-CYPOR primary antibody and biotin-goat anti-rabbit antibody in combination with the fluorescent streptavidin conjugate (WesternDot 625 Western Blot Kit; Invitrogen). Western blot signals were analyzed with LabWorks 4.6 software (UVP, Cambridge, UK).
Enzyme Assays.
The electron transfer capacity of the CYPOR variants was measured using the NADPH-cytochrome c reduction assay as described previously (Kranendonk et al., 2008). Catalytic activity of CYP3A4, sustained by the different CYPOR variants was evaluated by its capacity to mediate the O-debenzylation of DBF, which has been used as a CYP3A-specific reporter reaction (Ghosal et al., 2003). The debenzylation activity was measured as the rate of the increase in fluorescein fluorescence (excitation 485 nm and emission 535 nm). All measurements were performed in triplicate, in a 96-well format using a Zenyth 3100 microplate reader (Anthos, Wals, Austria) and rates [picomoles of fluorescein formed per (picomoles of CYP3A4 per minute)] were calculated by using a standard curve of the product fluorescein. The reaction was performed with either bacterial membranes or pooled human liver microsomes (n = 200) in 100 mM potassium phosphate buffer (pH 7.6) with 3 mM MgCl2 and 200 μM NADPH in a total volume of 200 μl, using a final well concentration of 25 nM CYP3A4 (for liver microsomes, assuming that ∼30% of total human liver P450 is CYP3A) (Shimada et al., 1994). Stock solutions of DBF were prepared in acetonitrile (ACN), with the solvent maintained at a final concentration of 0.1% (v/v) throughout the experiment, to prevent CYP3A4 inhibition by the solvent (Busby et al., 1999). The reaction was followed for 30 min at 37°C, and rates were derived from the linear part of the kinetic traces. To maintain a constant NADPH concentration, an NADPH-generating system was used, which consisted of 0.5 mM glucose 6-phosphate and 40 mU/ml glucose-6-phosphate dehydrogenase. Initial assays with human liver microsomes and BTC3A4_PORWT membranes were performed with DBF gradients up to 1.25 μM. Subsequent assays with bacterial membranes were performed with a DBF gradient up to 5 μM, except for membranes containing the Q153R variant for which a maximum DBF concentration of 10 μM was applied. Plots of velocity traces versus DBF concentration were fitted according to the Michaelis-Menten equation to determine kinetic parameters kcat and Km. BTC3A4 membranes containing CYPOR variants WT, Q153R, or A287P were also tested with the Vivid CYP450 substrates BOMCC and DBOMF with assay conditions as described above, adapted according to the indications of the manufacturer. The substrate BOMCC was used up to 100 μM, copying conditions described in the study of Flück et al. (2010). Because of low solubility, the concentration gradient of BOMCC could only be performed with final solvent (ACN) concentrations greater than 2%, thus inhibiting CYP3A4 by more than 30% (Busby et al., 1999). Therefore, a different Vivid CYP450 substrate (DBOMF) was used in the present studies, with a concentration gradient up to 10 μM, keeping the final ACN concentration at 0.5%. The final P450 concentration for this assay was 37.5 nM. Velocity traces were plotted, and Michaelis-Menten parameters could be derived appropriately.
Results
Human CYP3A4 and CYPOR Contents in the BTC3A4_POR Membrane Fractions.
All membrane fractions of the BTC3A4_POR strains were characterized with respect to CYPOR and CYP3A4 contents (Table 1). Human CYP3A4 expression levels range from 50 to 127 pmol/mg protein, as determined by CO-difference spectrophotometry. CYPOR protein contents were determined by immunodetection (Supplemental Fig. 1). All membrane fractions demonstrated a major signal at around 77 kDa except that derived from the POR mock strain (BTC 3A4_PORNull: no CYPOR-specific signal) and the one containing the CYPOR variant R616X (Supplemental Fig. 1). This latter variant was expressed in a truncated form as demonstrated by its CYPOR-specific signal at a lower molecular mass (approximately 70 kDa).
TABLE 1.
CYP3A4 and CYPOR contents and NADPH-cytochrome c reductase activities of BTC3A4_POR membrane fractions
| BTC3A4_POR | CYP3A4 | CYPORa | Ratio CYPOR/CYP3A4 | Vmax Cytochrome cred | Electron Transfer Efficiencyb |
|---|---|---|---|---|---|
| pmol/mg protein | nmol · min−1 · mg protein−1 | ||||
| Null | 129 ± 2 | 0 | 2.1 ± 0.7 | ||
| WT | 77 ± 3 | 30.3 ± 1.5 | 1:3 | 97.1 ± 5.1 | 100 |
| P228L | 105 ± 28 | 37.8 ± 2.6 | 1:3 | 68.9 ± 2.9 | 55 (76) |
| M263V | 111 ± 11 | 24.4 ± 1.4 | 1:5 | 84.0 ± 0.4 | 105 (104) |
| A287P | 77 ± 8 | 16.3 ± 1.2 | 1:5 | 44.4 ± 5.3 | 85 (41) |
| G413S | 85 ± 13 | 24.0 ± 1.1 | 1:4 | 84.5 ± 5.2 | 108 (71) |
| Q153R | 58 ± 3 | 26.0 ± 2.7 | 1:2 | 84.2 ± 1.9 | 100 (23) |
| R457H | 53 ± 6 | 26.9 ± 1.3 | 1:2 | 27.1 ± 1.0 | 30 (8) |
| G539R | 127 ± 11 | 24.5 ± 1.0 | 1:5 | 5.0 ± 0.4 | 4 (41) |
| Y181D | 81 ± 6 | 36.2 ± 0.1 | 1:2 | 4.1 ± 0.7 | 2 (0) |
| Y459H | 75 ± 6 | 21.2 ± 0.7 | 1:4 | 10.1 ± 2.5 | 12 (7) |
| V492E | 50 ± 9 | 24.8 ± 1.6 | 1:2 | 3.5 ± 0.1 | 2 (7) |
| L565P | 120 ± 12 | 8.1 ± 0.6 | 1:14 | 3.0 ± 0.2 | 4 (29) |
| R616X | 104 ± 9 | 6.9 ± 0.3 | 1:14 | 2.5 ± 0.2 | 3 (0) |
| Human liver microsomes | 1:3 [1:2–1:13]c | ||||
CYPOR content of membranes, based on immunoblot analysis.
Percentage in electron transfer measured as NADPH cytochrome c reduction (Vmax) relative to the expected, calculated reduction activity based on the CYPOR protein content (immunodetection) using kcat of 3200 min−1 (Parikh et al., 1997). Numbers in parentheses indicate the same percentage, calculated by using the Vmax values, reported by Huang et al. (2005), with use of N-terminal (27 amino acids)-deleted CYPOR expression relatively to their CYPOR_WT form.
CYPOR/CYP3A ratio (average and interval) (Venkatakrishnan et al., 2000), assuming a ∼30% CYP3A content of human liver (Guengerich, 2005).
Densitometry analysis using a standard curve of purified full-length wild-type protein allowed the quantification of CYPOR protein in the membrane fractions. The CYPOR_ABS variants all demonstrated similar contents of CYPOR protein compared with that of the WT (Table 1), indicating that the respective mutations did not affect protein expression and stability in the BTC3A4_POR cell model, except for variants L565P and R616X. These variants showed lower levels, namely 8 and 7 pmol/mg membrane protein, respectively, indicating lower efficiency in expression or protein stability, as was already noted for the R616X variant (see above, this section).
These results indicate that the ABS-related CYPOR alleles were coexpressed with CYP3A4 in stoichiometries approximating those of the wild-type CYPOR (ratio CYPOR/CYP3A4 = 1:3) (Table 1) in the BTC3A4_PORWT system and that observed in human liver microsomes (average = 1:3; range = 1:2–1:13) (Paine et al., 1997; Venkatakrishnan et al., 2000) (Table 1). Exceptions to these ratios were observed in membrane fractions derived from BTC3A4_PORL565P and BT3A4_PORR616X, which demonstrated substantially lower ratios as a result of the lower expression levels of their CYPOR variants (see above, this section) but not as a result of altered CYP3A4 levels.
Electron Transfer Capacities of CYPOR Alleles in BTC3A4_POR Membranes.
Initial studies of the electron transfer capacity of the CYPOR variants in the different BTC3A4_POR membrane fractions, measured as the maximum velocity of NADPH-cytochrome c reduction, were performed taking into account the expression level of the CYPOR variants (Table 1). This capacity was considerably different among mutants. Several were apparently capable of reducing cytochrome c with a wild-type capability (Q153R, M263V, and G413S). Other variants had very low or almost no capacity for this reaction, such as Y181D, Y459H, V492E, G539H, L565P, and R616X. Variant A287P seemed not to be severely hampered (∼85% residual) and variants P228L and R457H demonstrated approximately 55 and 30% of wild-type capacity, respectively (Table 1).
CYPOR Variants Sustaining CYP3A4-Mediated Debenzylation of Dibenzylfluorescein.
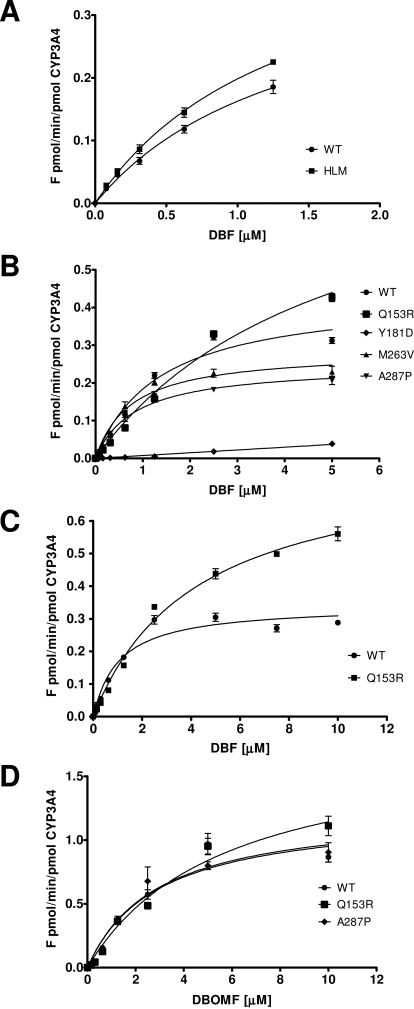
The fluorogenic substrate, DBF (Stresser et al., 2000), was selected for the purpose of determining the effects of the ABS-related CYPOR variants on CYP3A4 activity. DBF was previously shown to be debenzylated by CYP3A4/5 and CYP2C8, but when applied in low concentrations (<2 μM), it can be used as a CYP3A-specific substrate (Ghosal et al., 2003). In our first experimental setup using DBF concentrations up to 2.5 μM DBF, the human liver microsomes showed non-Michaelis-Menten velocity traces (data not shown), probably due to interference by CYP2C8-mediated metabolism (see above, this section). Therefore, the use of the BTC3A4_POR membranes was validated against human liver microsomes (pooled, n = 200) using a restricted substrate gradient with a maximum of 1.25 μM. The velocities obtained for the BTC3A4_PORWT membranes and human liver microsomes were very similar (Fig. 1A) and could be plotted according to the Michaelis-Menten equation, with high confidence (r2 = 0.97 and 0.99, respectively). The results demonstrated equal debenzylation activities of the BTC3A4_PORWT membranes compared with the liver microsomes, with kcat (0.34 ± 0.04 versus 0.47 ± 0.04 min−1) and Km (1.0 ± 0.2 versus 1.4 ± 0.2 μM) values, respectively. Although these parameters were derived using a suboptimal concentration-range of the substrate DBF (0–1.25 μM) to prevent CYP2C8 interference in the human liver microsomes, the velocity trace and kinetic parameters of the BTC3A4_POR system very closely resembled those of human liver microsomes in the CYP3A4-mediated DBF biotransformation. Then, velocity traces of the bacterial CYP3A4 membranes containing ABS-CYPOR variants were measured. For several membrane preparations, in particular those containing debilitated CYPORs, it was necessary to use a substrate concentration variation up to 5 μM to be able to plot the velocity traces with high confidence (Fig. 1B). Results are presented in Table 2, including those for the WT variant under these conditions.
Fig. 1.
Reaction velocity plots of BTC3A4_POR membranes. A, velocity plots of human liver microsomes versus BTC3A4_PORWT membranes with DBF up to 1.25 μM. B, velocity plots of BTC3A4_POR membranes containing CYPOR variants WT, Q153R, Y181D, M263V, or A287P with DBF up to 5 μM. C, velocity plots of BTC3A4_POR membranes containing CYPOR variants WT or Q153R, using DBF gradient up to 10 μM. D, velocity plots of BTC3A4 membranes containing CYPOR variants WT, Q153R, or A287P with DBOMF.
TABLE 2.
Dibenzylfluorescein O-debenzylation activities
| BTC3A4_PORa | kcat | Km | R2 | kcat/Km |
|---|---|---|---|---|
| fluorescein-formed pmol · min−1 · pmol CYP3A4−1 | μM | % WT | ||
| Null | —b | — | — | — |
| WT | 0.43 ± 0.03 | 1.37 ± 0.22 | 0.97 | 0.32 (100) |
| P228L | 0.23 ± 0.01 | 0.78 ± 0.14 | 0.95 | 0.30 (95) |
| M263V | 0.28 ± 0.02 | 0.76 ± 0.12 | 0.95 | 0.38 (119) |
| A287P | 0.25 ± 0.01 | 0.84 ± 0.10 | 0.97 | 0.29 (93) |
| G413S | 0.33 ± 0.02 | 0.89 ± 0.15 | 0.96 | 0.37 (115) |
| Q153R | 0.80 ± 0.04 | 4.24 ± 0.45 | 0.99 | 0.19 (60) |
| R457H | 0.052 ± 0.001 | 1.70 ± 0.07 | 0.99 | 0.03 (10) |
| G539R | 0.059 ± 0.002 | 0.62 ± 0.06 | 0.98 | 0.10 (30) |
| Y181D | — | — | — | — |
| Y459H | — | — | — | — |
| V492E | — | — | — | — |
| L565P | — | — | — | — |
| R616X | — | — | — | — |
| Human liver microsomes | 0.47 ± 0.04 | 1.36 ± 0.17 | 0.99 | 0.34 (108) |
Velocity parameters obtained using a DBF gradient up to 5 μM, except for human liver microsomes for which a gradient up to 1.25 μM was applied to prevent interference with DBF metabolism by CYP2C8 (Ghosal et al., 2003) and for CYPOR variant Q153R for which a gradient up to 10 μM was applied.
—, not measurable.
For the BTC3A4_POR membranes containing the severely debilitated CYPOR forms Y181D, Y459H, or V492E, no activity parameters could be determined. This was also the case for the CYPOR forms L565P or R616X, which were expressed in very low quantities relative to CYP3A4 (see above). The catalytic efficiency (kcat/Km) of CYP3A4 was substantially compromised when supported by CYPOR variants R457H or G539R, demonstrating 10 or 30%, respectively, of the efficiency calculated for the wild-type CYPOR. This result was mainly due to a severe drop in maximum velocities (kcat), but maintenance of a wild-type-like apparent affinity (Km) for DBF for these two variants (Table 2). The domain perturbations producing these lowered activities have been reported in X-ray structures for R457H and V492E in collaborative studies between the Kim and Masters laboratories (Xia et al., 2011). Membrane fractions containing variants P228L, M263V, A287P, and G413S showed WT capacity in sustaining CYP3A4, all of which demonstrated similar maximum reaction velocities and similar apparent affinities for DBF. The velocity plot of the O-debenzylation of DBF by CYP3A4, when supported by the CYPOR variant Q153R, demonstrated a substantially different trace compared with that supported by the wild-type enzyme (Fig. 1B). For this variant, it was necessary to extend the gradient to 10 μM to obtain a velocity trace with high confidence (Fig. 1C). The maximum velocity showed an increase of approximately 2-fold relative to the wild-type CYPOR. However, the apparent Km for DBF increased 3 times, leading to a decrease of 40% in catalytic efficiency (kcat/Km). The effect of several of the CYPOR variants on the CYP3A4 investigated here were described in two recent studies, with several variants demonstrating substantially different efficacy outcomes (Table 3). In particular, opposing results were obtained for the effects of variants Q153R and A287P on CYP3A4 (noted in bold type). For clarification of these discrepancies, the WT, Q153R, and A287P variants were tested with the Vivid CYP450 substrate BOMCC, the substrate used by Flück et al. (2010). With application of their concentration gradients up to 100 μM, serious BOMCC solubility problems were encountered, which could be surpassed only by using final solvent (ACN) concentrations greater than 2%. Such high solvent concentrations are known to inhibit P450s substantially, in particular with CYP3A4, in which case 2% ACN-inhibited activity is greater than 30% (Busby et al., 1999). This level of inhibition is incompatible with the objective of accurately determining effects of ABS-related CYPOR variants on CYP3A4 activity. Further, in a persist search for clarifications for the described discrepancy, a different Vivid CYP450 substrate was used, specifically DBOMF, for variants WT, Q153R, and AP278P, with a final ACN concentration of 0.5%. Accurate velocity traces for the BTC3A4 membranes of these CYPOR variants could be plotted (Fig. 1D). Maximum velocities were 1.2 ± 0.1, 1.8 ± 0.2, and 1.2 ± 0.1 min−1 for WT, Q153R, and A278P, respectively. The corresponding apparent affinities (Km) were 2.9 ± 0.6, 5.6 ± 1.0, and 2.9 ± 0.7 μM, respectively. The efficacies (kcat/Km) relative to CYPOR_WT could thus be calculated, demonstrating an almost 25% drop for CYP3A4 when sustained by variant Q153R and an equal (WT) efficacy when supported by variant A287P.
TABLE 3.
Comparison of CYP3A4 efficiency when sustained by ABS-CYPORs, using different in vitro systems and substrates
Bold indicates CYPOR variants of this present study with most deviating results compared with those of the Fluc̈k et al. (2010) and Agrawal et al. (2010) studies.
| BTC3A4_POR |
kcat/Kma |
||||||
|---|---|---|---|---|---|---|---|
| Debenzylation DBFb | Debenzylation BOMCCc | 6β-Hydroxylation Testosteroned | 1-Hydroxylation Midazolamd | 4-Hydroxylation Midazolamd | 3-Hydroxylation Quinidined | N-Demethylation Erythromycind | |
| WT | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Q153R | 60 | 119 | 129 | 94 | 92 | 150 | 76 |
| Y181D | N.D. | N.D. | —e | — | — | — | — |
| D211N | 107 | — | — | — | — | — | — |
| P228L | 95 | 101 | — | — | — | — | — |
| M263V | 119 | — | — | — | — | — | — |
| A287P | 93 | 26 | 17 | 17 | 14 | 3 | N.D. |
| G413S | 115 | 110 | — | — | — | — | — |
| R457H | 10 | N.D. | N.D. | N.D. | N.D. | — | N.D. |
| Y459H | N.D. | N.D. | — | — | — | — | — |
| V492E | N.D. | N.D. | — | — | — | — | — |
| G539R | 30 | — | — | — | — | — | — |
| L565P | N.D. | — | — | — | — | — | — |
| R616X | N.D. | N.D. | — | — | — | — | — |
N.D., no activity detected.
Percentage kcat/Km values relative to WT.
Present study.
Data from Flück et al. (2010).
Data from Agrawal et al. (2010).
—, CYPOR variant not evaluated.
Discussion
Variation in CYP3A-mediated drug metabolism is considered to be one of the main reasons for observed differences in the therapeutic efficacy and toxicity of drugs (Evans and McLeod, 2003). The current study explored the importance of POR variation in CYP3A4 metabolism, particularly of haplotypes encountered in individuals with perturbed steroidogenesis, by evaluating the influence of 12 different CYPOR variants on CYP3A4 activity, using an engineered bacterial cell model. The CYPOR/CYP3A4 ratios of the variants approximated those found in human liver (1:3–1:5; Table 1) (Paine et al., 1997; Venkatakrishnan et al., 2000), except for variants L565P and R616X, which were expressed in substantially lower amounts, because of apparent instability. Characterization of the membranes (Table 1) demonstrated the expression of full-length CYPOR variants in the BTC3A4_POR model system, except that R616X was expressed in a truncated form, ascribed previously to a truncation of the C-terminal end (Huang et al., 2005).
DBF was shown previously to be a CYP3A-specific model substrate when used in low micromolar concentrations (Ghosal et al., 2003) and was used for detailed analysis of the CYPOR variants to determine their capacities to sustain CYP3A4-mediated activity. Under these conditions CYP3A4 and its congener CYP3A5 debenzylate DBF with very similar kcat and Km values (Ghosal et al., 2003). Our bacterial membrane fractions, combining CYP3A4 with wild-type CYPOR demonstrated: 1) kinetic parameters almost identical to those previously described for (recombinant) CYP3A4 (kcat 0.55 ± 0.08 min−1; Km 1.03 ± 0.42 μM) (Ghosal et al., 2003) and 2) parameters very similar to those for pooled human liver microsomes (Fig. 1A). These data validate the BTC3A4_POR system for in vitro studies to evaluate CYP3A4-mediated biotransformation.
On the basis of DBF debenzylation catalytic efficiencies (kcat/Km), the BTC3A4_PORABS membranes may be grouped (Table 2). One group comprises CYPOR variants containing mutations directly involved in the binding of the prosthetic flavins (Y181D, Y459H, or V492E), for which no CYP3A4 activity could be detected. These results correlate with their severe decline in electron transfer efficiency to background levels, measured as cytochrome c reductase activity (Table 1). In previous studies, we described the deficiency of these variants in sustaining CYP1A2 activity, which could be substantially recovered by exogenous FMN (Y181D) or FAD (Y459H and V492E) (Marohnic et al., 2006; Kranendonk et al., 2008). The second group, P228L, M263V, A287P, and G413S, metabolized DBF similarly to WT CYPOR, reflected by their kcat and Km parameters (Table 2). This finding is corroborated by their wild-type-like cytochrome c reduction efficiency (Table 1), except for variant P228L, which demonstrated 45% loss of this activity. Variants classified in this second group have been identified in individuals with altered steroidogenesis (catalyzed by CYP17A1, CY19A1, and CYP21A1) (Huang et al., 2005; Dhir et al., 2007; Pandey et al., 2007). Our results with these variants indicate that these mutations may produce P450-dependent effects, explainable by the location of the P228L mutation (interaction domain with protein redox partners), but to a lesser extent for the M263V, A287P, and G413S mutations (hinge region/connecting domain). In previous studies, variant A287P was found to influence differentially CYP17A1 (decreased activity) relative to CYP19A1 and CYP21A1 (normal activity) (Dhir et al., 2007; Pandey et al., 2007). Both kcat and Km values for this mutant CYPOR were lower (Table 2, present study), resulting in only slightly lower catalytic efficiency. The third group is composed of variants R457H and G539R. These variants demonstrated low CYP3A4 catalytic efficiencies, i.e., 10 and 30% relative to those for the WT, respectively, mainly due to a severe drop in their maximum velocities (Table 2). These low kcat values correlated with low cytochrome c reduction capacities, 30 and 4%, respectively (Table 1). The R457H and the G539R mutations are located in the FAD- and NADPH-binding domains, respectively, explaining their effects on electron transfer and CYP3A4 maximum velocity. Variant Q153R demonstrated quite different kinetics compared with CYP3A4 sustained by WT CYPOR (Fig. 1C), with a 40% drop in DBF debenzylation catalytic efficiency (Table 2). Of interest, there was not a corresponding drop in electron transfer capability to cytochrome c, which was equal (100%) relative to that for the WT (Table 1). The Q153R mutation is located in the FMN binding domain, which forms the interface for interaction with the redox partners of CYPOR, including cytochrome c (Vermilion and Coon, 1978), suggesting that protein-protein interactions and/or electron transfer in CYPOR-CYP3A4 or CYPOR-cytochrome c complexes occur differentially. No DBF metabolism could be detected with variants L565P or R616X, precluding conclusions concerning their effects on CYP3A4 DBF biotransformation.
The effects of ABS_CYPOR variants on CYP3A4 activity have been described in recent studies (Agrawal et al., 2010; Flück et al., 2010). Many of the ABS_CYPOR variants we presented here were included in these reports; most noteworthy were the Q153R and A287P variants. Neither the Flück et al. (2010) nor the Agrawal et al. (2010) study indicated decreases in CYP3A4 efficiency (kcat/Km) for variant Q153R versus the ∼40% loss we describe here for DBF. In addition, in these two studies variant A287P produced a very severe defect in CYP3A4 efficiency, contradicting our data showing that this variant sustained CYP3A4 activity similar to WT CYPOR, with the interesting exception of erythromycin, a sterically large CYP3A4 substrate (Table 3). Some discrepancies between our data and these earlier studies can be expected for particular ABS CYPOR variants on CYP3A4 when different substrates were used. However, results for the second CYP3A4 substrate DBOMF herein confirm our data for variants Q153R and A287P. Moreover, our observation that substrate BOMCC used in the study of Flück et al., (2010) can only be used with high acetonitrile concentrations, leading to severe inhibition of CYP3A4, suggests some compromise of their CYP3A4 activity data due to solvent effects. Although not clinically used, DBF is defined as a drug-like compound because of its CYP3A4 specificity.
We have used full-length constructs of CYPOR in our studies to recapitulate the activities observed in human liver microsomes and based our calculations on the generally accepted turnover of 3000 to 3200 min−1 (Parikh et al., 1997; Guengerich et al., 2009) for CYPOR-catalyzed cytochrome c reduction. However, the studies by Agrawal et al. (2010) and Flück et al. (2010) made use of N-terminally truncated CYPOR variants in their CYP3A4/ABS_CYPOR in vitro systems. Comparison of the electron transfer efficiency of the full-length variants (Table 1) with the same variants expressed in the N(27)-deleted form (values in parentheses) reveals substantial differences.
A second possibility concerns the CYPOR/P450 stoichiometry in these two studies. Their experiments used ratios strongly favoring CYPOR (5-fold excess). Although empirical comparisons can be made using higher CYPOR/P450 ratios, nonphysiological ratios in favor of CYPOR could produce protein-protein interactions via nonphysiological equilibria. This was demonstrated for CYP3A biotransformation, producing different results, depending on the relative ratio of CYPOR over CYP3A4/5 (Emoto et al., 2008; Christensen et al., 2011).
Differences could also result from the in vitro systems applied in the studies of Flück et al. (2010) and Agrawal et al. (2010) compared with those used in the present system. These former studies involved in vitro reconstitution combining purified and membrane-bound proteins, specific phospholipids, and a detergent with the objective of forming artificial lipid vesicles. This method requires specific procedures to ensure their proper application, including size exclusion chromatography, to remove adventitious nonincorporated proteins and the subsequent analysis of lipid vesicles to obtain their actual CYPOR/P450 stoichiometry. Such characterization of the lipid vesicles as well as a rigorous validation of their use for a detailed study of the effect of these variants on CYP3A4 were not performed in the studies of Flück et al. (2010) and Agrawal et al. (2010). Of interest, in a previous study by Flück et al. (Pandey et al., 2007) in which ABS-related variants were expressed as full-length proteins, using more in vivo-like CYPOR:P450 stoichiometries, variant A287P showed no diminished efficiency to sustain CYP19A1 activity.
In conclusion, we have evaluated the effect of 12 CYPOR variants on human CYP3A4, using an engineered bacterial model system, validated as a robust in vitro system for the purpose of CYP3A4-mediated metabolism studies. Substantial differences were found for several CYPOR variants, when our data are compared with two previous studies. Comparison of the experimental approaches of the current study with previous approaches highlights the importance of the use of well defined, validated in vitro systems, with full-length proteins simulating in vivo-like stoichiometric quantities of protein partners for enzymatic evaluations to assist in making pharmacogenetic predictions in biotransformation. It is tempting to predict that each CYPOR variant will differentially affect interactions with various redox partners. Evidence derived from studies performed in our laboratories with various redox partners (Kranendonk et al., 2008; Marohnic et al., 2011), as well as those of Flück et al. (2010), Agrawal et al. (2010), and Dhir et al. (2007), has strongly suggested these differences. Finally, although there is no perfect system that recapitulates the in vivo environment, it is important to maintain biochemical principles in measuring catalytic parameters and to attempt to mimic the in vivo system to the extent possible. Proper validation of in vitro systems remains an imperative control for their correct application, such as the use of pooled human microsomes in the present study.
Supplementary Material
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM081568] (to B.S.M.); the Robert A. Welch Foundation [Endowed Chair AQ-0012] (to B.S.M.); and the Fundação para a Ciência e a Tecnologia (Portugal) [Grant PTDC/SAU-GMG/71911/2006] (to D.M., R.F., and M.K.).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- P450
- cytochrome P450
- CYPOR
- NADPH cytochrome P450 oxidoreductase
- FAD
- flavin adenine dinucleotide
- FNM
- flavin mononucleotide
- ABS
- Antley-Bixler syndrome
- DBF
- dibenzylfluorescein
- BOMCC
- benzyl-O-methyl-cyanocoumarin
- DBOMF
- di-[benzyl-O-methyl]-fluorescein
- WT
- wild type
- ACN
- acetonitrile.
Authorship Contributions
Participated in research design: Moutinho, Marohnic, Panda, and Kranendonk.
Conducted experiments: Moutinho, Marohnic, Panda, and Kranendonk.
Performed data analysis: Moutinho, Marohnic, Panda, Rueff, Masters, and Kranendonk.
Wrote or contributed to the writing of the manuscript: Moutinho, Marohnic, Panda, Rueff, Masters, and Kranendonk.
References
- Agrawal V, Choi JH, Giacomini KM, Miller WL. (2010) Substrate-specific modulation of CYP3A4 activity by genetic variants of cytochrome P450 oxidoreductase. Pharmacogenet Genomics 20:611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal V, Huang N, Miller WL. (2008) Pharmacogenetics of P450 oxidoreductase: effect of sequence variants on activities of CYP1A2 and CYP2C19. Pharmacogenet Genomics 18:569–576 [DOI] [PubMed] [Google Scholar]
- Aigrain L, Pompon D, Moréra S, Truan G. (2009) Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase. EMBO Rep 10:742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antley R, Bixler D. (1975) Trapezoidocephaly, midfacial hypoplasia and cartilage abnormalities with multiple synostoses and skeletal fractures. Birth Defects Orig Artic Ser 11:397–401 [PubMed] [Google Scholar]
- Arlt W, Walker EA, Draper N, Ivison HE, Ride JP, Hammer F, Chalder SM, Borucka-Mankiewicz M, Hauffa BP, Malunowicz EM, et al. (2004) Congenital adrenal hyperplasia caused by mutant P450 oxidoreductase and human androgen synthesis: analytical study. Lancet 363:2128–2135 [DOI] [PubMed] [Google Scholar]
- Black SD, Coon MJ. (1982) Structural features of liver microsomal NADPH-cytochrome P-450 reductase. Hydrophobic domain, hydrophilic domain, and connecting region. J Biol Chem 257:5929–5938 [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Busby WF, Jr, Ackermann JM, Crespi CL. (1999) Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab Dispos 27:246–249 [PubMed] [Google Scholar]
- Christensen H, Hestad AL, Molden E, Mathiesen L. (2011) CYP3A5-mediated metabolism of midazolam in recombinant systems is highly sensitive to NADPH-cytochrome P450 reductase activity. Xenobiotica 41:1–5 [DOI] [PubMed] [Google Scholar]
- Dhir V, Ivison HE, Krone N, Shackleton CH, Doherty AJ, Stewart PM, Arlt W. (2007) Differential inhibition of CYP17A1 and CYP21A2 activities by the P450 oxidoreductase mutant A287P. Mol Endocrinol 21:1958–1968 [DOI] [PubMed] [Google Scholar]
- Duarte MP, Palma BB, Laires A, Oliveira JS, Rueff J, Kranendonk M. (2005) Escherichia coli BTC, a human cytochrome P450 competent tester strain with a high sensitivity towards alkylating agents: involvement of alkyltransferases in the repair of DNA damage induced by aromatic amines. Mutagenesis 20:199–208 [DOI] [PubMed] [Google Scholar]
- Ellis J, Gutierrez A, Barsukov IL, Huang WC, Grossmann JG, Roberts GC. (2009) Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J Biol Chem 284:36628–36637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto C, Murayama N, Yamazaki H. (2008) Effects of enzyme sources on midazolam 1′-hydroxylation activity catalyzed by recombinant cytochrome P450 3A4 in combination with NADPH-cytochrome P450 reductase. Drug Metab Lett 2:190–192 [DOI] [PubMed] [Google Scholar]
- Evans WE, McLeod HL. (2003) Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med 348:538–549 [DOI] [PubMed] [Google Scholar]
- Flück CE, Mullis PE, Pandey AV. (2010) Reduction in hepatic drug metabolizing CYP3A4 activities caused by P450 oxidoreductase mutations identified in patients with disordered steroid metabolism. Biochem Biophys Res Commun 401:149–153 [DOI] [PubMed] [Google Scholar]
- Fukami M, Horikawa R, Nagai T, Tanaka T, Naiki Y, Sato N, Okuyama T, Nakai H, Soneda S, Tachibana K, et al. (2005) Cytochrome P450 oxidoreductase gene mutations and Antley-Bixler syndrome with abnormal genitalia and/or impaired steroidogenesis: molecular and clinical studies in 10 patients. J Clin Endocrinol Metab 90:414–426 [DOI] [PubMed] [Google Scholar]
- Ghosal A, Hapangama N, Yuan Y, Lu X, Horne D, Patrick JE, Zbaida S. (2003) Rapid determination of enzyme activities of recombinant human cytochromes P450, human liver microsomes and hepatocytes. Biopharm Drug Dispos 24:375–384 [DOI] [PubMed] [Google Scholar]
- Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM. (2009) Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10:579–599 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2005) Human cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism and Biochemistry (Ortiz de Montellano PR. ed) pp 377–530, Plenum Publishers, New York [Google Scholar]
- Guengerich FP, Martin MV, Sohl CD, Cheng Q. (2009) Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc 4:1245–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L. (2009) Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J Biol Chem 284:11374–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. (2008) Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet Genomics 18:11–24 [DOI] [PubMed] [Google Scholar]
- Huang N, Pandey AV, Agrawal V, Reardon W, Lapunzina PD, Mowat D, Jabs EW, Van Vliet G, Sack J, Flück CE, et al. (2005) Diversity and function of mutations in p450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am J Hum Genet 76:729–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranendonk M, Fisher CW, Roda R, Carreira F, Theisen P, Laires A, Rueff J, Vermeulen NP, Estabrook RW. (1999) Escherichia coli MTC, a NADPH cytochrome P450 reductase competent mutagenicity tester strain for the expression of human cytochrome P450: comparison of three types of expression systems. Mutat Res 439:287–300 [DOI] [PubMed] [Google Scholar]
- Kranendonk M, Marohnic CC, Panda SP, Duarte MP, Oliveira JS, Masters BS, Rueff J. (2008) Impairment of human CYP1A2-mediated xenobiotic metabolism by Antley-Bixler syndrome variants of cytochrome P450 oxidoreductase. Arch Biochem Biophys 475:93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranendonk M, Mesquita P, Laires A, Vermeulen NP, Rueff J. (1998) Expression of human cytochrome P450 1A2 in Escherichia coli: a system for biotransformation and genotoxicity studies of chemical carcinogens. Mutagenesis 13:263–269 [DOI] [PubMed] [Google Scholar]
- Marohnic CC, Huber WJ, III, Connick JP, Reed JR, McCammon K, Panda SP, Martásek P, Backes WL, Masters BS. (2011) Mutations of human cytochrome P450 reductase differentially modulate heme oxygenase-1 activity and oligomerization. Arch Biochem Biophys 513:42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marohnic CC, Panda SP, Martásek P, Masters BS. (2006) Diminished FAD binding in the Y459H and V492E Antley-Bixler syndrome mutants of human cytochrome P450 reductase. J Biol Chem 281:35975–35982 [DOI] [PubMed] [Google Scholar]
- Marohnic CC, Panda SP, McCammon K, Rueff J, Masters BS, Kranendonk M. (2010) Human cytochrome P450 oxidoreductase deficiency caused by the Y181D mutation: molecular consequences and rescue of defect. Drug Metab Dispos 38:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. (1997) Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther 283:1552–1562 [PubMed] [Google Scholar]
- Pandey AV, Kempná P, Hofer G, Mullis PE, Flück CE. (2007) Modulation of human CYP19A1 activity by mutant NADPH P450 oxidoreductase. Mol Endocrinol 21:2579–2595 [DOI] [PubMed] [Google Scholar]
- Parikh A, Gillam EM, Guengerich FP. (1997) Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol 15:784–788 [DOI] [PubMed] [Google Scholar]
- Porter TD, Beck TW, Kasper CB. (1990) NADPH-cytochrome P-450 oxidoreductase gene organization correlates with structural domains of the protein. Biochemistry 29:9814–9818 [DOI] [PubMed] [Google Scholar]
- Porter TD, Kasper CB. (1986) NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry 25:1682–1687 [DOI] [PubMed] [Google Scholar]
- Sandee D, Morrissey K, Agrawal V, Tam HK, Kramer MA, Tracy TS, Giacomini KM, Miller WL. (2010) Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro. Pharmacogenet Genomics 20:677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270:414–423 [PubMed] [Google Scholar]
- Stresser DM, Blanchard AP, Turner SD, Erve JC, Dandeneau AA, Miller VP, Crespi CL. (2000) Substrate-dependent modulation of CYP3A4 catalytic activity: analysis of 27 test compounds with four fluorometric substrates. Drug Metab Dispos 28:1440–1448 [PubMed] [Google Scholar]
- Venkatakrishnan K, von Moltke LL, Court MH, Harmatz JS, Crespi CL, Greenblatt DJ. (2000) Comparison between cytochrome P450 (CYP) content and relative activity approaches to scaling from cDNA-expressed CYPs to human liver microsomes: ratios of accessory proteins as sources of discrepancies between the approaches. Drug Metab Dispos 28:1493–1504 [PubMed] [Google Scholar]
- Vermilion JL, Coon MJ. (1978) Identification of the high and low potential flavins of liver microsomal NADPH-cytochrome P-450 reductase. J Biol Chem 253:8812–8819 [PubMed] [Google Scholar]
- Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. (1997) Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA 94:8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Panda SP, Marohnic CC, Martásek P, Masters BS, Kim JJ. (2011) Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc Natl Acad Sci USA 108:13486–13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.