Abstract
The mechanism by which CYP2B6*6 allele alters drug metabolism in vitro and in vivo is not fully understood. To test the hypothesis that altered substrate binding and/or catalytic properties contribute to its functional consequences, efavirenz 8-hydroxylation and bupropion 4-hydroxylation were determined in CYP2B6.1 and CYP2B6.6 proteins expressed without and with cytochrome b5 (Cyt b5) and in human liver microsomes (HLMs) obtained from liver tissues genotyped for the CYP2B6*6 allele. The susceptibility of the variant protein to inhibition was also tested in HLMs. Significantly higher Vmax and Km values for 8-hydroxyefavirenz formation and ∼2-fold lower intrinsic clearance (Clint) were noted in expressed CYP2B6.6 protein (−b5) compared with that of CYP2B6.1 protein (−b5); this effect was abolished by Cyt b5. The Vmax and Clint values for 4-hydroxybupropion formation were significantly higher in CYP2B6.6 than in CYP2B6.1 protein, with no difference in Km, whereas coexpression with Cyt b5 reversed the genetic effect on these kinetic parameters. In HLMs, CYP2B6*6/*6 genotype was associated with markedly lower Vmax (and moderate increase in Km) and thus lower Clint values for efavirenz and bupropion metabolism, but no difference in catalytic properties was noted between CYP2B6*1/*1 and CYP2B6*1/*6 genotypes. Inhibition of efavirenz 8-hydroxylation by voriconazole was significantly greater in HLMs with the CYP2B6*6 allele (Ki = 1.6 ± 0.8 μM) than HLMs with CYP2B6*1/*1 genotype (Ki = 3.0 ± 1.1 μM). In conclusion, our data suggest the CYP2B6*6 allele influences metabolic activity by altering substrate binding and catalytic activity in a substrate- and Cyt b5-dependent manner. It may also confer susceptibility to inhibition.
Introduction
The CYP2B6 enzyme catalyzes several clinically important drugs, other xenobiotics, and endogenous compounds (Mo et al., 2009). Studies in human liver tissues indicate that the expression and activity of CYP2B6 are highly variable in part due to genetic polymorphisms of the gene coding CYP2B6 protein (Lang et al., 2001; Lamba et al., 2003; Hesse et al., 2004; Desta et al., 2007). The CYP2B6 gene is highly polymorphic (Zanger et al., 2007) as reflected by 29 associated alleles, many suballeles, and single nucleotide polymorphisms (SNPs) (http://www.imm.ki.se/CYPalleles/cyp2b6.htm). Of the variants identified so far, the CYP2B6*6 haplotype defined by two nonsynonymous SNPs, 516G>T (Q172H) and 785A>G (K262R), is clinically important because this allele or the SNP tagging it (G516T) occurs at high frequency in all ethnic populations [14–62% (Zanger et al., 2007)] and has been associated with functional consequences in expressed systems (Ariyoshi et al., 2001; Jinno et al., 2003; Bumpus and Hollenberg, 2008; Watanabe et al., 2010; Ariyoshi et al., 2011; Zhang et al., 2011) and in human liver microsomes (HLMs) (Lang et al., 2001; Lamba et al., 2003; Xie et al., 2003; Hesse et al., 2004; Desta et al., 2007). Subsequent to the demonstration that CYP2B6 is the principal clearance mechanism of efavirenz in vitro (Ward et al., 2003), several studies have documented that the CYP2B6*6 allele or its tagging SNP is at increased risk for higher efavirenz exposure and/or adverse effects (Haas et al., 2004; Tsuchiya et al., 2004; Zanger et al., 2007). In addition, this variant has also been associated with the elimination and/or response of clinically relevant drugs, which include nevirapine (Rotger et al., 2005), cyclophosphamide (Nakajima et al., 2007), and methadone (Eap et al., 2007).
In HLMs, the CYP2B6*6 allele is associated with reduced total amount of CYP2B6 protein (Xie et al., 2003; Hesse et al., 2004; Desta et al., 2007). The G516T SNP was predicted to disrupt an exonic splicing enhancer in silico (Lamba et al., 2003). Subsequently, Hofmann et al. (2008) provided evidence that this variant affects splicing and thereby reduces CYP2B6 expression and activity. However, mounting evidence indicates that reduced protein expression alone may not explain the functional consequences of this allele. For substrates that include cyclophosphamide, this allele is associated with enhanced metabolism despite reduced protein expression (Xie et al., 2003), which appears due to substantially lower Km in the variant versus wild-type protein (Ariyoshi et al., 2011).
Other in vitro studies, mostly in expression systems, have also reported that the CYP2B6*6 allele or the amino acids harbored in it influence catalytic properties, although the extent and direction of effect appears to depend on the substrate and the enzyme sources used (Ariyoshi et al., 2001; Jinno et al., 2003; Bumpus and Hollenberg, 2008; Watanabe et al., 2010; Zhang et al., 2011). Therefore, in addition to reduced protein expression, altered protein structure due to amino acid changes may contribute to altered substrate metabolism.
The first purpose of this study was to examine the influence of the CYP2B6*6 allele on catalytic properties measured by efavirenz 8-hydroxylation (Ward et al., 2003) and bupropion 4-hydroxylation (Faucette et al., 2000) as probes of activity using expressed enzymes and HLMs. As has been shown for other cytochromes P450 (P450s) such as CYP2C8 (Kaspera et al., 2011) and CYP2C9 (Kumar et al., 2006), several factors inherent to specific enzyme sources that include differences in cytochrome b5 (Cyt b5) contents may influence in vitro kinetic parameters and inhibition constants in a substrate-dependent manner. Cyt b5 has been reported to activate several P450s including CYP2B6 (Reed and Hollenberg, 2003; Jushchyshyn et al., 2005), but its influence on the catalytic properties of CYP2B6.6 protein has not been studied. Therefore, the second purpose was to test the influence of Cyt b5 on metabolic activities of expressed CYP2B6.1 and CYP2B6.6 proteins. In addition, it has been shown that amino acid substitutions, such as those found in the variant of CYP2B6*6 allele, may also alter the degree of susceptibility to competing metabolic inhibitors for certain CYP2B6 variants (Bumpus et al., 2006; Bumpus and Hollenberg, 2008; Talakad et al., 2009). Therefore, the third aim was to test whether the variant protein is more or less susceptible to metabolic inhibition.
Materials and Methods
Chemicals.
Efavirenz, 8-hydroxyefavirenz, bupropion, 4-hydroxybupropion, nevirapine, ritonavir, voriconazole, and clopidogrel were obtained from Toronto Research Chemicals Inc. (North York, ON, Canada). Glucose 6-phosphate, glucose-6-phosphate dehydrogenase, and NADP were purchased from Sigma-Aldrich (St. Louis, MO). All of the other chemicals were of high-performance liquid chromatography (HPLC) grade.
Microsomal Preparations.
Expressed CYP2B6.1 and CYP2B6.6 proteins.
CYP2B6.1 and CYP2B6.6 proteins with coexpression of human P450 oxidoreductase (POR) (without and with coexpression of Cyt b5) and plasmid-transfected negative controls were produced by BD Biosciences (Woburn, MA) and kindly provided by Dr. Guo (Eli Lilly and Company, Indianapolis, IN). In those proteins expressed without Cyt b5, the protein content, P450 content using spectral assay, and Cyt c reductase activity were 26.6 mg/ml, 1451 pmol/ml, and 1739 nmol/(min · mg protein) for CYP2B6.1, and 33.5 mg/ml, 1582 pmol/ml, and 1489 nmol/(min · mg protein) for CYP2B6.6. Assuming that a specific activity of 3.0 micromoles of Cyt c reduced per minute per nanomole of reductase (Parikh et al., 1997), the molar ratios of P450:reductase for CYP2B6.1 and CYP2B6.6 were 1:10.6 and 1:10.5, respectively. In those proteins coexpressed with Cyt b5, the protein content, P450 content, Cyt c reductase activity, and Cyt b5 content were 9.0 mg/ml, 1000 pmol/ml, 1900 nmol/(min · mg protein), and 220 pmol/mg protein for CYP2B6.1, whereas they were 2.7 mg/ml, 1000 pmol/ml, 851 nmol/(min · mg protein), and 370 pmol/mg protein for CYP2B6.6. The molar ratio of P450:reductase:Cyt b5 of CYP2B6.1 was 1:5.4:2, and that of CYP2B6.6 was 1:0.73:1.
Human liver microsomes.
HLMs obtained from liver tissues with CYP2B6*1/*1, CYP2*1/*6, and CYP2*6/*6 genotypes were used for the metabolism and inhibition studies. Two HLM sources were used. HLMs that were obtained from the Medical College of Wisconsin (Milwaukee, WI), Medical College of Virginia (Richmond, VA), Indiana University School of Medicine (Indianapolis, IN), and University of Pittsburgh (Pittsburgh, PA) under protocols approved by the appropriate committees for the conduct of human research were prepared by Eli Lilly and Company (Indianapolis, IN) and kindly provided by Dr. Guo. Liver microsomes were prepared by differential centrifugation (van der Hoeven and Coon, 1974). Additional HLMs for inhibition study were obtained from in-house human liver tissues, which were medically unsuitable for transplantation and were prospectively collected in the Division of Clinical Pharmacology by Dr. Hall through the liver transplantation units of Indiana University hospitals. HLMs were prepared from these liver tissues by ultracentrifugation, and protein concentrations were determined using standard protocols (Desta et al., 1998). Genotyping for the CYP2B6*6 allele was performed in those human liver tissues from which HLMs were prepared. Liver samples were homogenized, and genomic DNA was isolated using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. The concentration of DNA was determined using the PicoGreen assay, and the quality of DNA was checked by agarose gel and polymerase chain reaction. The DNA samples were stored at −80°C until analysis. The two SNPs tagging CYP2B6*6 allele, 516G>T and 785A>G, were genotyped using either the Affymetrix DMET Premier Pack (Affymetrix, Santa Clara, CA) or TaqMan SNP-genotyping assays (Applied Biosystems, Foster City, CA) according to the respective manufacturer's protocols. CYP2B6*1 was designated as the allele without these two tagging SNPs. Other microsomal preparations (HLMs, expressed enzymes, and plasmid-transfected negative controls) were obtained from BD Biosciences (San Jose, CA). All microsomal preparations were stored at −80°C until analysis.
General Incubation Conditions in Expressed Enzymes and HLMs.
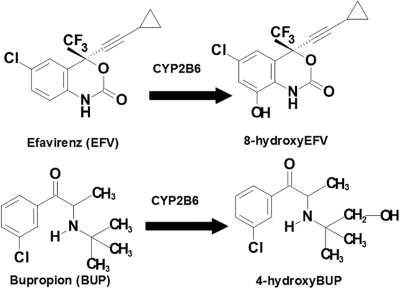
Efavirenz 8-hydroxylation and bupropion 4-hydroxylation (Fig. 1) have been shown to be mainly catalyzed by CYP2B6 (Faucette et al., 2000; Ward et al., 2003). Therefore, we used these two probe reactions to determine CYP2B6 activity in expressed CYP2B6 proteins (CYP2B6.1 and CYP2B6.6) and HLMs obtained from human liver tissues genotyped for the CYP2B6*6 allele. Efavirenz and bupropion were dissolved and diluted in methanol to the required concentrations (1–200 μM efavirenz and 10–1000 μM bupropion), and methanol was removed by drying in a speed vacuum before the addition of the incubation components. The reaction components contain 200 mM potassium phosphate buffer (pH 7.4), expressed CYP2B6 (10–15 pmol) or 25 μl of HLMs (2.5 mg/ml), and a substrate (efavirenz or bupropion) (total incubation volume of 250 μl). The incubation mixture was prewarmed for 5 min at 37°C. The reaction was initiated by adding a NADPH-regenerating system (1.3 mM NADP, 3.3 mM glucose 6-phosphate, 3.3 mM MgCl2, and 1 μl/ml glucose-6-phosphate dehydrogenase). Reaction was allowed to proceed for 15 min and then was terminated by placing tubes on ice and immediate addition of 500 μl of acetonitrile. After an internal standard was added, the sample was vortex-mixed and centrifuged at 14,000 rpm for 5 min. Ritonavir (50 μl of 0.01 mg/ml) and nevirapine (50 μl of 500 ng/ml) were used as an internal standard for 8-hydroxyefavirenz assay by HPLC and liquid chromatography/tandem mass spectrometry (LC/MS/MS) methods, respectively. For 4-hydroxybupropion assay, nevirapine (25 μl of 50 μM) was used as an internal standard for the HPLC assay and 25 μl of 5 μM nevirapine was used as an internal standard for the LC/MS/MS assay. The supernatant layer was extracted with 500 μl of 0.5 ml glycine/NaOH buffer (pH 11.3) and 6 ml of ethyl acetate and then centrifuged at 36,000 rpm for 15 min. The organic layer was dried and reconstituted with mobile phase, and an aliquot was injected into an HPLC or LC/MS/MS (see Quantification of Efavirenz Metabolites).
Fig. 1.
Structure of probe substrate reactions: CYP2B6-mediated 8-hydroxylation of efavirenz and CYP2B6-mediated 4-hydroxylation of bupropion.
Quantification of Efavirenz Metabolites.
8-Hydroxyefavirenz formed from efavirenz incubations in expressed CYP2B6.1 and CYP2B6.6 was quantified by an HPLC/UV system as described previously (Ward et al., 2003). Due to the slow formation rates of 8-hydroxyefavirenz in HLMs samples, particularly in those with CYP2B6*6/*6 genotype, a sensitive and selective LC/MS/MS method was developed to assay 8-hydroxyefavirenz from HLMs incubation and was implemented as described in our previous publication (Ogburn et al., 2010). The MS/MS system was an API 2000 MS/MS triple quadruple system (Applied Biosystems, Foster City, CA) equipped with a turbo ion spray and was coupled with a Shimadzu (Columbia, MD) HPLC system consisting of an LC-20AB pump and SIL-20A HT autosampler, all controlled by Analyst 1.4.2 software (Applied Biosystems/MDS Sciex, Foster City, CA) in conjunction with Windows 2000 (Microsoft, Redmond, WA). Hydroxyefavirenz and nevirapine were detected using multiple reactions monitoring at a m/z of 332.2/248.3 and 267.1/226.4 in positive ion mode, respectively.
Quantification of 4-Hydroxybupropion.
An HPLC assay method with UV detection was developed for the quantification of 4-hydroxybupropion from bupropion incubation in expressed enzymes. The HPLC system consisted of a Shimadzu LC-10AT pump, SIL-10AD autosampler, SCL-10A system controller, and SPD-10A UV-VIS detector. The separation system consisted of a Zorbax SB-C18 column (150 × 4.6 mm, 3.5-μm particle size; Phenomenex, Torrance, CA), a Luna C18 guard column (30 × 4.6 mm, 5 μm; Phenomenex), and a mobile phase composed of 85% 10 mM KH2PO4 (adjusted to pH 3 with 85% phosphoric acid) and 15% (v/v) acetonitrile (flow rate, 1 ml/min). The column elute was monitored by UV detection at 214 nm for 4-hydroxybupropion and 282 nm for internal standard (nevirapine). A LC/MS/MS assay was developed for the quantification of bupropion metabolite in HLM incubation samples. The MS/MS system was the same as that for efavirenz metabolites quantification described above. In brief, bupropion, 4-hydroxybupropion, and the internal standard (nevirapine) were separated using Zorbax SB-C18 column (100 × 2.00 mm, 3-μm particle size), a Luna C18 guard column (30 × 4.6 mm, 5 μm), and an isocratic mobile phase that consisted of 75% formic acid (0.1% in H2O) and 25% acetonitrile (flow rate, 0.3 ml/min). 4-Hydroxybupropion and nevirapine were detected using multiple reactions monitoring at a m/z of 256.1/238.0 and 267.2/224.4 in positive ion mode, respectively.
Inhibition of CYP2B6 by Voriconazole and Clopidogrel in HLMs.
To test the impact of the CYP2B6*6 allele on metabolic inhibition of CYP2B6, inhibition experiments were performed in HLMs obtained from human liver tissues genotyped for the CYP2B6*6 allele. IC50 values for the inhibition of CYP2B6 by voriconazole and clopidogrel were determined by incubating efavirenz (10 μM) with a NADPH-generating system and 25 μl of HLMs (2.5 mg/ml) at 37°C for 15 min in the absence or presence of voriconazole (0.01–4 μM) and clopidogrel (0.003–2.5 μM) (total incubation volume of 250 μl). Dixon plots for the inhibition of CYP2B6 by voriconazole were determined by incubating efavirenz (10–100 μM) with a NADPH-generating system and 25 μl of HLMs (2.5 mg/ml) at 37°C for 15 min in the absence or presence of voriconazole (0.1–10 μM) (total incubation volume of 250 μl). The samples were processed, and formation of 8-hydroxyefavirenz was quantified by LC/MS/MS as described above.
Data Analysis.
Apparent kinetic constants (Km and Vmax) were estimated by fitting formation rates of metabolites versus substrate concentrations to simple single-site Michaelis-Menten equation by nonlinear regression analysis using Prism version 5.0 software (GraphPad Software Inc., San Diego, CA). In vitro intrinsic clearance (Clint) was given as Vmax/Km. Inhibition constants (Ki values) were calculated by fitting the inhibition data to different models of enzyme inhibition (competitive, noncompetitive, and uncompetitive) using nonlinear least-squares regression analysis. The appropriate type of inhibition or metabolism model for each data set was selected on the basis of visual inspection of the plots, the size of the sum of squares of residuals, the Akaike information criterion, and the 95% confidence interval of the parameter estimates.
Statistical Analysis.
Statistical comparisons of metabolism and inhibition kinetic parameters among genotypes were performed using one-way analysis of variance with Dunn's post hoc test for multiple comparison correction. Independent t test was used to compare parametric data from two groups. Mann-Whitney U test or Wilcoxon test was performed for nonparametric data. Correlation analysis was performed by a nonparametric test (Spearman's rank correlation test). All statistical tests were performed using GraphPad. P < 0.05 was considered statistically significant.
Results
Efavirenz 8-Hydroxylation and Bupropion 4-Hydroxylation by CYP2B6.1 and CYP2B6.6 Proteins without Coexpression of Cyt b5.
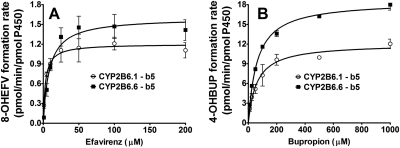
The catalytic properties of expressed CYP2B6.1 and CYP2B6.6 proteins were determined using efavirenz and bupropion as probes. The kinetic profiles for formation of 8-hydroxyefavirenz from efavirenz and 4-hydroxybupropion from bupropion in these proteins are depicted in Fig. 2. The kinetic parameters estimated are summarized in Table 1. The Km and Vmax values for the formation of 8-hydroxye were significantly higher in the CYP2B6.6 than in the CYP2B6.1 protein (Fig. 2A; Table 1). The in vitro intrinsic clearance (Vmax/Km or Clint) in the CYP2B6.6 protein was significantly lower than in the CYP2B6.1 protein (Table 1). As shown in Fig. 2B and Table 1, Vmax value for the formation of 4-hydroxybupropion in the CYP2B6.6 protein was significantly higher than that estimated from CYP2B6.1 protein, whereas there was no statistically significant difference in the Km values between the variant and the wild-type proteins. Accordingly, the Clint for the formation of 4-hydroxybupropion was significantly increased in the CYP2B6.6 protein compared with that of the CYP2B6.1 protein (Table 1).
Fig. 2.
Efavirenz concentrations versus formation rate of 8-hydroxyefavirenz (A) and bupropion concentrations versus formation rate of 4-hydroxybupropion (B) in microsomes containing cDNA-expressed CYP2B6.1 and CYP2B6.6 without coexpression of Cyt b5. Efavirenz (1–200 μM) or bupropion (10–1000 μM) was incubated with reconstituted systems containing either CYP2B6.1 or CYP2B6.6 (15 pmol) and an NADPH-generating system for 15 min at 37°C. Formation rate of 8-hydroxyefavirenz and 4-hydroxybupropion (pmol/min/pmol P450) versus substrate concentrations were fit to the simple single-site Michaelis-Menten equation. Each point represents mean ± S.D. of three replicates. 8-OHEFV, 8-hydroxyefavirenz; 4-OHBUP, 4-hydroxybupropion.
TABLE 1.
Kinetic parameters for the formation of 8-hydroxyefavirenz from efavirenz and 4-hydroxybupropion from bupropion in expressed CYP2B6.1 and CYP2B6.6 without and with coexpression of Cyt b5
Kinetic data are presented as mean ± S.D. (n = 3 incubations were performed in duplicate). In vitro Clint was calculated as Vmax/Km. Kinetic parameters for the formation of 8-hydroxyefavirenz and 4-hydroxybupropion were estimated by fitting the velocity versus substrate concentrations to the simple single-site Michaelis-Menten equation.
| Kinetic Parameters | Without Cyt b5 |
With Cyt b5 |
P Value (Comparing with vs. without b5)† |
|||
|---|---|---|---|---|---|---|
| CYP2B6.1 | CYP2B6.6 | CYP2B6.1 | CYP2B6.6 | CYP2B6.1 | CYP2B6.6 | |
| Efavirenz 8-hydroxylation | ||||||
| Vmax, pmol · min−1 · pmol−1 | 1.21 ± 0.15 | 1.61 ± 0.11** | 1.02 ± 0.11 | 0.95 ± 0.02 | 0.14 | 0.00002 |
| Km, μM | 3.2 ± 1.0 | 8.8 ± 1.6*** | 3.4 ± 0.6 | 6.2 ± 2.6 | 0.74 | 0.14 |
| Clint, μl · min−1 · pmol− | 0.39 ± 0.08 | 0.19 ± 0.04** | 0.30 ± 0.06 | 0.17 ± 0.07 | 0.18 | 0.69 |
| Bupropion 4-hydroxylation | ||||||
| Vmax, pmol · min−1 · pmol−1 | 13.37 ± 0.97 | 18.55 ± 0.82** | 15.18 ± 0.99 | 11.77 ± 1.46** | 0.04 | 0.0002 |
| Km, μM | 64.2 ± 13.4 | 62.6 ± 7.3 | 90.6 ± 10.3 | 110.0 ± 29.8 | 0.02 | 0.02 |
| Clint, μl · min−1 · pmol−1 | 0.21 ± 0.03 | 0.30 ± 0.03** | 0.17 ± 0.02 | 0.11 ± 0.04* | 0.07 | 0.0002 |
P < 0.05,
P < 0.01, and
P < 0.001 compared CYP2B6.6 to CYP2B6.1 without and with coexpression of Cyt b5, respectively.
Kinetic parameters of CYP2B6.1 with and without Cyt b5 as well as CYP2B6.6 with and without Cyt b5 were also compared.
Efavirenz 8-Hydroxylation and Bupropion 4-Hydroxylation by CYP2B6.1 and CYP2B6.6 Proteins Coexpressed with Cyt b5.
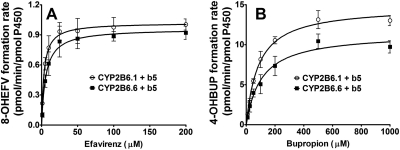
To evaluate the potential effect of Cyt b5 on catalytic properties of the variant versus wild-type protein, kinetic parameters for the formation of 8-hydroxyefavirenz and 4-hydroxybupropion were determined in CYP2B6.1 and CYP2B6.6 proteins that were coexpressed with Cyt b5, and the results were compared with those data obtained from CYP2B6.1 and CYP2B6.6 proteins without coexpression of Cyt b5. Kinetic profiles for the formation of 8-hydroxyefavirenz and 4-hydroxybupropion in CYP2B6.1 and CYP2B6.6 proteins are shown in Fig. 3. The corresponding kinetic parameters are summarized in Table 1. In contrast to the significant changes by the CYP2B6*6 allele observed in expressed system without coexpression of Cyt b5, the Vmax value for the formation of 8-hydroxyefavirenz was not significantly different (P = 0.20) between CYP2B6.1 and CYP2B6.6 proteins coexpressed with Cyt b5 (Fig. 3A; Table 1). Although the Km value in CYP2B6.6 protein was higher by 81% than that in the CYP2B6.1 protein and the Clint value was decreased by 43% consistent with the data obtained from system without Cyt b5 (Table 1), the differences did not reach a statistically significant level (Fig. 3A; Table 1) (P = 0.059). When bupropion 4-hydroxylation was used as a reaction probe, a significantly lower Vmax value was observed in CYP2B6.6 protein compared with the value obtained from the CYP2B6.1 protein (Fig. 3B; Table 1). The Km value was increased from 90.6 μM in the CYP2B6.1 protein to 110.0 μM in the variant protein, but this did not reach a statistically significant difference (P = 0.27). A significant decrease in Clint was observed in the CYP2B6.6 protein compared with the CYP2B6.1 protein.
Fig. 3.
Efavirenz concentrations versus formation rate of 8-hydroxyefavirenz (A) and bupropion concentrations versus formation rate of 4-hydroxybupropion (B) in microsomes containing cDNA-expressed CYP2B6.1 and CYP2B6.6 with coexpression of Cyt b5. Efavirenz (1–200 μM) or bupropion (10–1000 μM) was incubated with reconstituted systems containing either CYP2B6.1 or CYP2B6.6 with coexpression of Cyt b5 (15 pmol) and a NADPH-generating system for 15 min at 37°C. The formation rate of 8-hydroxyefavirenz and 4-hydroxybupropion (pmol/min/pmol P450) versus substrate concentrations were fit to the simple single-site Michaelis-Menten equation. Each point represents mean ± S.D. of three replicates. 8-OHEFV, 8-hydroxyefavirenz; 4-OHBUP, 4-hydroxybupropion.
As described in the preceding paragraph, the catalytic properties of CYP2B6 seemed to be genotype- and Cyt b5-dependent. To gain further insight regarding the differential effect of Cyt b5 on CYP2B6.1 versus CYP2B6.6 protein, kinetic parameters for efavirenz 8-hydroxylation and bupropion 4-hydroxylation obtained in the presence of Cyt b5 were compared with those values obtained in P450 proteins expressed without Cyt b5 (Table 1). In the CYP2B6.1 protein, none of the kinetic parameters of efavirenz 8-hydroxylation were significantly different compared with values obtained from CYP2B6.1 protein without coexpression of Cyt b5. Coexpression of Cyt b5 with the CYP2B6.6 protein significantly decreased the Vmax value for the formation of 8-hydroxyefavirenz compared with the value obtained from CYP2B6.6 protein without coexpression of Cyt b5. However, because the Km value also tended toward decrease in the CYP2B6.6 protein, the Clint value in the CYP2B6.6 protein coexpressed with Cyt b5 was not significantly different than that obtained from the CYP2B6.6 protein without Cyt b5 (Table 1). Similar to the findings with efavirenz 8-hydroxylation, differential effects of Cyt b5 on the catalytic properties of CYP2B6.1 and CYP2B6.6 were observed with bupropion 4-hydroxylation (Table 1). Compared with the CYP2B6.1 protein without Cyt b5, the CYP2B6.1 protein coexpressed with Cyt b5 exhibited modest increases in the Vmax and Km values for the formation of 4-hydroxybupropion, the Clint tended toward decrease (Table 1). The presence of Cyt b5 with CYP2B6.6 protein decreased the Vmax value to 11.77 pmol · min−1 · pmol P450−1 from 18.55 pmol · min−1 · pmol P450−1 in the CYP2B6.6 protein without Cyt b5 (P = 0.0002). A significant increase was observed in Km value of CYP2B6.6 protein coexpressed with Cyt b5 compared with that of CYP2B6.6 protein coexpressed without Cyt b5. As a result, Clint for the formation of 4-hydroxybupropion was significantly decreased in CYP2B6.6 coexpressed with Cyt b5 (Table 1).
We recognized that recombinant protein systems have limitations, which include differences in cofactor expression between variants and wild type or even between batches of the same protein. In the expressed enzymes used in this experiment, the amounts of Cyt b5 expressed in CYP2B6.1 and CYP2B6.6 proteins were slightly different (220 and 370 pmol/mg protein, respectively). However, we have observed substrate-dependent effects for the CYP2B6*6 allele in the presence of Cyt b5, i.e., no significant differences were found in Vmax and Km comparing CYP2B6.1 to CYP2B6.6 using efavirenz as substrate, whereas Vmax was significantly lower in CYP2B6.6 using bupropion. These data suggest that expression differences in Cyt b5 do not seem to significantly contribute to the differences in kinetic parameters observed.
Catalytic Properties of CYP2B6 in HLMs Genotyped for the CYP2B6*6 Allele.
To further evaluate the effect of CYP2B6*6 allele on catalytic properties, the kinetics of efavirenz 8-hydroxylation and bupropion 4-hydroxylation were characterized in 15 HLM samples with CYP2B6*1/*1, CYP2B*1/*6, and CYP2B*6/*6 genotypes (n = 5 for each genotype).
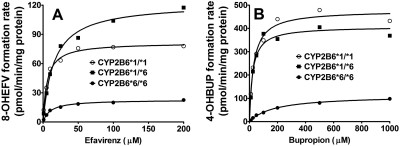
In Fig. 4A, efavirenz concentrations versus formation rate of 8-hydroxyefavirenz in the different genotypes are shown. Formation rates of 8-hydroxyefavirenz versus efavirenz concentrations were fit into a Michaelis-Menten equation to estimate kinetic parameters. The mean ± S.D. of these parameters for each genotype are listed in Table 2. The kinetic parameters for individual HLMs are summarized in Supplemental Table 1. None of the kinetic parameters were statistically different among the three genotypes, probably due to the high inter-HLMs variability in Vmax and Clint for the formation of 8-hydroxyefavirenz, particularly in HLMs with CYP2B6*1/*1 and CYP2B6*1/*6 [coefficient of variation (CV) of more than 100%]; this variability was smaller in HLMs with CYP2B*6/*6 genotype (CV of approximately 30% for Vmax and 65% for Clint). Despite this lack of statistical significance, it is noteworthy that the average Vmax in HLMs with CYP2B6*6/*6 genotype were lower by 71 and 75% compared with values in HLMs with CYP2B6*1/*1 and CYP2B6*1/*6 genotypes, respectively. The Km values in CYP2B6*6/*6 genotype were higher on average by 114 and 58% than that in CYP2B6*1/*1 and CYP2B6*1/*6 genotypes, respectively. Accordingly, the Clint values in HLMs with CYP2B6*6/*6 were 83% lower compared with that of HLMs with CYP2B6*1/*1 and 62% lower compared with that of HLMs with CYP2B6*1/*6 genotype.
Fig. 4.
Efavirenz concentrations versus formation rate of 8-hydroxyefavirenz (A) and bupropion concentrations versus formation rate of 4-hydroxybupropion (B) in 15 human liver microsomal samples with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotypes (n = 5 HLMs for each genotype). Efavirenz (1–200 μM) or bupropion (10–1000 μM) was incubated with human liver microsomal samples (0.25 mg/ml) with CYP2B6*1/*1, CYP2B*1/*6, and CYP2B*6/*6 genotypes (n = 5 HLMs for each genotype) and a NADPH-generating system for 15 min at 37°C in duplicate. The formation rate of 8-hydroxyefavirenz and 4-hydroxybupropion (pmol/min/mg protein) versus substrate concentrations were fit to the simple single-site Michaelis-Menten equation. Each point represented as the average of five individual incubations in human liver microsomal samples with the same CYP2B6 genotype. 8-OHEFV, 8-hydroxyefavirenz; 4-OHBUP, 4-hydroxybupropion.
TABLE 2.
Kinetic parameters (mean ± S.D.) for the formation of 8-hydroxyefavirenz from efavirenz in 15 human liver microsomal samples with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotypes (n = 5 HLMs for each genotype)
Efavirenz (1–200 μM) was incubated with human liver microsomal samples (0.25 mg/ml) with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotypes (n = 5 HLM for each genotype) and a NADPH-generating system at 37°C for 15 min in duplicate. Kinetic parameters (Vmax and Km) for the formation of 8-hydroxyefavirenz were estimated by fitting the velocity versus efavirenz concentrations to the simple single-site Michaelis-Menten equation. In vitro Clint was calculated as Vmax/Km. The kinetic parameters (Vmax, Km, and Clint) for each genotype group are listed in Supplemental Table 1. The data presented here are mean ± S.D. calculated from five individual HLM values for each genotype.
| HLMs | 8-Hydroxyefavirenz |
||
|---|---|---|---|
| Vmax | Km | Clint | |
| pmol · min−1 · mg protein−1 | μM | μl · min−1 · mg protein−1 | |
| CYP2B6*1/*1 | 87.1 ± 87.4 | 11.2 ± 6.7 | 14.5 ± 22.6 |
| CYP2B6*1/*6 | 100.6 ± 143.7 | 15.2 ± 8.0 | 6.4 ± 7.8 |
| CYP2B6*6/*6 | 25.0 ± 6.7 | 24.0 ± 31.3 | 2.4 ± 1.6 |
Kinetic analyses for the formation of 4-hydroxybupropion were also performed in the same 15 HLM samples that were used for the characterization of efavirenz metabolism. In Fig. 4B, bupropion concentrations versus formation rate of 4-hydroxybupropion in the different genotypes are shown. Formation rates of 4-hydroxybupropion versus bupropion concentrations were fit into a Michaelis-Menten equation to estimate kinetic parameters. The mean ± S.D. of these parameters for each genotype are listed in Table 3. The kinetic parameters for individual HLMs are summarized in Supplemental Table 2. Similar to that observed for the kinetics of 8-hydroxyefavirenz, the values of Vmax and Clint for the formation of 4-hydroxybupropion in HLMs also exhibited a large variability. HLMs with CYP2B6*1/*1 and CYP2B6*1/*6 genotypes showed higher variability (CV of more than 100%) than that in HLMs with CYP2B6*6/*6 (CV of approximately 60 and 65%, respectively). The average Vmax values for the formation of 4-hydroxybupropion were also much lower in HLMs with CYP2B6*6/*6 genotype than that in wild type and heterozygotes. The HLMs with CYP2B6*1/*1 exhibited lower average Km values compared with HLMs with CYP2B6*1/*6 and CYP2B6*6/*6, although this difference did not reach statistical significance (Table 3). The Clint in CYP2B6*6/*6 genotype was decreased by over 95% compared with CYP2B6*1/*1 and CYP2B6*1/*6 (Table 3).
TABLE 3.
Kinetic parameters (mean ± S.D.) for the formation of 4-hydroxybupropion from bupropion in 15 human liver microsomal samples with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotypes (n = 5 HLMs for each genotype)
Bupropion (10–1000 μM) were incubated with HLM samples (0.25 mg/ml) with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotypes (n = 5 HLMs for each genotype) and a NADPH-generating system at 37°C for 15 min in duplicate. Kinetic parameters (Vmax and Km) for the formation of 4-hydroxybupropion were estimated by fitting the velocity versus bupropion concentrations to the simple single-site Michaelis-Menten. In vitro Clint was calculated as Vmax/Km. The kinetic parameters (Vmax, Km, and Clint) for each genotype group are listed in Supplemental Table 2. The data presented here are mean ± S.D. calculated from five individual HLM values for each genotype.
| HLMs | 4-Hydroxybupropion |
||
|---|---|---|---|
| Vmax | Km | Clint | |
| pmol · min−1 · mg protein−1 | μM | μl · min−1 · mg protein−1 | |
| CYP2B6*1/*1 | 492.8 ± 427.9 | 86.0 ± 75.7 | 18.8 ± 26.3 |
| CYP2B6*1/*6 | 441.6 ± 583.0 | 212.1 ± 221.1 | 21.8 ± 44.2 |
| CYP2B6*6/*6 | 112.9 ± 66.7 | 204.2 ± 66.1 | 0.6 ± 0.5 |
Inhibition of 8-Hydroxyefavirenz Formation in HLMs Genotyped for the CYP2B6*6 Allele.
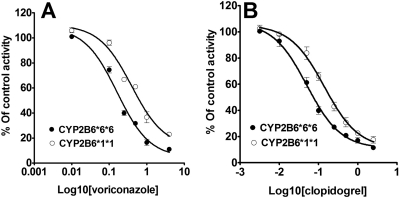
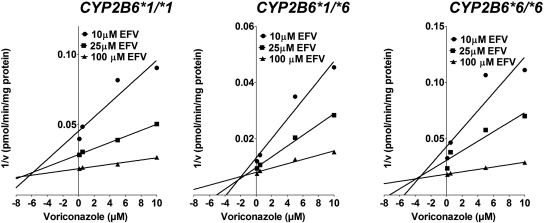
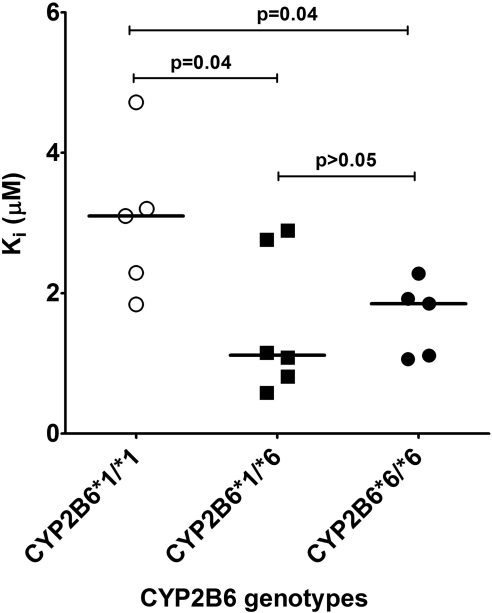
To test whether susceptibility to metabolic inhibitors differs between HLMs that carry the CYP2B6*6 allele and the wild type, inhibition potency of voriconazole and clopidogrel towards efavirenz 8-hydroxylation was determined in HLMs obtained from tissues genotyped for the CYP2B6*6 allele. Inhibition of CYP2B6 by voriconazole and clopidogrel in HLMs with CYP2B6*1/*1 and CYP2B6*6/*6 is shown in Fig. 5. IC50 values for voriconazole inhibition in HLMs with CYP2B6*1/*1 and CYP2B6*6/*6 were 0.40 and 0.16 μM, respectively. IC50 value for clopidogrel inhibition of efavirenz 8-hydroxylation in HLMs with CYP2B6*1/*1 (IC50 = 0.14 μM) was also higher than that in HLMs with CYP2B6*6/*6 (IC50 = 0.05 μM). Representative Dixon plots for the inhibition of efavirenz 8-hydroxylation in the HLMs with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotypes are shown in Fig. 6. As shown in Fig. 7, there was a statistically significant difference among the Ki values estimated from the three genotypes (P = 0.04). There was no significant difference between the Ki values estimated from CYP2B6*1/*6 (average Ki value = 1.55 μM) and CYP2B6*6/*6 (average Ki value = 1.64 μM) (P = 0.85). However, the Ki values estimated from HLMs with CYP2B6*6/*6 (P = 0.04) and CYP2B6*1/*6 (P = 0.04) genotypes were both significantly lower than that estimated from CYP2B6*1/*1 genotype (average Ki value = 3.03 μM) (Fig. 7). When the data from HLMs with CYP2B6*6/*6 and CYP2B6*1/*6 genotypes were combined and compared against HLMs with the CYP2B6*1/*1 genotype, the Ki values for the inhibition of efavirenz 8-hydroxylation by voriconazole in the HLMs with CYP2B6*1/*6 + CYP2B6*6/*6 genotypes was significantly lower (P = 0.009) than those observed in HLMs with CYP2B6*1/*1 genotype (data not shown).
Fig. 5.
Inhibition of CYP2B6 by voriconazole (A) and clopidogrel (B) in HLMs with CYP2B6*1/*1 and CYP2B6*6/*6. Efavirenz (10 μM) was incubated with HLMs (0.25 mg/ml) and the NADPH-generating system for 15 min without or with voriconazole (0–4 μM) and clopidogrel (0–2.5 μM). Each point represents the mean of duplicate.
Fig. 6.
Representative Dixon plots for the inhibition on 8-hydroxylation of efavirenz by voriconazole in HLMs with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6. Efavirenz (10–100 μM) was incubated with HLMs (0.25 mg/ml; IU 5, IU 73, and HL-G) and the NADPH-generating system at 37°C for 15 min without or with voriconazole (0.1–10 μM). Each point represents the mean of duplicate.
Fig. 7.
The Ki values for the formation of 8-hydroxyefavirenz by voriconazole in the HLMs with CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 genotype. The horizontal line indicates the median Ki value. Dots represent the Ki values generated using each individual human liver microsomal samples.
Discussion
In this study, we have shown that the CYP2B6*6 allele is associated with altered binding affinity and/or catalytic activity. Cyt b5 affects the kinetic profiles of CYP2B6 in genotype- and substrate-dependent manner. Our data also suggest that the variant protein is more susceptible to metabolic inhibition. These findings suggest that the mechanism by which the CYP2B6*6 allele is associated with altered substrate metabolism and drug interaction may be in part due to amino acid changes that modify catalytic properties of the variant versus wild-type protein.
Our data suggest that the amino acid changes harbored in CYP2B6*6 allele may influence substrate binding with pronounced effect on efavirenz than bupropion. The average Km value for the formation of 8-hydroxyefavirenz in CYP2B6.6 protein expressed without b5 was increased by 175% and in HLMs with CYP2B6*6/*6 genotype was also increased by 58 and 114% than in HLMs with CYP2B6*1/*6 and CYP2B6*1/*1 genotypes, respectively. These data concur with a recent report by Zhang et al. (2011). Km value for bupropion hydroxylation did not differ between expressed variant and wild-type proteins. A slight increase in Km for bupropion 4-hydroxylation was noted in HLMs with CYP2B6*1/*6 or CYP2B6*6/*6 genotypes than HLMs with CYP2B6*1/*1 genotype. However, because some Km values derived from HLMs with CYP2B6*1/*1 and CYP2B6*1/*6 were outliers and may have skewed the average data, these data should be interpreted with caution.
Consistent with previous reports in various expressed systems (Ariyoshi et al., 2001; Jinno et al., 2003; Bumpus et al., 2006), we noted that the Vmax values for the formation of 8-hydroxyefavirenz and 4-hydroxybupropion were significantly higher in CYP2B6.6 than in CYP2B6.1 proteins expressed without Cyt b5. However, Vmax values for the formation of 8-hydroxyefavirenz and 4-hydroxybupropion were substantially decreased (by ∼70%) in HLMs with CYP2B6*6/*6 genotype versus HLMs with CYP2B6*1/*6 and CYP2B6*1/*1 genotypes. Our interpretation is that the expressed variant protein inherently increases catalytic activity for most substrates, whereas the decreased Vmax value in HLMs is probably mainly due to reduced protein expression by the CYP2B6*6/*6 genotype (Hesse et al., 2004; Desta et al., 2007; Hofmann et al., 2008).
In vivo, the CYP2B6*6/*6 genotype is associated with >3-fold increase in efavirenz exposure compared with CYP2B6*1/*1 genotype (Rotger et al., 2007), but its effect on plasma exposure of bupropion or 4-hydroxybupropion was marginal (Kirchheiner et al., 2003). Our in vitro data mirror these clinical observations. It is well recognized that variants in other P450s that change amino acids affect metabolic activity in a substrate-dependent manner. However, the CYP2B6*6 allele seems unique in that its effect on catalytic activity is not only substrate-dependent but also results in opposite effects. This variant has been associated with enhanced cyclophosphamide metabolism in vitro (Xie et al., 2003; Ariyoshi et al., 2011) and in vivo (Nakajima et al., 2007), which seems to be primarily driven by the significantly lower Km for cyclophosphamide 4-hydroxylation in CYP2B6.6 than in CYP2B6.1 proteins (Ariyoshi et al., 2011). The CYP2B6*6 allele appears to alter substrate metabolism in two ways: 1) by decreasing (e.g., cyclophosphamide) or increasing (e.g., efavirenz) substrate binding (Ariyoshi et al., 2011; Zhang et al., 2011; present data) probably due to changes in the three-dimensional structures of the protein; and 2) by reducing catalytic efficiency secondary to reduced protein expression (Hofmann et al., 2008). Overall, altered substrate binding and/or catalytic activity as a result of amino acid changes seem to play a critical role in determining the substrate-dependent functional consequences of the CYP2B6*6 allele.
Our data show that Cyt b5 affects catalytic properties in a genotype- and substrate-dependent manner and highlight the fact that interpretation of in vitro studies performed with expressed proteins may vary depending on the presence or absence of Cyt b5, substrate used, and underlying genotype. The ability of Cyt b5 to influence P450-mediated drug oxidation (increase, inhibit, or no effect) has been described for multiple P450s (Schenkman and Jansson, 2003). The mechanisms by which Cyt b5 might alter substrate metabolism include the following: providing the second electron during the catalytic cycle of P450s; interacting physically with P450s and thus modifying conformation of the protein, which, in turn, influences interaction with the substrate or reductase; or by competing for same binding site with P450 reductase, thereby preventing reduction of ferric P450 and initiation of the catalytic cycle (Zhang et al., 2008). In this study, Cyt b5 had no impact on CYP2B6.1-catalyzed efavirenz 8-hydroxylation. However, in sharp contrast to the results obtained from CYP2B6.6 without Cyt b5, Vmax for efavirenz 8-hydroxylation was significantly reduced by Cyt b5 (with no effect on Km) in CYP2B6.6 protein, effectively abolishing the genotype-dependent effect observed in CYP2B6.6 protein expressed without Cyt b5. The kinetic properties were different for bupropion 4-hydroxylation. Cyt b5 significantly increased the Vmax and Km values for bupropion 4-hydroxylation in CYP2B6.1 protein compared with CYP2B6.1 without Cyt b5. In CYP2B6.6, the Km for bupropion 4-hydroxylation was significantly increased and Vmax was significantly reduced by Cyt b5, leading to marked reduction in Clint in the CYP2B6.6 protein (Table 1). For both substrates, CYP2B6.1 exhibited similar or increased catalytic activities with coexpression of Cyt b5 compared with that without Cyt b5, whereas Cyt b5 significantly decreased Vmax values in CYP2B6.6. These data suggest an overlapping binding site between P450 reductase and Cyt b5 in CYP2B6.6 but probably not in CYP2B6.1. The possibility that the observed effect of Cyt b5 could be due to differences in the expression of Cyt b5 or POR among the genotypes cannot be excluded. In our study, the POR level was relatively lower in CYP2B6.6 with coexpression of Cyt b5 than that without coexpression of Cyt b5. Thus, the possibility that the lower expression of POR in the CYP2B6.6 protein may influence the magnitude of effect of Cyt b5 among the genotypes and substrates cannot be fully excluded. However, variation in kinetic parameters were observed even when the POR level was balanced between the variant and wild-type protein (Ariyoshi et al., 2011; Zhang et al., 2011). However, POR is much less functionally variable in general population than hepatic drug-oxidation P450s (Venkatakrishnan et al., 2000; Huang et al., 2004). Although some POR SNPs have been found to affect activities of CYP1A2, CYP2C8, CYP2C19, and CYP3A4, no POR SNP has been identified to significantly influence CYP2B6 activity to date (Gomes et al., 2009). Therefore, we believe that the differences in kinetics we observed are most likely due to the effect of Cyt b5. Further studies are warranted to identify the mechanism of substrate-dependent effect of Cyt b5 and to provide insight into the topology of the variant.
The two SNPs (K262R and Q172H) harbored in the CYP2B6*6 allele are not within the active site of the enzyme. Therefore, the mechanism by which binding affinity and/or catalytic efficiency is altered by the CYP2B6*6 allele is not fully understood. The two amino acid mutations harbored by the CYP2B6*6 allele may indirectly involve in the ligand binding and substrate catalysis. A recent publication that characterized the crystal structure of CYP2B6 genetic variant (Y226H, K262R) indicates that the side chain of residue 172 may interact with the residues at active site and thus could affect binding affinity (Gay et al., 2010). It is noteworthy that the other mutated amino acid carried by CYP2B6*6 allele, K262R, is located at the G/H loop, which may be involved in the interaction between the enzyme and its redox partner, P450 reductase (Bumpus and Hollenberg, 2008; Gay et al., 2010). The oxidation reaction catalyzed by P450s requires transferring of two electrons from NADPH. The first electron is generally thought to be transferred by P450 reductase, whereas the second can be transferred by either P450 reductase or Cyt b5. That altered electron transfer from P450 reductase to CYP2B6 variant proteins may influence substrate metabolism was suggested by a recent study (Zhang et al., 2011). Therefore, it is reasonable to suggest that amino acid changes may influence the interaction between the P450s and electron transfer proteins and thus alter the catalysis of substrates in a Cyt b5- and substrate-dependent manner.
The same property of the variant that influences substrate metabolism may also influence inhibition drug interactions. The fact that the Ki values for CYP2B6 inhibition by voriconazole was significantly lower in HLMs with CYP2B6*6 allele than in those with the CYP2B6*1/*1 genotype suggests that the variant protein is more susceptible to metabolic inhibition than the wild type. This suggestion is further supported by our data using clopidogrel as an inhibitor (2.8-fold lower IC50). Our data are in contrast to a previous study that reported decreased susceptibility of CYP2B6.6 protein to metabolic inhibition (Talakad et al., 2009). However, the different type and composition of the proteins, substrates, and inhibitors used in our study versus the other study preclude direct comparison of the data.
In summary, we have provided in vitro evidence that amino acid changes harbored in the CYP2B6*6 allele alter substrate binding and/or catalytic activity. In addition to reduced total enzyme pool, this variant allele may alter drug clearance and drug interaction via changes in three-dimensional protein structures. This in vitro suggestion is further supported by in vivo observation that showed association of CYP2B6*6 allele with reduced clearance [e.g., efavirenz (Zanger et al., 2007)], increased metabolism [e.g., cyclophosphamide (Nakajima et al., 2007)], or no effect [e.g., bupropion (Kirchheiner et al., 2003)]. Our data also showed that CYP2B6*6 allele may influence susceptibility to metabolic inhibition. In conclusion, predicting functional consequences of the CYP2B6*6 allele seems complex and depends on the substrate (or inhibitor) and enzyme sources used. These factors should be taken in to account when predicting the influence of the CYP2B6*6 allele on substrate metabolism and drug interactions.
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences [Grants GM078501, GM078501-04S1, 2R56-GM067308-09A1].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- SNP
- single nucleotide polymorphism
- HLMs
- human liver microsomes
- P450
- cytochrome P450
- Cyt b5
- cytochrome b5
- HPLC
- high-performance liquid chromatography
- POR
- P450 oxidoreductase
- LC/MS/MS
- liquid chromatography/tandem mass spectrometry
- Clint
- in vitro intrinsic clearance
- CV
- coefficient of variation.
Authorship Contributions
Participated in research design: Xu and Desta.
Conducted experiments: Xu and Ogburn.
Contributed new reagents or analytic tools: Guo.
Performed data analysis: Xu and Desta.
Wrote or contributed to the writing of the manuscript: Xu, Guo, and Desta.
References
- Ariyoshi N, Miyazaki M, Toide K, Sawamura Y, Kamataki T. (2001) A single nucleotide polymorphism of CYP2b6 found in Japanese enhances catalytic activity by autoactivation. Biochem Biophys Res Commun 281:1256–1260 [DOI] [PubMed] [Google Scholar]
- Ariyoshi N, Ohara M, Kaneko M, Afuso S, Kumamoto T, Nakamura H, Ishii I, Ishikawa T, Kitada M. (2011) Q172H replacement overcomes effects on the metabolism of cyclophosfamide and efavirenz caused by CYP2B6 variant with Arg262. Drug Metab Dispos 39:2045–2048 [DOI] [PubMed] [Google Scholar]
- Bumpus NN, Hollenberg PF. (2008) Investigation of the mechanisms underlying the differential effects of the K262R mutation of P450 2B6 on catalytic activity. Mol Pharmacol 74:990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus NN, Kent UM, Hollenberg PF. (2006) Metabolism of efavirenz and 8-hydroxyefavirenz by P450 2B6 leads to inactivation by two distinct mechanisms. J Pharmacol Exp Ther 318:345–351 [DOI] [PubMed] [Google Scholar]
- Desta Z, Kerbusch T, Soukhova N, Richard E, Ko JW, Flockhart DA. (1998) Identification and characterization of human cytochrome P450 isoforms interacting with pimozide. J Pharmacol Exp Ther 285:428–437 [PubMed] [Google Scholar]
- Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, Flockhart DA, Zanger UM. (2007) Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8:547–558 [DOI] [PubMed] [Google Scholar]
- Eap CB, Crettol S, Rougier JS, Schläpfer J, Sintra Grilo L, Déglon JJ, Besson J, Croquette-Krokar M, Carrupt PA, Abriel H. (2007) Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin Pharmacol Ther 81:719–728 [DOI] [PubMed] [Google Scholar]
- Faucette SR, Hawke RL, Lecluyse EL, Shord SS, Yan B, Laethem RM, Lindley CM. (2000) Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28:1222–1230 [PubMed] [Google Scholar]
- Gay SC, Shah MB, Talakad JC, Maekawa K, Roberts AG, Wilderman PR, Sun L, Yang JY, Huelga SC, Hong WX, et al. (2010) Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution. Mol Pharmacol 77:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM. (2009) Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10:579–599 [DOI] [PubMed] [Google Scholar]
- Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, Clifford DB, Hulgan T, Marzolini C, Acosta EP. (2004) Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 18:2391–2400 [PubMed] [Google Scholar]
- Hesse LM, He P, Krishnaswamy S, Hao Q, Hogan K, von Moltke LL, Greenblatt DJ, Court MH. (2004) Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics 14:225–238 [DOI] [PubMed] [Google Scholar]
- Hofmann MH, Blievernicht JK, Klein K, Saussele T, Schaeffeler E, Schwab M, Zanger UM. (2008) Aberrant splicing caused by single nucleotide polymorphism c. 516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther 325:284–292 [DOI] [PubMed] [Google Scholar]
- Huang W, Lin YS, McConn DJ, 2nd, Calamia JC, Totah RA, Isoherranen N, Glodowski M, Thummel KE. (2004) Evidence of significant contribution from CYP3A5 to hepatic drug metabolism. Drug Metab Dispos 32:1434–1445 [DOI] [PubMed] [Google Scholar]
- Jinno H, Tanaka-Kagawa T, Ohno A, Makino Y, Matsushima E, Hanioka N, Ando M. (2003) Functional characterization of cytochrome P450 2B6 allelic variants. Drug Metab Dispos 31:398–403 [DOI] [PubMed] [Google Scholar]
- Jushchyshyn MI, Hutzler JM, Schrag ML, Wienkers LC. (2005) Catalytic turnover of pyrene by CYP3A4: evidence that cytochrome b5 directly induces positive cooperativity. Arch Biochem Biophys 438:21–28 [DOI] [PubMed] [Google Scholar]
- Kaspera R, Naraharisetti SB, Evangelista EA, Marciante KD, Psaty BM, Totah RA. (2011) Drug metabolism by CYP2C8.3 is determined by substrate dependent interactions with cytochrome P450 reductase and cytochrome b5. Biochem Pharmacol 82:681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchheiner J, Klein C, Meineke I, Sasse J, Zanger UM, Mürdter TE, Roots I, Brockmöller J. (2003) Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 13:619–626 [DOI] [PubMed] [Google Scholar]
- Kumar V, Rock DA, Warren CJ, Tracy TS, Wahlstrom JL. (2006) Enzyme source effects on CYP2C9 kinetics and inhibition. Drug Metab Dispos 34:1903–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, Fackenthal JD, Rogan PK, Ring B, Wrighton SA, et al. (2003) Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther 307:906–922 [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM. (2001) Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11:399–415 [DOI] [PubMed] [Google Scholar]
- Mo SL, Liu YH, Duan W, Wei MQ, Kanwar JR, Zhou SF. (2009) Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab 10:730–753 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Komagata S, Fujiki Y, Kanada Y, Ebi H, Itoh K, Mukai H, Yokoi T, Minami H. (2007) Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenet Genomics 17:431–445 [DOI] [PubMed] [Google Scholar]
- Ogburn ET, Jones DR, Masters AR, Xu C, Guo Y, Desta Z. (2010) Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 2A6 as the principal catalyst of efavirenz 7-hydroxylation. Drug Metab Dispos 38:1218–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A, Gillam EM, Guengerich FP. (1997) Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol 15:784–788 [DOI] [PubMed] [Google Scholar]
- Reed JR, Hollenberg PF. (2003) Comparison of substrate metabolism by cytochromes P450 2B1, 2B4, and 2B6: relationship of heme spin state, catalysis, and the effects of cytochrome b5. J Inorg Biochem 93:152–160 [DOI] [PubMed] [Google Scholar]
- Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, Keiser O, Biollaz J, Décosterd L, Telenti A, et al. (2005) Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics 15:1–5 [DOI] [PubMed] [Google Scholar]
- Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Décosterd L, Blievernicht J, Saussele T, Günthard HF, Schwab M, et al. (2007) Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther 81:557–566 [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I. (2003) The many roles of cytochrome b5. Pharmacol Ther 97:139–152 [DOI] [PubMed] [Google Scholar]
- Talakad JC, Kumar S, Halpert JR. (2009) Decreased susceptibility of the cytochrome P450 2B6 variant K262R to inhibition by several clinically important drugs. Drug Metab Dispos 37:644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya K, Gatanaga H, Tachikawa N, Teruya K, Kikuchi Y, Yoshino M, Kuwahara T, Shirasaka T, Kimura S, Oka S. (2004) Homozygous CYP2B6*6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun 319:1322–1326 [DOI] [PubMed] [Google Scholar]
- van der Hoeven TA, Coon MJ. (1974) Preparation and properties of partially purified cytochrome P-450 and reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase from rabbit liver microsomes. J Biol Chem 249:6302–6310 [PubMed] [Google Scholar]
- Venkatakrishnan K, von Moltke LL, Court MH, Harmatz JS, Crespi CL, Greenblatt DJ. (2000) Comparison between cytochrome P450 (CYP) content and relative activity approaches to scaling from cDNA-expressed CYPs to human liver microsomes: ratios of accessory proteins as sources of discrepancies between the approaches. Drug Metab Dispos 28:1493–1504 [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306:287–300 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sakuyama K, Sasaki T, Ishii Y, Ishikawa M, Hirasawa N, Hiratsuka M. (2010) Functional characterization of 26 CYP2B6 allelic variants (CYP2B6.2-CYP2B6.28, except CYP2B6.22). Pharmacogenet Genomics 20:459–462 [DOI] [PubMed] [Google Scholar]
- Xie HJ, Yasar U, Lundgren S, Griskevicius L, Terelius Y, Hassan M, Rane A. (2003) Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J 3:53–61 [DOI] [PubMed] [Google Scholar]
- Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. (2007) Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics 8:743–759 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hamdane D, Im SC, Waskell L. (2008) Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J Biol Chem 283:5217–5225 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, Hollenberg PF. (2011) Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional P450-reductase complex. J Pharmacol Exp Ther 338:803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]