Abstract
Background and Aims
The production of triploid banana and plantain (Musa spp.) cultivars with improved characteristics (e.g. greater disease resistance or higher yield), while still preserving the main features of current popular cultivars (e.g. taste and cooking quality), remains a major challenge for Musa breeders. In this regard, breeders require a sound knowledge of the lineage of the current sterile triploid cultivars, to select diploid parents that are able to transmit desirable traits, together with a breeding strategy ensuring final triploidization and sterility. Highly polymorphic single sequence repeats (SSRs) are valuable markers for investigating phylogenetic relationships.
Methods
Here, the allelic distribution of each of 22 SSR loci across 561 Musa accessions is analysed.
Key Results and Conclusions
We determine the closest diploid progenitors of the triploid ‘Cavendish’ and ‘Gros Michel’ subgroups, valuable information for breeding programmes. Nevertheless, in establishing the likely monoclonal origin of the main edible triploid banana subgroups (i.e. ‘Cavendish’, ‘Plantain’ and ‘Mutika-Lujugira’), we postulated that the huge phenotypic diversity observed within these subgroups did not result from gamete recombination, but rather from epigenetic regulations. This emphasizes the need to investigate the regulatory mechanisms of genome expression on a unique model in the plant kingdom. We also propose experimental standards to compare additional and independent genotyping data for reference.
Keywords: ‘Cavendish’, lineage, Musa acuminata, Musa balbisiana, ‘Mutika-Lujugira’, ‘Plantain’, phylogeny, polyploidy, SSR, triploid
INTRODUCTION
Banana (Musa spp.) is the number one tropical fruit in production, exceeding 100 million metric tonnes worldwide in 2009, with the ‘Cavendish’ variety comprising over 50 % of this (Loeillet et al., 2011). Banana provides a staple food for more than 400 million people (Loeillet, 2008). The genus Musa is divided into four sections: Callimusa and Australimusa have a chromosome number of 2n = 20, while Eumusa and Rhodochlamys have a chromosome number of 2n = 22 (Bakry et al., 2009; Christelová et al., 2011).
Most edible cultivars derived from two species of the around 30 in sect. Eumusa, namely Musa acuminata and Musa balbisiana, contributing the A and B genome, respectively. The naturally occurring genotypes are classified in six groups (AA, AAA, AB, AAB, ABB and ABBB) on the basis of their ploidy level and on the basis of a taxonomic scoring method encompassing 15 morphological characters (Simmonds and Shepherd, 1955). The S genome from Musa schizocarpa has also been shown in some edible cultivars (Carreel et al., 1994). The T genome from species in sect. Australimusa might also be part of rare cultivated accessions.
Wild diploid accessions are seeded whereas edible bananas are seedless, parthenocarpic and vegetatively propagated. Most of these cultivars are triploid, even if some edible AAs are cultivated in Asia, the origin of Musa species. These triploid cultivars were clustered in subgroups such as ‘Plantain’ (AAB), ‘Cavendish’ (AAA) or ‘Mutika-Lujugira’ (AAA), based on typical morphological traits (IPGRI-INIBAP(Bioversity)/CIRAD, 1996).
Work on Musa has to deal with the complexity of the different ploidy levels of the genotypes, the mixture of intra- and inter-specific hybrids, the sterility which prevents allele shuffling and the vegetative multiplication which fixes some selected genetic events.
Diversity has been analysed over the past 70 years using agro-morphological traits, and these have been standardized in the Musa descriptors reference list (IPGRI-INIBAP(Bioversity)/CIRAD, 1996). Data on agro-morphological characterizations of accessions in Musa collections are published in the Musa Germplasm Information System (IPGRI-INIBAP(Bioversity), 2003). Based on these characteristics, around 1200 cultivars are currently distinguished. Wild diploids are also used for other purposes, such as a source of fibre and feed (Lescot, 2008).
More recently, studies based on polyphenols, isozymes, molecular markers on nuclear and cytoplasmic DNAs, AFLP (amplified fragment length polymorphism), RAPD (random amplification of polymorphic DNA), RFLP (restriction fragment length polymorphism), STMS (sequence tagged microsatellite sites), IRAP (inter-retrotransposon amplified polymorphism), DArT (diversity arrays technology), rRNA, SRAP (sequence-related amplified polymorphism) or retroelement markers and molecular cytogenetics (Gawel et al., 1992; Lanaud et al., 1992; Horry et al., 1997; Carreel et al., 2002; Ude et al., 2002; Wong et al., 2002; D'hont, 2005; Raboin et al., 2005; Heslop-Harrison and Schwarzacher, 2007; Risterucci et al., 2009; Hribova et al., 2011; Youssef et al., 2011) have sustained, and sometimes refined, the agro-morphological classification.
In the last decade, single sequence repeat (SSR) markers have been tested to analyse Musa diversity for their properties of genetic co-dominance, high reproducibility, high overall mutation rate and high polymorphism (Weber and Wong, 1993; Ellegren, 2002; Vigouroux et al., 2002). Despite the economic importance of Musa, the development of these markers remains limited; until recently, fewer than 100 SSR markers were available (Kaemmer et al., 1997; Crouch et al., 1998; Lagoda et al., 1998; Buhariwalla et al., 2005). Only some of these have been used in diversity analysis, and the analyses have been conducted on a limited number of banana samples (Grapin et al., 1998; Creste et al., 2003, 2004; Ning et al., 2007). The present study represents the first attempt to obtain an overview of Musa diversity, with more than 500 genotyped accessions. The large amount of data were analysed for different purposes such as understanding the domestication process (Perrier et al., 2009, 2011). Beside these synthetic analyses, which aggregate SSR marker results in a single overall similarity between accessions, here we investigated the resolving power of each marker at the species, subspecies, subgroup and accession level. We analysed the information provided by 22 SSR markers at the interspecific level between the A and B genomes. We compared allelic patterns within and between the main triploid subgroups. Lineages between diploid and triploid accessions were also investigated.
MATERIAL AND METHODS
Plant material
In total, 561 Musa accessions were genotyped. The initial germplasm sample consisted of 547 accessions of cultivated and wild bananas classified in section Eumusa, which are currently conserved within three significant field collections. Of these accessions, 236 were obtained from CIRAD Neufchateau (Guadeloupe), 192 from IITA (Ibadan, Nigeria) and 119 from CARBAP (Cameroon) (Hippolyte et al., 2011). Each genotype was documented with the genome constitution and subgroup classification according to the current agro-morphological classification (IPGRI-INIBAP(Bioversity), 2003) as well as the ploidy level identified by flow cytometry (Dolezel et al., 1997). In the sample, no duplicated accession (ITC code) was found within a collection or between collections.
The germplasm sample included 186 M. acuminata, 12 M. balbisiana, 16 interspecific diploid accessions (AB, AS, AT), 287 triploid bananas (AAA, AAB, ABB, BBB, AAS, AAT) and 30 tetraploid accessions (AAAA, AAAB,AABB, ABBT) (Hippolyte et al., 2011). In the genus Musa, other wild species from section Eumusa (two Musa basjoo, one Musa schizocarpa), from section Callimusa (one Musa beccarii, one Musa coccinea), from section Rhodoclamys (one Musa laterita) and from section Australimusa (one Musa jackeyi) were also included.
Ten diploid and triploid accessions from the Comoros islands were added to the initial sampling (ID 550–558 and 563; Hippolyte et al., 2011), as well as three M. balbisiana originating from China (ID 560–562) and one Ensete superbum (ID 559). They were genotyped independently of the other experiments.
To check the reliability of the genotyping, four ‘Cavendish’ (germplasm ID: 179, 183, 306, 508), five ‘Mutika-Lujugira’ (germplasm ID: 109, 115, 116, 487, 508) and 12 ‘Plantain’ (germplasm ID: 91, 96, 113, 195, 312, 356, 359, 419, 428, 429, 430, 491) accessions were genotyped twice with 16 SSR markers (Ma3_90, mMaCir01, mMaCir03, mMaCir07, mMaCir08, mMaCir13, mMaCir152, mMaCir164, mMaCir195, mMaCir196, mMaCir214, mMaCir264, mMaCir27, mMaCir307, mMaCir39, mMaCir40). These experiments were conducted independently of the main analysis.
DNA isolation
Three grams of frozen leaves were ground in liquid nitrogen using a mortar and pestle. Leaf DNA was extracted using the modified Matab method (Risterucci et al., 2000). DNA was re-suspended in PCR-grade water after isopropanol evaporation. DNA of samples from the CIRAD and CARBAP were extracted at CIRAD, while DNA from IITA was extracted there.
SSR markers
Twenty-two SSR primer pairs (Table 1) were selected to analyse the accessions. Twelve SSRs were identified from M. acuminata ‘Gobusik’ (Lagoda et al., 1998), while the ten others were newly defined from M. balbisiana ‘Pisang Klutuk Wulung’ (Hippolyte et al., 2010). The 22 SSRs were distributed across ten of the 11 linkage groups (Hippolyte et al., 2010) (Table 1).
Table 1.
Characteristics of the 22 SSR markers used for genotyping
| Origin | SSR | EMBL1 | No. of indels2 | Min.–max. size (bp) | Al < 1 %4 | Main allele frequency5 | Total alleles | LG6 | Motif |
|---|---|---|---|---|---|---|---|---|---|
| AA Gobusik | Ma1-32 | (Crouch et al. 1998) | 3x; 1 bp | 208–251 | 7 | 0·53 | 20 | 4 | (GA)17AA(GA)8AA(GA)2 |
| Ma3-90 | (Crouch et al. 1998) | – | 123–157 | 4 | 0·54 | 18 | 3 | (CT)11 | |
| mMaCIR01 | X872623 | 1x; 16 bp | 219–295 | 5 | 0·39 | 22 | 2 | (GA)20 | |
| mMaCIR03 | X872633 | 3x; 1 bp | 91–119 | 4 | 0·71 | 14 | 1 | (GA)10 | |
| mMaCIR07 | X872583 | – | 127–165 | 3 | 0·58 | 18 | 1 | (GA)13 | |
| mMaCIR08 | X872643 | – | 233–279 | 3 | 0·85 | 12 | 1 | (TC)6N24(TC)7 | |
| mMaCIR13 | X907453 | – | 251–279 | 0 | 0·82 | 12 | 3 | (GA)16N76(GA)8 | |
| mMaCIR24 | Z859723 | – | 218–278 | 9 | 0·84 | 19 | 5 | (TC)7 | |
| mMaCIR27 | Z859623 | – | 212–240 | 4 | 067 | 12 | 5 | (GA)9 | |
| mMaCIR39 | Z859703 | – | 310–350 | 7 | 0·77 | 20 | 2 | (CA)5GATA(GA)5 | |
| mMaCIR40 | Z859773 | 1 bp | 149–187 | 8 | 0·62 | 17 | 8 | (GA)13 | |
| mMaCIR45 | Z859683 | 4x; 1 bp | 253–275 | 1 | 0·84 | 9 | 10 | (TA)4CA(CTCGA)4 | |
| BB Pisang Klutuk Wulung | MMaCIR150 | AM950440 | 1 bp | 238–251 | 0 | 0·84 | 05 | 6 | (CA)10 |
| MMaCIR152 | AM950442 | 1 bp | 139–175 | 1 | 0·41 | 12 | 4 | (CTT)18 | |
| MMaCIR164 | AM950454 | 1x; 52 bp + 1x 1 bp + 1x; 38 bp | 236–390 | 7 | 0·37 | 17 | 4 | (AC)14 | |
| mMaCIR195 | AM950461 | – | 239–295 | 11 | 0·55 | 21 | 5 | (GA)17 | |
| mMaCIR196 | AM950462 | 2x; 1 bp | 147–173 | 3 | 0·70 | 12 | 7 | (TA)4(TC)17(TC)3 | |
| mMaCIR214 | AM950480 | 1x; 1 bp | 96–116 | 2 | 0·69 | 6 | – | (AC)7 | |
| mMaCIR231 | AM950497 | 1x; 1 bp | 219–267 | 5 | 0·53 | 19 | – | (TC)10 | |
| mMaCIR260 | AM950515 | – | 175–211 | 6 | 0·69 | 10 | 9 | (TG)8 | |
| mMaCIR264 | AM950519 | 3x; 1 bp | 215–273 | 7 | 0·44 | 18 | 4 | (CT)17 | |
| mMaCIR307 | AM950533 | – | 141–153 | 0 | 0·82 | 6 | – | (CA)6 |
1 EMBL, registration number on EMBL database or publication reference.
2 Number of alleles deviating from stepwise model (number x); size of the observed indels (bp).
4 Rare alleles with a frequency lower than 1 %.
5 Highest frequency of an allele observed at this locus.
6 Linkage groups (Hippolyte et al., 2010).
For all SSR loci, the forward primer was designed with a 5′-end M13 extension (5′-CACGACGTTGTAAAACGAC-3′). This extension enabled the generation of fluorescent amplicons after fluorescent dye hybridization.
Ten nanograms of Musa DNA was PCR amplified in a 384-well Eppendorf mastercycler with 10 µL final volume of buffer [10 mm Tris-HCl (pH 8), 100 mm KCl, 0·05 % (w/v) gelatin and 2·0 mm MgCl2] containing 0·08 µm of the M13-labelled primer, 0·1 µm of the other primer, 160 µm dNTP, 1 U Taq DNA polymerase (Life Technologies, Foster City, CA, USA) and 0·06 µm M13 primer-fluorescent dye IRD700 or IRD800 (Eurofins MWG Operon, Ebersberg, Germany).
The amplification was performed on a 384-well plate under touchdown PCR conditions: after an initial denaturation at 94 °C for 60 s, touchdown cycles were performed at a rate of –1 °C per cycle. These initial cycles were followed by 35 cycles at 94 °C for 30 s (lowest Tm –1 °C) for 1 min and 72 °C for 2 min, and a final elongation stage at 72 °C for 5 min.
Gel standards
A classical ladder covering a range of 71–367 bp was added to each gel. We refined the calibration of the allele sizes using accessions within the sample. These accessions belonged to the three triploid subgroups largely represented in the study: AAA ‘Cavendish’, AAB ‘Plantain’ and AAA ‘Mutika-Lujugira’. We defined these accessions as CPM (‘Cavendish’, ‘Plantain’, ‘Mutika-Lujugira’) standards in the text. These numerous accessions were distributed on the different migration gels of the analysis. They were also added later for genotyping the additional accessions (ID 550–560, ID 562 and 563) (Hippolyte et al., 2011).
Data analysis
The gel pictures were analysed using AFLP-Quantar Pro software (Keygene, Wageningen, the Netherlands), which can record more than two alleles for one individual. Each genotype was evaluated according to the presence (1) and absence (0) of an allele. Two independent readings were performed for each individual.
We observed differences in band intensity, which disturbed the recording of the data, especially in the case of extremely weak bands compared with other band(s) at the same locus. We chose to allocate a presence score to any clearly detectable band. When the intensity of a band at a locus was too weak to assume its presence, the marker was scored as missing data. This decision reinforced the robustness of our data, but consequently increased the number of missing data.
Eleven per cent (1361/12 342) of the data were missing (no amplification, unreadable pattern and null alleles). Accessions displaying more than six missing loci (45) were removed from the analyses (diversity tree, lineage determination and comparison within and between triploid subgroups) and are indicated in the ‘accessions’ sheet, column missing data (Hippolyte et al., 2011). This brought the proportion of missing data to 8 % (907/11 352 loci).
In addition, accessions displaying doubtful passport data were also excluded from the analyses (Hippolyte et al., 2011).
All genotyping data from the 22 SSR are available on the Generation Challenge Program registry (Hippolyte et al., 2011, ‘data_list’ sheet). Allele sizes (bp) are provided for each locus and each accession. The independent genotyping data of some new accessions using CPM standards (‘Cavendish’, ‘Mutika-Lujugira’ and ‘Plantain’) are also presented on the GCP registry website (Sample ID B5 is Ensente superbum, B6–B8 are M. balbisiana accessions and samples B9–B18 are diploid and triploid Comorian samples).
Diversity tree construction
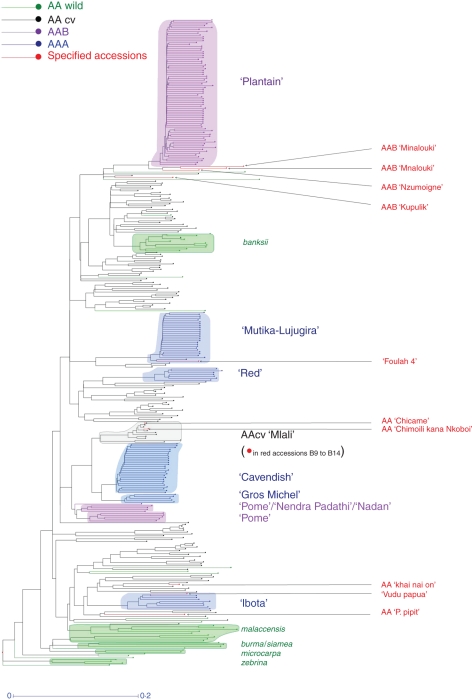
A diversity tree (see Fig. 2 below) was constructed using the Darwin software (Perrier and Jacquemoud-Collet, 2006). To deal with the mixture of several levels of ploidy, a specific measure of dissimilarity was defined as the probability of parentage between two accessions, regardless of ploidy level. Based on this dissimilarity, a first tree was build on the subset of AA diploids, using the neighbour-joining (NJ) algorithm (Saitou and Nei, 1987). A tree built on the AA diploids and our target triploids was constructed using a modified version of the NJ tree proposed by the Darwin software. These triploid accessions belong to the subgroups reported in this paper: AAA ‘Cavendish’, ‘Gros Michel’, ‘Mutika-Lujugira’, ‘Ibota’ and ‘Red’ subgroups and AAB ‘Plantain’, ‘Pome’, ‘Nendra Padathi’ and ‘Nadan’ subgroups. This modified version exhibits a solution in the NJ sense, but such that the a priori known topology of a subset is forced. In this case, the topology observed on the AA diploids was used as constraint in order to insert the triploid subgroups in the structure of their parental diploids.
Fig. 2.
NJ diversity tree constructed with accessions from AAA subgroups (‘Cavendish’, ‘Gros Michel’, ‘Ibota’, ‘Mutika-Lujugira’ and ‘Red’) and from AAB subgroups (‘Nadan’, ‘Nendra padathy’, ‘Pome’ and ‘Plantain’) under constraint of 172 diploid M. acuminata accessions. AA wild, M. acuminata wild-type accessions; AAcv, M. acuminata cultivars. Accessions in red in the diploid Mlaly group are from the Comoros islands and have been genotyped independently of the others. Accessions names on the right in red are accessions discussed in the text.
Parental lineage determination
In addition to indices based on allelic frequencies observed in the defined groups, a specific method was developed to detect direct affiliations between triploids and their diploid parents. For a target triploid tested against a pair of potential diploid parents, a marker was regarded as positive when two alleles were found in the first putative parent, regarded as the 2n gamete donor, and the third one was in the second parent. Each triploid was successively taken as a target and a kinship score was calculated for each pair of diploids as the proportion of positive markers. High scores indicated that the two diploids were potential parents or, more exactly, were closely related to these parents.
RESULTS
SSR characteristics
Twelve markers displayed dinucleotide motifs. Nine markers exhibited imperfect dinucleotide motifs and one an imperfect trinucleotide motif (Tables 1 and 2).
Table 2.
Allele sizes of each of the 22 SSR markers
| SSR | Alleles (bp) |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ma 1–32 | 227 | 229 | 232 | 233 | 235 | 237 | 239 | 242 | 245 | 247 | 249 | 251 | 252 | 254 | 258 | 260 | 262 | 264 | 266 | 270 | |||
| Ma 3–90 | 142 | 144 | 146 | 148 | 150 | 152 | 154 | 156 | 158 | 160 | 162 | 164 | 166 | 168 | 170 | 172 | 174 | 176 | |||||
| mMaCIR01 | 238 | 240 | 248 | 250 | 252 | 254 | 256 | 258 | 260 | 262 | 264 | 266 | 268 | 270 | 274 | 276 | 290 | 292 | 298 | 300 | 304 | 310 | 314 |
| mMaCir03 | 110 | 114 | 116 | 119 | 120 | 121 | 122 | 124 | 126 | 127 | 128 | 130 | 134 | 138 | |||||||||
| mMaCir07 | 146 | 152 | 154 | 156 | 154 | 156 | 154 | 158 | 160 | 162 | 164 | 166 | 168 | 170 | 172 | 174 | 176 | 178 | 180 | 184 | |||
| mMaCir08 | 255 | 257 | 259 | 261 | 263 | 265 | 267 | 269 | 271 | 275 | 279 | 285 | |||||||||||
| mMaCir13 | 274 | 276 | 278 | 282 | 284 | 286 | 290 | 292 | 294 | 296 | 298 | ||||||||||||
| mMaCir150 | 257 | 259 | 261 | 263 | 267 | ||||||||||||||||||
| mMaCIR152 | 158 | 161 | 164 | 165 | 167 | 170 | 173 | 176 | 179 | 182 | 185 | 191 | 194 | ||||||||||
| mMaCIR164 | 255 | 293 | 298 | 313 | 315 | 323 | 329 | 339 | 347 | 401 | 403 | 405 | 407 | 409 | 413 | ||||||||
| mMaCIR195 | 258 | 262 | 268 | 270 | 276 | 284 | 286 | 288 | 290 | 292 | 294 | 296 | 298 | 300 | 302 | 304 | 308 | 310 | 312 | 314 | |||
| mMaCIR196 | 166 | 168 | 170 | 172 | 174 | 176 | 177 | 179 | 180 | 182 | 184 | 192 | |||||||||||
| mMaCIR214 | 115 | 119 | 123 | 125 | 128 | 135 | |||||||||||||||||
| mMaCIR231 | 238 | 242 | 244 | 246 | 248 | 249 | 250 | 252 | 254 | 256 | 260 | 262 | 264 | 270 | 274 | 276 | 278 | 282 | 286 | ||||
| mMaCIR24 | 235 | 237 | 239 | 241 | 243 | 247 | 249 | 251 | 253 | 255 | 257 | 261 | 267 | 269 | 271 | 273 | 281 | 285 | 289 | 297 | |||
| mMaCIR260 | 194 | 200 | 204 | 206 | 208 | 210 | 212 | 214 | 216 | 218 | 220 | 222 | 226 | 230 | |||||||||
| mMaCIR264 | 234 | 236 | 238 | 240 | 242 | 244 | 246 | 248 | 250 | 251 | 252 | 254 | 256 | 257 | 258 | 260 | 262 | 263 | 264 | 266 | 268 | 270 | 274 |
| mMaCIR27 | 231 | 235 | 237 | 239 | 241 | 243 | 245 | 247 | 249 | 251 | 253 | 259 | |||||||||||
| mMaCIR307 | 160 | 162 | 164 | 166 | 168 | 172 | |||||||||||||||||
| mMaCIR39 | 329 | 331 | 333 | 335 | 337 | 339 | 341 | 343 | 345 | 347 | 349 | 351 | 353 | 355 | 357 | 359 | 361 | 363 | 365 | 369 | |||
| mMaCIR40 | 168 | 172 | 174 | 176 | 177 | 178 | 180 | 182 | 184 | 186 | 190 | 192 | 196 | 200 | 202 | 204 | 206 | ||||||
| mMaCIR45 | 272 | 274 | 277 | 279 | 281 | 283 | 284 | 289 | 294 | ||||||||||||||
Bold type indicates alleles differing from the expected stepwise model.
The number of alleles per marker ranged from five to 22, with a mean of 14. Rare alleles, those present on less than 1 % of the sample (Kimura, 1983), ranged from zero to 11 per marker. The frequency of the most frequent allele for each locus ranged from 0·37 to 0·85 (Table 1).
From the 22 SSR markers, 12 generated the expected pattern, with allele sizes following strictly stepwise the repeated motif (2 bp) (Table 2). Ten other SSR markers displayed both stepwise alleles and alleles with a shift of 1 bp from stepwise alleles, possibly an indel in the flanking or repeated regions. These differences of 1 bp were efficient and in several cases had a clear evolutionary interpretation. For example, the alleles mMaCir03 of 121 and 127 bp were specific to M. balbisiana (these alleles have also been recorded independently with the capillary systems genotyper, C. Billot, CIRAD, 2011, pers. comm.), while the alleles of 120, 122, 126 and 128 bp were found in accessions containing acuminata genomes. Among these ten markers, two also displayed larger indels (54 bp for mMaCIR164 and 16 bp for mMaCIR01).
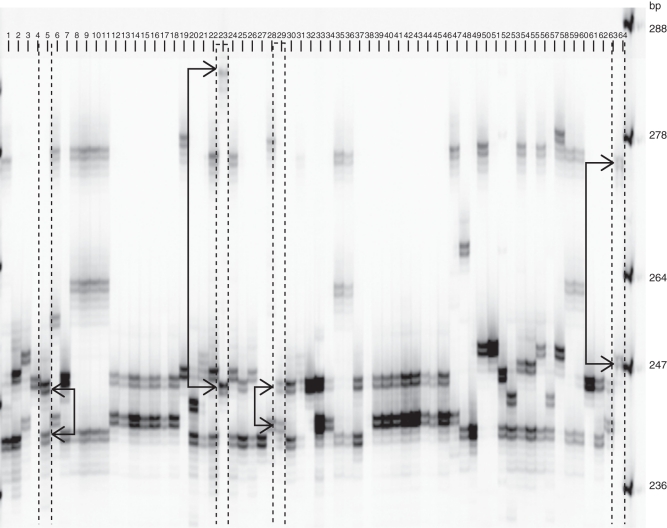
The migration profile of amplicons generated by mMaCIR231 on different accessions showed several unexpected features (Fig. 1). For example, the same band intensity expected for a heterozygous diploid was often not verified, as illustrated by the diploid accessions M. acuminata ‘Zebrina maia oa’ (Fig. 1, lane 23), for which the intensity of the 286-bp allele was weaker than for the 248-bp allele, and by the accession M. acuminata ‘Pisang madu’ (Fig. 1, lane 5), for which the intensity of the 242-bp allele was weaker than for the 248-bp allele. On the other hand, the triploids AAB ‘Laknau’ (Fig. 1, lane 29) and AAA ‘Red’ (Fig. 1, lane 64) displayed two bands of the same intensity, preventing us from determining which of the alleles is double dose or simple dose. Because the intensity of an allele cannot be read in number of doses, the allelic distribution within the triploids displaying two bands remained partly undetermined. This inability to estimate the number of doses of each allele at a locus also hampered estimation of null alleles within the dataset.
Fig. 1.
Migration profile of amplicons generated by mMaCIR231 on different accessions. Lane 1–64 are samples: 1, T1; 2, S. F. (265) (AA); 3, Uwati (AA); 4, Maduranga (ABB); 6, Ouro Mel (AAA); 7, Blue Torres Strait (ABB); 19, No 110 (AA); 20, P. Gigi Buaya (AA); 21, Klue Roi Wee (AAB); 25, Kinkala (ABB); 26, Pitu (AA); 27, P. Tongat (AA); 28, Guyod (11–33) (AA); 30, Ney Poovan (AB); 31, Oura da Mata (AAAB); 32, Singapuri (BB); 37, Kamaramasenge (AB); 47, Gwanhour (AA); 48, Selangor (AA); 49, Hy (302) (AA); 50, Lai THA002 (AAA); 51, Malaccensis Holotype (AA); 52, Sabra (ABB); 53, Morong Princesa (AA); 55, Galeo (AA); 56, Highgate (AAA); 57, Saing Todloh (AA); 58, P. Mulik (AA); 61, Los Banos (BB); 62, T1. ‘Cavendish’ accessions (AAA) (C standard): 22, Robusta 133; 24, P. Masak Hijau; 54, Valery. ‘Mutika-Lujira’ accessions (AAA) (M standard): 8, Indemera y' Ymbihire; 9, Kibungo; 10, Makara; 11, Kitawira; 35, Imbogo; 36, Igisahira Gisanzwe; 38, Igihuni; 59, Igitsiri; 60, Bakurura. ‘Plantain’ accessions (AAB) (P standard): 12, Dominico Rojo (641); 13, Nazika; 14, Motouka 1; 15, Apem Onniaba; 16, Zue Ekon; 17, Harton Maqueno (628); 18, Purple Plantain; 33, Banane Serpent; 34, Currare; 39, O. Ntanga G. M; 40, Mbi Egome; 41, Gabon 4; 42, Diby 2 off-type; 43, Atali Kiogo; 44, Red Plantain Hembra; 45, Moto Ebanga; 46, Msisa; 63, Agbagba. Bold numbers and dotted lines highlight profiles exhibiting unexpected band intensities; alleles are indicated with bold arrows: 5, P. madu (AA); 23, Maia Oa (AA); 29, Laknau (AV-66) (AAB); 64, Red (AAA); L, ladder 98–364 bp.
Gel standards
Because the classical standard ladder used in the experiments, covering broadly 100–400 bp of the genotyped SSRs, generated very large gaps between successive amplicons and hampered precise reading, we calibrated the allele sizes by directly using accessions from the sample itself. Three triploid subgroups, AAA ‘Cavendish’, AAB ‘Plantain’ and AAA ‘Mutika-Lujugira’, were over-represented in the study. Within these triploid subgroups the allelic polymorphism between the accessions was very low, with a main pattern present on more than 99 % of the accessions at every locus (see below). The accessions belonging to these subgroups, spread among the different gels, could thus be used for allele size calibration. The CPM standards covered a broad part of the Musa diversity because the CPM subgroups originate from different M. acuminata subspecies and include the M. balbisiana genome. Using these references, a scoring precision of 1 bp appeared to be of phylogenetic significance (see above).
These CPM accessions are publicly available upon request from the International Transit Center of Bioversity International hosted by the Katholieke Universiteit (Leuven, Belgium).
Analysis of main edible triploid subgroups
Investigation of the common alleles at each locus between the AA diploids and the accessions from the triploid ‘Cavendish’, ‘Gros Michel’, ‘Mutika Lujugira’, ‘Red’, ‘Ibota’, ‘Pome’ and ‘Plantain’ subgroups provided contrasting results.
The results of putative lineages of ‘Cavendish’, ‘Gros Michel’ and ‘Pome’ subgroups are shown in Table 3. For the ‘Mutika Lujugira’, ‘Red’, ‘Ibota’ and ‘Plantain’ subgroups, such likely ancestors were not found.
Table 3.
Putative diploid parents (2n and n gamete donors) of ‘Cavendish’, ‘Gros Michel’ and ‘Pome’ triploid subgroups
| Ma1_32 | Ma3_90 | mMa CIR01 | mMa CIR03 | mMa CIR07 | mMa CIR08 | mMa CIR13 | mMa Cir150 | mMa CIR152 | mMa CIR164 | mMa CIR195 | mMa CIR196 | mMa CIR214 | mMa CIR231 | mMa CIR24 | mMa CIR260 | mMa CIR264 | mMa CIR27 | mMa CIR307 | mMa CIR39 | mMa CIR40 | mMa CIR45 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ‘Cavendish’ AAA | 235 | 150 | 254 | 122 | 158 | 261 | 286 | 257 | 164 | 401 | 298 | 168 | 119 | 242 | 237 | 212 | 250 | 235 | 162 | 331 | 176 | 284 |
| 245 | 162 | 258 | 124 | 170 | 265 | 261 | 407 | 180 | 123 | 250 | 247 | 258 | 243 | 164 | 335 | 178 | 289 | |||||
| 168 | 264 | 276 | 253 | 245 | 180 | |||||||||||||||||
| ‘Chimoili Kana Nkoboï’ AA 2n donor | 245 | 150 | 254 | 122 | 158 | 261 | 286 | 261 | 164 | 401 | 298 | 180 | 119 | 242 | 247 | 210 | 258 | 243 | 162 | 335 | 178 | 284 |
| 168 | 258 | 124 | 170 | 265 | 257 | 123 | 276 | 253 | 212 | 245 | 164 | 180 | 289 | |||||||||
| P. pipit AA n donor | 235 | x | 250 | 120 | 170 | 261 | 286 | 257 | 167 | 407 | 258 | 168 | 119 | 242 | x | 208 | 250 | 235 | 160 | 341 | 176 | 284 |
| 254 | 264 | 124 | 172 | 271 | 257 | 164 | 128 | 270 | 212 | 264 | 243 | 162 | 349 | 178 | 289 | |||||||
| ‘Gros Michel’ AAA | 235 | 150 | 254 | 122 | 158 | 261 | 286 | 257 | 164 | 401 | 258 | 176 | 119 | 242 | 247 | 212 | 258 | 243 | 162 | 329 | 176 | 284 |
| 168 | 258 | 124 | 170 | 265 | 261 | 298 | 180 | 123 | 252 | 253 | 266 | 164 | 335 | 178 | 289 | |||||||
| 270 | 276 | 180 | ||||||||||||||||||||
| ‘Chimoili Kana Nkoboï’ AA 2n donor | 245 | 150 | 254 | 122 | 158 | 261 | 286 | 257 | 164 | 401 | 298 | 180 | 119 | 242 | 247 | 210 | 258 | 243 | 162 | 335 | 178 | 284 |
| 168 | 258 | 124 | 170 | 265 | 261 | 123 | 276 | 253 | 212 | 245 | 164 | 180 | 289 | |||||||||
| ‘Khai nai on’ AA n donor | 235 | 162 | 250 | 120 | 170 | 261 | 286 | 257 | 161 | 403 | 258 | 168 | 115 | 250 | x | 210 | 252 | 235 | 162 | x | 176 | 284 |
| 254 | 168 | 270 | 124 | 269 | 165 | 176 | 119 | 266 | 241 | 178 | 289 | |||||||||||
| ‘Pome’ AAB | 239 | 150 | 254 | 122 | 158 | 261 | 286 | 257 | 164 | 298 | 298 | 177 | 119 | 242 | 247 | 210 | 242 | 245 | 164 | 335 | 178 | 274 |
| 245 | 168 | 258 | 124 | 166 | 265 | 261 | 182 | 401 | 123 | 252 | 253 | 258 | 243 | 168 | 357 | 180 | 284 | |||||
| 290 | 170 | 125 | 276 | 289 | ||||||||||||||||||
| ‘Samba Nkundre’/Chicame' AA 2n donor | 245 | 150 | 254 | 122 | 158 | 261 | 286 | 257 | 164 | 401 | 298 | 180 | 119 | 242 | 247 | 210 | 258 | 243 | 164 | 335 | 178 | 284 |
| 168 | 258 | 124 | 170 | 265 | 261 | 123 | 276 | 253 | 245 | 180 | 289 | |||||||||||
| ‘Lal velchi’ BB n donor | 239 | 152 | 290 | 121 | 166 | 257 | 282 | 257 | 173 | 298 | 296 | 177 | 123 | 249 | 241 | 212 | 242 | 235 | 168 | 357 | 180 | 274 |
| 310 | 261 | 290 | 261 | 182 | 313 | 182 | 243 |
For each locus, allele sizes are given in base pairs. The allele pattern given for each triploid subgroup is the most common pattern observed within these subgroups at the considered locus.
X, missing data; italic indicates allelic discrepancy between the major pattern observed within the subgroup and the putative ancestor pattern; bold indicates allelic identity between the major pattern within the subgroup and the putative ancestor pattern.
The M. balbisiana-specific alleles and their presence in ‘B’-containing accessions are shown in Table 4. From a total of 329 alleles defined (Table 2), four alleles recorded on B-containing genomes were absent from all AA and AAA genotypes (mMaCIR01, 298 bp; mMaCIR01, 310 bp; mMaCIR03, 121 bp; mMaCIR03, 127 bp). Six other alleles were highly infrequent (<2 %) in these AA or AAA accessions (Table 4).
Table 4.
Number of ‘B-specific alleles’ encountered in accessions of the sampling in each group of the Simmonds and Shepherd's classification (1955) (AA, AAA, AAB, AAB ‘Plantain's, AB, ABB, BB).
| Locus | ‘B allele’ (bp) | AA, 186 | AAA, 100 | AAB ‘Plantain’, 78 | AAB others, 63 | AB, 9 | ABB, 7 | BB, 12 |
|---|---|---|---|---|---|---|---|---|
| mMaCIR01 | 298 | – | – | 56 | 20 | 2 | 12 | 1 |
| mMaCIR03 | 127 | – | – | 76 | 34 | 2 | 20 | 7 |
| mMaCIR01 | 310 | – | – | – | – | – | 21 | 8 |
| mMaCIR03 | 121 | – | – | – | 10 | 5 | 24 | 10 |
| Ma1_32 | 239 | – | 3 | – | 21 | 7 | 30 | 10 |
| mMaCIR39 | 355 | 1 | – | – | 8 | 1 | 6 | 6 |
| mMaCIR214 | 125 | 2 | 1 | 73 | 30 | 7 | 25 | 9 |
| mMaCIR260 | 225 | 3 | – | – | 11 | 2 | 10 | 2 |
| mMaCIR164 | 298 | 3 | – | – | 20 | 5 | 17 | 3 |
| mMaCIR39 | 357 | 4 | – | 76 | 43 | 6 | 24 | 7 |
| mMaCIR264 | 242 | 4 | 2 | 75 | 51 | 7 | 36 | 12 |
For the triploid subgroups, we identified loci with triallelic combinations, which were specific to a triploid subgroup (Table 5).
Table 5.
SSR markers displaying specific allelic combinations for accessions belonging to the same triploid subgroup
| Subgroup | SSR marker | Allele 1 | Allele 2 | Allele 3 | Out of subgroup* |
|---|---|---|---|---|---|
| ‘Plantain’, AAB | mMaCIR01 | 258 | 266 | 298 | Nzumoigne, Kupulik indet |
| mMaCIR264 | 242 | 251 | 258 | M009 | |
| ‘Mutika-Lujugira’, AAA | mMaCIR242 | 231 | 260 | 276 | Foulah 4 |
| mMaCIR307 | 160 | 164 | 172 | Foulah 4 | |
| Ma 3_90 | 156 | 166 | 170 | Foulah 4 | |
| ‘Cavendish’, AAA | mMaCIR01 | 254 | 258 | 264 | |
| mMaCIR231 | 242 | 250 | 276 | Pisang bakar, Hom Thong Mokho (Ambon) | |
| mMaCIR 24 | 237 | 247 | 253 | ||
| ‘Gros Michel’, AAA | mMaCIR01 | 254 | 258 | 270 | |
| ‘Pome’, AAB | mMaCIR01 | 254 | 258 | 290 | Lady finger |
| mMaCIR45 | 274 | 284 | 289 | Lady finger Rajapuri india | |
| ‘Red’, AAA | mMaCIR264 | 250 | 264 | 266 | Mata kun |
| mMaCIR 39 | 329 | 331 | 339 | ||
| mMaCIR40 | 176 | 180 | 196 | Leite (Rio) | |
| ‘Ibota’, AAA | mMaCIR03 | 110 | 120 | 124 | Vudu papua |
| mMaCIR07 | 152 | 162 | 170 | Vudu papua beccarii | |
| mMaCIR27 | 235 | 241 | 245 | Vudu papua |
* Accessions displaying the same SSR triallelic allelic pattern of the subgroup, but not belonging to the subgroup.
Polymorphism within triploid subgroups
The analysis was restricted to the triploid subgroups represented by more than 20 exploitable accessions: AAA ‘Cavendish’ (27), AAA ‘Mutika-Lujugira’ (25) and AAB ‘Plantain’ (78) (Hippolyte et al., 2011). From the 22 SSRs analysed, the number of loci with an identical allelic pattern for all the accessions ranged from 14 in ‘Plantain’ to 20 in ‘Cavendish’ and ‘Mutika-Lujugira’ (Table 6). The variations around this main pattern were spread between different accessions and were not concentrated in just a few accessions. From more than 2800 allelic profiles within the three subgroups, we recorded 19 deviations from the main pattern, which could be divided into two classes: loss of main alleles (11) or emergence of new alleles in addition or as substitution to the main alleles (eight).
Table 6.
Accessions from the ‘Plantain’, ‘Cavendish’ and ‘Mutika-Lujugira’ subgroups deviating from common patterns of their subgroup at 22 SSR loci
| ‘Plantain’ (78) |
‘Cavendish’ (27) |
Mutika/Lujugira (25) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Main profile | Other profiles |
Main profile | Other profiles |
Main profile | Other profiles |
||||
| bp | bp | accessions | bp | bp | accessions | bp | bp | accessions | |
| Ma 1_32 | 245–251 | 235–245 | 260 | ||||||
| Ma3_90 | 152–162–166 | 152–166 | Bobby Tanap | 150–162–168 | 156–166–170 | ||||
| mMaCir01 | 258–266–298 | 254–258–264 | 256–266 | ||||||
| mMaCIR03 | 122–127 | 122 | Okoyo Ukom | 122–124 | 122 | Chinese Cavendish | 120–122 | ||
| mMaCIR07 | 158–160–172 | 158–160–174 | Atali Kiogo | 158–170 | 152–154–160 | ||||
| mMaCIR08 | 261 | 261–265 | 261–263–267 | 261–267 | Ingumba y' inyamunyo-Ingoromoka-Indemera y' imbihire | ||||
| mMaCIR13 | 286 | 274–286–298 274–286 286–298 | Haton Tigre mbouroukou kelong mekintu | 286 | 286–298 | ||||
| mMaCIR24 | 247 | 237–247–253 | 241–247 | ||||||
| mMaCIR27 | 235–243 | 235–243–245 | 235–241 | 901 | 235–243 | ||||
| mMaCIR39 | 335–357 | 335 | Bungaoisan | 331–335 | 335–345 | ||||
| mMaCIR40 | 182 | 176–178–180 | 180 | ||||||
| mMaCIR45 | 284–289 | 284–289 | 284–289 | ||||||
| mMaCIR150 | 257 | 257–261 | 257–263 | ||||||
| mMaCIR152 | 194 | 185 182–194 | Mbi Egome 1 Okyo ukom | 164 | 164 | ||||
| mMaCIR164 | 313–407 | 313 | Nselouka Obino L'ewai Njock Kon | 401–407 | 409 | ||||
| mMaCIR195 | 290–294 | 298 | 298 | ||||||
| mMaCIR196 | 177–180 | 168–180 | 180–184 | ||||||
| mMaCIR214 | 125–128 | 119–123–125 | Ndingo Liko | 119–123 | 123 | ||||
| mMaCIR231 | 244–249 | 242–250–276 | 242–260–276 | ||||||
| mMaCIR260 | 210 | 212 | 210 | 206–210 | Bolo Bigouyo | ||||
| mMaCIR264 | 242–251–58 | 250–258 | 256–266 | ||||||
| mMaCIR307 | 164–168 | 162–164 | 160–164–172 | ||||||
DISCUSSION
Diversity in the genus Musa has previously been analysed based on levels of dissimilarity calculated between accessions in these 22 SSR loci (Perrier et al., 2009). The resulting phylogenetic structure of Musa species was congruent with previous results using molecular markers (Grapin et al., 1998; Creste et al., 2004; Risterucci et al., 2009). Furthermore, the large degree of polymorphism of these markers enabled us to refine the typology, particularly for genetically close subgroups, and to elucidate the relationships between wild diploids and cultivated diploids, and also between edible diploids and the main triploid subgroups (Perrier et al., 2009). In this paper, we investigated the resolving power of each marker at the species, subspecies and subgroup and accession levels in order to refine the lineage.
SSR characteristics
The observed allelic ranges were derived from a typical SSR stepwise mutation model, with some additional events (often indel), as observed in other species (Colson and Goldstein, 1999). Variations in band intensities have been observed previously for Musa (Creste et al., 2004) (Fig. 1). They probably resulted from preferential amplification of shorter alleles (Wattier et al., 1998) as seen in the diploid M. acuminata ‘Zebrina maia oa’, i which the intensity of the 248-bp allele was higher than that of the 286-bp allele (Fig. 1). These intensity variations could also result from mutations in annealing sequences (Ishibashi et al., 1996; Colson and Goldstein, 1999) leading to a null allele (Dakin and Avise, 2004; Chapuis and Estoup, 2007) or competitive amplification, if annealing still occurs.
Paying particular attention to these scoring difficulties, we showed that the use of the CPM standards allowed the recording of reliable independent data. For example, when the additional Comorian accessions were added to the initial diversity tree, they co-localized with the ‘Mlali’ diploid subgroup originating from these islands, and the triploid ‘Minalouki’ also co-localized with the ‘Mnalouki’ accessions of the initial sampling (Fig. 2).
Therefore, using the protocol defined here and the publicly available genotyping data of this analysis (Hippolyte et al., 2011), there is an opportunity to compare local collections with this broad Musa sample as reference.
Lineage between triploid subgroups and AA diploids
The analysis confirmed that the ‘Mlali’ subgroup was the closest 2n gamete donor for the ‘Cavendish’ and ‘Gros Michel’ subgroups, as previously proposed (Raboin et al., 2005). We identified M. acuminata ‘Chimoili Kana Nkoboï’, a Comorian diploid, as the best 2n gamete donor to ‘Cavendish’ and ‘Gros Michel’ accessions (Table 3). This accession was a better candidate than M. acuminata ‘Akondro mainty’, originating from Madagascar, which has been suggested previously (Raboin et al., 2005), although the accessions within the ‘Mlali’ subgroup are roughly genetically homogeneous. We also defined M. acuminata ‘Pisang pipit’ as the putative n gamete donor for the ‘Cavendish’ accessions and M. acuminata ‘Khai nai on’ as n gamete donor for the ‘Gros Michel’ subgroup (Table 3). This analysis also demonstrated that the ‘Mlali’ subgroup is probably the M. acuminata 2n gamete donor of the AAB ‘Pome’ subgroup, M. acuminata ‘Samba kundre’ or M. acuminata ‘Chicame’ being the best candidates (Table 3). These relationships between triploid subgroups and the ‘Mlali’ accessions are also clear from the diversity tree (Fig. 2)
For the ‘Mutika-Lujugira’, ‘Red’, ‘Ibota’ and ‘Plantain’ subgroups, we did not find any convincing lineage with the diploid M. acuminata accessions sampled. Nevertheless, analysis based on allele frequencies clearly showed that the ‘Plantain’ subgroup has a dominant banksii subspecies origin, while the ‘Mutika-Lujugira’ accessions have a binary banksii-zebrina subspecies origin. While the ‘Ibota’ subgroup displays a main malaccensis subspecies origin, the putative origin of the ‘Red’ subgroup is much less clear (results not shown). This information has been useful for refining the geographical origin (Perrier et al., 2009, 2011), but is not detailed enough to assist with breeding strategy.
Specificity of balbisiana alleles
Four alleles from two loci were fully discriminating between A- and B-genome-containing accessions (Table 4). For those displaying highly imbalanced occurrence between A and B accessions (Table 4), the introgression of B alleles into the A genome is very unlikely and could only be cautiously hypothesized for ‘Calcutta 4’ (mMaCIR39_355), a wild diploid from M. acuminata subspecies burmanicoides, which is sympatric with M. balbisiana in Burma and north Thailand. For all other cases, the most likely explanation is that alleles of the same length (bp) arose from different alleles with convergent evolution (i.e. homoplasy), which is known to be frequent in microsatellite evolution (Estoup et al., 2002).
The distribution of these B-specific alleles was not homogeneous between wild BB accessions and interspecific cultivars (AB, AAB, ABB). For example, the 298-bp allele from mMaCIR01 was only found in M. balbisiana ‘ITC 0626’ while it was widely recorded within interspecific cultivars, most of which were of ‘Plantain’ genotypes (Table 4). Nevertheless, only 50 % of the loci of M. balbisiana ‘ITC 0626’ displayed alleles also recorded in AAB ‘Plantain’, excluding it as the potential M. balbisiana parent of this subgroup. Conversely, the 239-bp allele from MA1–32 was present on nearly all BBs (Table 4) and most AB and ABB cultivars, but it was absent from ‘Plantain’ and ‘Iholena’ AAB subgroups. For this locus, none of the alleles of ‘Plantain’ or ‘Iholena’ was found in any BB accession. We only found this for the AAB ‘Pome’ subgroup, a potential close donor of the balbisiana genome, which is M. balbisiana ‘Lal velchi’ (Table 3).
The balbisiana diversity provided by the interspecific cultivars seemed larger than the BB diploid diversity present in our BB sample, given that several alleles from the cultivars were not present in diploid M. balbisiana sampled at numerous loci, suggesting an under-representation of the whole balbisiana diversity in collections or extinction of the BB parents of the current hybrids. In fact, available BB genotypes are sparse and their origin is poorly documented (IPGRI-INIBAP(Bioversity), 2003). This gap could be related to the level of diversity in BB accessions, which is lower than in AA accessions (De Langhe and De Maret, 1999; Swangpol et al., 2007), with no subspecies clustering (Sotto and Rabara, 2000).
Nevertheless, M. balbisiana originated from a broad area ranging from India (Simmonds, 1962; Uma et al., 2006) to the south of China (Wang et al., 2007) and possibly the Philippines (Sotto and Rabara, 2000). Several studies have demonstrated the existence of a local diversity of these M. balbisiana (Uma et al., 2006; Ning et al., 2007; Swangpol et al., 2007; Wang et al., 2007). Unfortunately, differences in analytical methods, as well as the lack of accessions in common and the use of vernacular names prevented cross-analysis.
Analysis of a broad sample, enriched by additional collecting efforts, would be of significant importance to better characterize M. balbisiana genomes, particularly for lineage studies of the interspecific cultivars and consequently for breeding programmes. This would also provide information on the possible extinction of BB ancestors of the current hybrids.
From the present set of 22 SSR markers, several highlighted the divergence between acuminata and balbisiana species, which would have occurred between 4 Mya (Lescot et al., 2008) and 28 Mya (Christelová et al., 2011). The absence of discriminating alleles between M. acuminata subspecies meant that no specific allele has been generated since their divergence, which is assumed to have begun with the maximal geographical isolation of the south-east Asian islands, during the last interglacial period. SSR markers trace recent evolution events as their mutation rate is quite high, roughly between 10−3 and 10−6, depending on species and location on the genome (Vigouroux et al., 2002). Despite the absence of specific alleles discriminating between M. acuminata diploids, allele frequencies distinguished AA wild subspecies (banksii, zebrina, malaccensis and burmanica) (Perrier et al., 2009) and enabled the determination of the wild origin of AA cultivars.
Discrimination between triploid subgroups
The triploid accessions clustered in subgroups based on agromorphological characters (IPGRI-INIBAP(Bioversity)/CIRAD, 1996; Pollefeys et al., 2004), despite broad phenotypic diversity within these subgroups, as illustrated for ‘Plantain’ (Ortiz et al., 1998; Lescot et al., 2008) and ‘Cavendish’ (Simmonds, 1954). The genetic status of ‘Plantain’ (i.e. intra-subgroup homogeneity versus intra-subgroup heterogeneity) has been investigated using molecular markers. There was no clear outcome from these studies, as two studies using AFLP, SSR, MSAP or DArT markers (Noyer et al., 2005; Risterucci et al., 2009) found genetic homogeneity within this subgroup, whereas two other studies based on RAPD and AFLP markers (Crouch et al., 2000; Ude et al., 2003) found ‘Plantain’ subgroup genetic heterogeneity.
Our results suggested predominantly genetic homogeneity within six triploid subgroups (Table 5). The specific triallelic combinations, at fully heterozygous loci, were accurate both for the discrimination of a subgroup from the other subgroups and for the allocation (or rejection) of accessions to (from) a subgroup (Table 5). As an example of subgroup discrimination, the locus mMaCIR01 generated specific triallelic combinations for each of four subgroups: ‘Plantain’ (AAB), ‘Pome’ (AAB), ‘Cavendish’ (AAA) and ‘Gros Michel’ (AAA), although the last two are genetically very close (Table 3; Raboin et al., 2005). Regarding the triallelic combinations, the relationships between AAB Indian dessert bananas, classified within ‘Pome’, ‘Nadan’ or ‘Nendra padathi’, need to be clarified. Although they shared common triallelic combinations, Fig. 2 suggests clustering into two subgroups. These examples indicate that the triallelic combinations should conveniently help in defining or refining subgroup clustering, the NJ tree providing a graphical tool for investigating putative clustering.
Concerning allocation to a subgroup, the ‘Nzumoigne’ accession, classified as a ‘Plantain’ based on morphological characteristics, differs from the mMaCIR01 triallelic pattern of ‘Plantain’ and did not cluster with this subgroup (Fig. 2). In fact, it has only been found in the Comoros Islands other Indian Ocean islands, and probably has a different history (domestication period, human migration, etc.) than African ‘Plantain’. Most of the morphological traits of the ‘Kupulik’ accession fit the characteristics of the AAB ‘Plantain’ subgroup, but some others, such as rounded fruit apex, prevented its classification to this subgroup (C. Jenny, CIRAD, 2009, pers. comm.). This accession displayed some of the triallelic combinations characterizing ‘Plantain’, but the discrepancies at some loci prevented its genetic classification in the ‘Plantain’ subgroup (Fig. 2). The accession ‘Foulah 4’, classified as ABB, shared all specific allelic combinations of the AAA ‘Mutika-Lujugira’ subgroup. This probably resulted from a mislabelling, and according based on the diversity tree (Fig. 2) and its allelic pattern (Hippolyte et al., 2011, data sheet), differing only by two missing alleles, the sample analysed should be classified into the ‘Mutika-Lujugira’ subgroup of East Africa. Similarly, the AA accession ‘Vudu Papua, ITC0590’ should be included in the AAA ‘Ibota’ subgroup based on triallelic combinations and its location in Fig. 2. Checking all loci, ‘Vudu papua’ most closely matches ‘Ibota’ subgroup profiles and differs for one allele only. Therefore, the ‘Nzumoigne’, ‘Kupulik’, ‘Foulah’ and ‘Vudu papua’ cases reveal that the stringency of these triallelic combinations allows refinement of subgroup classification and that these allelic combinations might be used as easy keys for assigning accessions to subgroups.
Polymorphism within triploid subgroups
The genetic homogeneity within subgroups showed some exceptions. From more than 2800 allelic profiles obtained in this study with accessions belonging to the ‘Cavendish’, ‘Mutika-Lujugira’ and ‘Plantain’ subgroups, 19 deviations from the main allelic pattern were established: 11 were due to missing alleles (null alleles), and eight arose from the occurrence of an extra allele (Table 6). The occurrence and transmission of mutations in the flanking region were as frequent as the mutations occurring in repeated regions in the triploid samples.
Independently, by genotyping 21 accessions of the initial sample with 16 SSR markers, confirmed differences in band intensities at some loci, corroborating the previous hypothesis of allelic preferential amplification. This duplicated genotyping also confirmed deviations from the main profiles of these loci, such as the presence of the 274-bp extra allele of mMacir13 or the absence of the 407-bp allele from mMaCIR164 on some ‘Plantain’ accessions (Table 6).
According to the process of banana triploidization, resulting from the association of a non-reduced 2n gamete (gamete with sporophytic chromosome number) and an n gamete (Ortiz, 1997), two main hypotheses might explain this reduced intra-subgroup diversity: (1) all accessions in a subgroup were derived from the same initial clone and evolved by somatic mutations fixed through vegetative propagation; and (2) the genotypes of these subgroups arose from sexual events from the same parents or from genetically related parents. Most authors have suggested a mix of these two hypotheses, with a diversification of ‘Plantain’ by somatic mutations from a few introduced cultivars (Simmonds, 1966; De Langhe et al., 1994–1995; Crouch et al., 2000; Ude et al., 2003).
The widely different frequencies between main and extra alleles or null alleles within the ‘Cavendish’, ‘Mutika-Lujugira’ and ‘Plantain’ subgroups (Table 6) favour the first hypothesis. Based on RFLP markers (Raboin et al., 2005) and SSR markers (this study), the two AAA subgroups ‘Cavendish’ and ‘Gros Michel’ were found to derive from a common 2n gamete donor and probably two different, but genetically close, n donors. For these two much closer subgroups, nine SSR loci were different for at least one allele, while we never found more than one deviating loci per accession within the analysed triploid subgroups. It is therefore likely that each triploid subgroup arose from a unique clone, as hypothesized by Noyer et al. (2005), and that sparse somatic mutations have been ‘inherited’ (transmitted through clonal propagation), leading to new SSR alleles (or null allele from mutations in annealing sequences). The transmission of somatic mutations is possible through vegetative propagation, leading to non-chimeric or to chimeric plant structure (Marcotrigiano, 1997; Klekowski, 2003) and a mosaic state (i.e. initial cells associated with mutated cells) (Gill et al., 1995; Santelices, 1999; Pineda-Krch and Lehtilä, 2004).
Nevertheless, the low number and in most cases the absence of genetic differences between the accessions of a subgroup cannot explain the huge phenotypic diversity observed within these subgroups (Ortiz et al., 1998; Daniells et al., 2001; IPGRI-INIBAP(Bioversity), 2003). Therefore other possibilities, such as epigenetic regulation, need to be explored.
Human migration brought the ‘Cavendish’, ‘Mutika-Lujugira’ and ‘Plantain’ subgroups from Asian centres of origin to Africa in the case of ‘Plantain’ and ‘Mutika-Lujugira’ (De Langhe et al., 1994–1995), and more recently worldwide in the case of ‘Cavendish’. Several genetically close accessions were found in the centres of origin, but they could not be allocated to the three subgroups. They probably resulted from crosses of close diploid parents or the same parents (full-sibling). This is illustrated for the ‘Gros Michel’ and ‘Cavendish’ subgroups (half-sibling) or for a single accession such as ‘Kupulik’ or ‘Nzumoigne’ compared with the ‘Plantain’ subgroup. Human migrations introduced a drastic bottleneck in the diffusion of these triploids, with only some suckers of the same clonal origin being exported and then spread. This human influence therefore shaped the triploid diversity landscape, favouring an over-representation of sparse genotypes, which evolved phenotypically.
CONCLUSIONS
Using a broad sample, this study contributed to improving our understanding of Musa species diversity. The accuracy of the results depended greatly on experimental control, which limited the impact of preferential amplification, and thus misinterpretation. Moreover, the CPM standards enabled accurate scoring. Studying additional accessions, with these experimental procedures (including CPM standards), allowed us to add these independent data to previous diversity analysis.
By using the co-dominance of SSR markers for parentage analysis, we showed that the high polymorphism of the SSR markers was able to identify specific loci efficient for discrimination and assignment. At the interspecific level, some alleles discriminated genotypes containing the B genome from strictly M. acuminata genotypes. These specific alleles showed also that the balbisiana species diversity displayed through interspecific cultivars was broader than that of the available M. balbisiana diploids. Therefore, new exploration for and collection of balbisiana species is recommended, especially if M. balbisiana provides agronomic traits to important AAB cultivars, such as ‘Plantain’. We did not find any specific allele of M. acuminata subspecies, probably due to the more recent divergence within acuminata species (diploids and triploids).
The analysis of allelic distributions supported the monoclonal origin of the major triploid subgroups, ‘Cavendish’, ‘Mutika-Lujugira’ and ‘Plantain’, despite wide geographical distribution and huge phenotypic diversity. The current CIRAD's breeding strategy to develop triploid cultivars consists of crossing a diploid accession with an auto-tetraploid accession (2n = 4x), obtained through chromosome doubling using colchicine treatment (Bakry and Horry, 1994; Bakry et al., 2001, 2009). With this approach, using the identified putative parents, it should be possible to generate genotypes very close to those of ‘Cavendish’ or ‘Gros Michel’. For the other triploid subgroups, the lack of close ancestors of M. acuminata, as shown for the ‘Mutika-Lujugira’ subgroup, or the lack of both M. acuminata and M. balbisiana putative parents, as for the ‘Plantain’ subgroup, hampers this kind of process and other strategies have to be developed.
The ongoing full genome sequencing of the double haploid of M. acuminata ‘Pahang’ will provide data useful for the comparison between cultivars and probably also structural comparisons. Our results showed that the origin of the huge and valuable phenotypic diversity within the different triploid subgroups, which is essential for breeding programmes, will have to be investigated within epigenetic mechanisms in addition to genetic mechanisms and inherited somatic mutations. The genetically homogeneous cultivars within these triploid subgroups are well characterized phenotypically and differentiated from each other. They represent unique models to investigate and compare the influence of more than 1000 years of epigenetic regulation through mitosis on the same genomes, without any interference with meiosis. Furthermore, evolution and diversification processes mixing sexuality and clonality within Musa should be compared with current studies on other vegetatively propagated crops, such as grape, potato and fruit trees.
ACKNOWLEDGEMENTS
We thank the ‘Grand plateau régional de génotypage and robotique’ of CIRAD for accommodating the experiments. We also thank Markku Häkkinen for providing Ensete and additional M. balbisiana samples and Saïd Mohamed Mzemouigni for collecting Comorian accessions. This work was supported by the Generation Challenge Program (grant ‘Genotyping composite Musa germplasm sets’).
LITERATURE CITED
- Bakry F, Horry JP. Musa breeding at CIRAD-FLHOR. In: Jones DR, editor. The improvement and testing of Musa: a global partnership. Montpellier: INIBAP; 1994. pp. 169–175. [Google Scholar]
- Bakry F, Carreel F, Caruana M-L, Côte F, Jenny C, Tézenas du Montcel H. Banana. In: Charrier A, Jacquot M, Hamon S, Nicolas D, editors. Tropical plant breeding. Montpellier: CIRAD; 2001. pp. 1–29. [Google Scholar]
- Bakry F, Carreel F, Jenny C, Horry J-P. Genetic improvement of banana. In: Jain SM, Priyadarshan PM, editors. Breeding Plantation tree crops: tropical species. New York: Springer; 2009. pp. 3–51. [Google Scholar]
- Buhariwalla HK, Jarret RL, Jayashree B, Crouch JH, Ortiz R. Isolation and characterization of microsatellite markers from Musa balbisiana (Primer Note) Molecular Ecology Notes. 2005;5:327–330. [Google Scholar]
- Carreel F, Fauré S, León DGd, et al. Evaluation de la diversité génétique chez les bananiers diploïdes (Musa sp) Genetics Selection Evolution. 1994;26(Suppl.):125–136. [Google Scholar]
- Carreel F, Leon DGd, Lagoda P, et al. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome. 2002;45:679–692. doi: 10.1139/g02-033. [DOI] [PubMed] [Google Scholar]
- Chapuis M-P, Estoup A. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- Christelová P, Valárik M, Hribová E, De Langhe E, Dolezel J. A multi gene sequence-based phylogeny of the Musaceae (banana) family. BMC Evolutionary Biology. 2011;11(103) doi: 10.1186/1471-2148-11-103. http://dx.doi.org/10.1186/1471-2148-11-103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson I, Goldstein DB. Evidence for complex mutations at microsatellite loci in Drosophila. Genetics. 1999;152:617–627. doi: 10.1093/genetics/152.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creste S, Tulmann Neto A, de Oliveira Silva S, Figueira A. Genetic characterization of banana cultivars (Musa spp.) from Brazil using microsatellite markers. Euphytica. 2003;132:259–268. [Google Scholar]
- Creste S, Tulmann Neto A, Vencovsky R, de Oliveira Silva S, Figueira A. Genetic diversity of Musa diploid and triploid accessions from the Brazilian banana breeding program estimated by microsatellite markers. Genetic Resources and Crop Evolution. 2004;51:723–733. [Google Scholar]
- Crouch HK, Crouch JH, Jarret RL, Cregan PB, Ortiz R. Segregation at microsatellite loci in haploid and diploid gametes of Musa. Crop Science. 1998;38:211–214. [Google Scholar]
- Crouch HK, Crouch JH, Madsen S, Vuylsteke DR, Ortiz R. Comparative analysis of phenotypic and genotypic diversity among plantain landraces (Musa spp., AAB group) Theoretical and Applied Genetics. 2000;101:1056–1065. [Google Scholar]
- D'hont A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenetic and Genome Research. 2005;109:27–33. doi: 10.1159/000082378. [DOI] [PubMed] [Google Scholar]
- Dakin EE, Avise JC. Microsatellite null alleles in parentage analysis. Heredity. 2004;93:504–509. doi: 10.1038/sj.hdy.6800545. [DOI] [PubMed] [Google Scholar]
- Daniells J, Jenny C, Karamura D, Tomekpe K. Musalogue. Diversity in the genus Musa. A catalogue of Musa germplasm. Rome: IPGRI; 2001. , 202 pp. [Google Scholar]
- De Langhe E, De Maret P. Tracking the Banana: significance to early agriculture. In: Gosden C, Hather J, editors. The prehistory of food: appetites for change. London: Routledge; 1999. pp. 377–396. [Google Scholar]
- De Langhe E, Swennen RL, Vuylsteke D. ‘Plantain’ in early Bantu world. Azania. 29/30:147–160. 1994–1995. [Google Scholar]
- Dolezel J, Lysák MA, Van den Houwe I, Dolezelová M, Roux N. Use of flow cytometry for rapid ploidy determination in Musa species. InfoMusa. 1997;6:6–9. [Google Scholar]
- Ellegren H. Microsatellite evolution: a battle between replication slippage and point mutation. Trends in Genetics. 2002;18 70–70. [Google Scholar]
- Estoup A, Jarne P, Cornuet J-M. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Gawel NJ, Jarret RL, Whittemore AP. Restriction fragment length polymorphism (RFLP)-based phylogenetic analysis of Musa. Theoretical and Applied Genetics. 1992;84:286–290. doi: 10.1007/BF00229484. [DOI] [PubMed] [Google Scholar]
- Gill DE, Chao L, Perkins SL, Wolf JB. Genetic mosaicism in plants and clonal animals. Annual Review of Ecology and Systematics. 1995;26:423–444. [Google Scholar]
- Grapin A, Noyer J-L, Carreel F, et al. Diploid Musa acuminata genetic diversity assayed with sequence-tagged microsatellite sites. Electrophoresis. 1998;19:1374–1380. doi: 10.1002/elps.1150190829. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Annals of Botany. 2007;97:1–12. doi: 10.1093/aob/mcm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte I, Bakry F, Seguin M, et al. A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biology. 2010;10(65) doi: 10.1186/1471-2229-10-65. http://dx.doi.org/10.1186/1471-2229-10-65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte I, Rouard M, Gardes L, Pomies V, Perrier X. Datafile: list of 561 accessions characterized with 22 SSR v5.xls. GCP-Bioinformatics – Central Registry -Musa. 2011 Generation Challenge Program. http://gcpcr.grinfo.net/ . [Google Scholar]
- Horry JP, Ortiz R, Arnaud E, et al. Banana and Plantain. In: Fuccillo D, Sears L, Stapleton P, editors. Biodiversity in Trust. Cambridge: Cambridge University Press; 1997. pp. 67–81. [Google Scholar]
- Hribova E, Cizkova J, Christelová P, Taudien S, de Langhe E, Dolezel J. The ITS1-5·8S-ITS2 sequence region in the Musaceae: structure, diversity and use in molecular phylogeny. PLoS ONE. 2011;6:e17863. doi: 10.1371/journal.pone.0017863. http://dx.doi.org/10.1371/journal.pone.0017863 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPGRI-INIBAP(Bioversity) Musa Germplasm Information System (MGIS) 2003 http://mgis.inibap.org . [Google Scholar]
- IPGRI-INIBAP(Bioversity)/CIRAD. Descriptors for Banana (Musa spp.) Rome: IPGRI; 1996. Montpellier: INIBAP; Montpellier: CIRAD, 59 pp. [Google Scholar]
- Ishibashi Y, Saitoh T, Abe S, Yoshida MC. Null microsatellite alleles due to nucleotide sequence variation in the grey-sided vole Clethrionomys rufocanus. Molecular Ecology. 1996;5:589–590. [PubMed] [Google Scholar]
- Kaemmer D, Fischer D, Jarret RL, et al. Molecular breeding in the genus Musa: a strong case for STMS marker technology. Euphytica. 1997;96:49–63. [Google Scholar]
- Kimura M. Rare variant alleles in the light of the neutral theory. Molecular Biology and Evolution. 1983;1:84–93. doi: 10.1093/oxfordjournals.molbev.a040305. [DOI] [PubMed] [Google Scholar]
- Klekowski EJ. Plant clonality, mutation, diplontic selection and mutational meltdown. Biological Journal of the Linnean Society. 2003;79:61–67. [Google Scholar]
- Lagoda PJL, Noyer JL, Dambier D, Baurens F-C, Grapin A, Lanaud C. Sequence tagged microsatellite site (STMS) markers in the Musaceae. Molecular Ecology. 1998;7:657–666. [PubMed] [Google Scholar]
- Lanaud C, du Montcel HT, Jolivot MP, Glaszmann JC, de Leon DG. Variation of ribosomal gene spacer length among wild and cultivated banana. Heredity. 1992;68:147–156. doi: 10.1038/hdy.1992.23. [DOI] [PubMed] [Google Scholar]
- Lescot M, Piffanelli P, Ciampi A, et al. Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics. 2008;9(58) doi: 10.1186/1471-2164-9-58. http://dx.doi.org/10.1186/1471-2164-9-58 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot T. The genetic diversity of banana in figures. FruiTrop. 2008;155:29–33. [Google Scholar]
- Loeillet D. Close-up: banana. FruiTrop. 2008;155:3–39. [Google Scholar]
- Loeillet D, Imbert E, Dawson C, Fouré E, de Lapeyre L, Lescot T. Banana. FruiTrop. 2011;189:15–62. [Google Scholar]
- Marcotrigiano M. Chimeras and variegation: patterns of deceit. HortScience. 1997;32:773–784. [Google Scholar]
- Ning S-P, Xu L-B, Lu Y, Huang B-Z, Ge X-J. Genome composition and genetic diversity of Musa germplasm from China revealed by PCR-RFLP and SSR markers. Scientia Horticulturae. 2007;114:281–288. [Google Scholar]
- Noyer JL, Causse S, Tomekpe K, Bouet A, Baurens FC. A new image of plantain diversity assessed by SSR, AFLP and MSAP markers. Genetica. 2005;124:61–69. doi: 10.1007/s10709-004-7319-z. [DOI] [PubMed] [Google Scholar]
- Ortiz R. Occurrence and inheritance of 2n pollen in Musa. Annals of Botany. 1997;79:449–453. [Google Scholar]
- Ortiz R, Madsen S, Vuylsteke D. Classification of African plantain landraces and banana cultivars using a phenotypic distance index of quantitative descriptors. Theoretical and Applied Genetics. 1998;96:904–911. [Google Scholar]
- Perrier X, Jacquemoud-Collet JP. DARwin software Version 5·0·155. 2006 CIRAD: http://darwin.cirad.fr/darwin . [Google Scholar]
- Perrier X, Bakry F, Carreel F, et al. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobotany Research and Applications. 2009;7:199–216. [Google Scholar]
- Perrier X, De Langhe E, Donohue M, et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proceedings of National Academy of Science of the USA. 2011;108:11311–11318. doi: 10.1073/pnas.1102001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Krch M, Lehtilä K. Costs and benefits of genetic heterogeneity within organisms. Journal of Evolutionary Biology. 2004;17:1167–1177. doi: 10.1111/j.1420-9101.2004.00808.x. [DOI] [PubMed] [Google Scholar]
- Pollefeys P, Sharrock S, Arnaud E. Preliminary analysis of the literature on the distribution of wild Musa species using MGIS and DIVA-GIS. 2004 http://bananas.bioversityinternational.org/files/files/pdf/publications/wildspecies_pollefeys.pdf . [Google Scholar]
- Raboin L-M, Carreel F, Noyer J-L, et al. Diploid ancestors of triploid export Banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Molecular Breeding. 2005;16:333–341. [Google Scholar]
- Risterucci AM, Grivet L, N'Goran JAK, Pieretti I, Flament MH, Lanaud C. A high-density linkage map of Theobroma cacao L. Theoretical and Applied Genetics. 2000;101:948–955. doi: 10.1007/BF00223910. [DOI] [PubMed] [Google Scholar]
- Risterucci AM, Hippolyte I, Perrier X, et al. Development and assessment of diversity arrays technology for high-throughput DNA analyses in Musa. Theoretical and Applied Genetics. 2009;119:1093–1103. doi: 10.1007/s00122-009-1111-5. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santelices B. How many kinds of individual are there? Trends in Ecology & Evolution. 1999;14:152–155. doi: 10.1016/s0169-5347(98)01519-5. [DOI] [PubMed] [Google Scholar]
- Simmonds NW. Varietal identification in the ‘Cavendish’ group of bananas. Journal of Horticultural Science. 1954;29:81–88. [Google Scholar]
- Simmonds NW. The evolution of bananas. London: Longman, 170 pp; 1962. [Google Scholar]
- Simmonds NW. Bananas. London: Longmans, 512 pp; 1966. [Google Scholar]
- Simmonds NW, Shepherd K. Taxonomy and origins of cultivated Bananas. Journal of the Linnean Society of London, Botany. 1955;55:302–312. [Google Scholar]
- Sotto RC, Rabara RC. Morphological diversity of Musa balbisiana Colla in the Philippines. InfoMusa. 2000;9:28–30. [Google Scholar]
- Swangpol S, Volkaert H, Sotto RC, Seelanan T. Utility of selected non-coding chloroplast DNA sequences for lineage assessment of Musa interspecific hybrids. Journal of Biochemistry and Molecular Biology. 2007;40:577–587. doi: 10.5483/bmbrep.2007.40.4.577. [DOI] [PubMed] [Google Scholar]
- Ude G, Pillay M, Nwakanma D, Tenkouano A. Genetic diversity in Musa acuminata Colla and Musa balbisiana Colla and some of their natural hybrids using AFLP markers. Theoretical and Applied Genetics. 2002;104:1246–1252. doi: 10.1007/s00122-002-0914-4. [DOI] [PubMed] [Google Scholar]
- Ude G, Pillay M, Ogundiwin E, Tenkouano A. Genetic diversity in an African plantain core collection using AFLP and RAPD markers. Theoretical and Applied Genetics. 2003;107:248–255. doi: 10.1007/s00122-003-1246-8. [DOI] [PubMed] [Google Scholar]
- Uma S, Siva S, Saraswathi M, et al. Variation and intraspecific relationships in Indian wild Musa balbisiana (BB) population as evidenced by random amplified polymorphic DNA. Genetic Resources and Crop Evolution. 2006;53:349–355. [Google Scholar]
- Vigouroux Y, Jaqueth JS, Matsuoka Y, et al. Rate and pattern of mutation at microsatellite loci in maize. Molecular Biology and Evolution. 2002;19:1251–1260. doi: 10.1093/oxfordjournals.molbev.a004186. [DOI] [PubMed] [Google Scholar]
- Wang X-L, Chiang T-Y, Roux N, Hao G, Ge X-J. Genetic diversity of wild banana (Musa balbisiana Colla) in China as revealed by AFLP markers. Genetic Resources and Crop Evolution. 2007;54:1125–1132. [Google Scholar]
- Wattier R, Engel CR, Saumitou-Laprade P, Valero M. Short allele dominance as a source of heterozygote deficiency at microsatellite loci: experimental evidence at the dinucleotide locus Gv1CT in Gracilaria gracilis (Rhodophyta) Molecular Ecology Notes. 1998;7:1569–1573. [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Human Molecular Genetics. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Wong C, Kiew R, Argent G, Set OHN, Lee SK, Gan YY. Assessment of the validity of the sections in Musa (Musaceae) using AFLP. Annals of Botany. 2002;90:231–238. doi: 10.1093/aob/mcf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef M, James AC, Rivera-Madrid R, Ortiz R, Escobedo-Graciamedrano RM. Musa genetic diversity revealed by SRAP and AFLP. Molecular Biotechnology. 2011;47:189–199. doi: 10.1007/s12033-010-9328-8. [DOI] [PubMed] [Google Scholar]