Abstract
Background and Aims
Populations established by long-distance colonization are expected to show low levels of genetic variation per population, but strong genetic differentiation among populations. Whether isolated populations indeed show this genetic signature of isolation depends on the amount and diversity of diaspores arriving by long-distance dispersal, and time since colonization. For ferns, however, reliable estimates of long-distance dispersal rates remain largely unknown, and previous studies on fern population genetics often sampled older or non-isolated populations. Young populations in recent, disjunct habitats form a useful study system to improve our understanding of the genetic impact of long-distance dispersal.
Methods
Microsatellite markers were used to analyse the amount and distribution of genetic diversity in young populations of four widespread calcicole ferns (Asplenium scolopendrium, diploid; Asplenium trichomanes subsp. quadrivalens, tetraploid; Polystichum setiferum, diploid; and Polystichum aculeatum, tetraploid), which are rare in The Netherlands but established multiple populations in a forest (the Kuinderbos) on recently reclaimed Dutch polder land following long-distance dispersal. Reference samples from populations throughout Europe were used to assess how much of the existing variation was already present in the Kuinderbos.
Key Results
A large part of the Dutch and European genetic diversity in all four species was already found in the Kuinderbos. This diversity was strongly partitioned among populations. Most populations showed low genetic variation and high inbreeding coefficients, and were assigned to single, unique gene pools in cluster analyses. Evidence for interpopulational gene flow was low, except for the most abundant species.
Conclusions
The results show that all four species, diploids as well as polyploids, were capable of frequent long-distance colonization via single-spore establishment. This indicates that even isolated habitats receive dense and diverse spore rains, including genotypes capable of self-fertilization. Limited gene flow may conserve the genetic signature of multiple long-distance colonization events for several decades.
Keywords: Asplenium scolopendrium L., Asplenium trichomanes subsp., quadrivalens, colonization, founder effects, gene flow, microsatellites, multiple colonization events, Polystichum setiferum, Polystichum aculeatum, population differentiation, population genetics, selfing
INTRODUCTION
Long-distance dispersal (LDD) is of vital importance for the colonization of remote sites (Eriksson 1996), as well as for the dynamics and structure of remote populations (Bohrer et al., 2005). Immigration from distant sources plays a key role both in determining the levels of genetic diversity in such remote populations and in the way this diversity is partitioned among populations (Pannell and Dorken, 2006; Broquet et al., 2009). Understanding this role of LDD in shaping patterns of genetic diversity is essential, as the fragmentation and impoverishment of natural habitats continues and levels of genetic diversity are important for population viability (Lande and Shannon, 1996). Yet, reliable estimates of frequencies of diaspore arrival by LDD in relation to source distance remain largely unknown, as these are inherently difficult to measure (Cain et al., 2000), and the consequences of low immigration rates for genetic diversity and genetic structure in and among peripheral populations remain controversial (Eckert et al., 2008).
Generally, long-distance colonization is expected to result in low genetic variation within peripheral populations and strong differentiation among them, due to various mechanisms acting both during colonization and after initial establishment (Pannell and Dorken, 2006). Low genetic variation may result from genetic drift: strong founder effects during colonization and/or stochastic loss of alleles within small populations (Pannell and Dorken, 2006). Strong partitioning of diversity among populations may originate during colonization when different populations are founded by different genotypes (Whitlock and McCauley, 1999), or in the founded populations due to genetic drift or high inbreeding rates (Schaal and Leverich, 1996; Young et al., 1996). Many studies indeed detected reduced genetic diversity and/or increased spatial structure towards range edges (Maguire et al., 2000; Arnaud-Haond et al., 2006). However, differences among central and peripheral populations are generally not large (Eckert et al., 2008), and in various cases genetic variation among arriving colonizers or local adaptation seemed sufficient to maintain high diversity in peripheral populations (Barton, 2001; Sundberg, 2005).
Long-distance colonization is of particular interest in ferns, as all homosporous fern species can disperse their spores over large distances. Evidence from island studies has shown that spores of various fern species may be dispersed over thousands of kilometres to reach a new habitat (e.g. Tryon, 1966, 1970; Perrie et al., 2010). Although only part of the spores produced will be dispersed over such large distances (Peck et al., 1990; Sundberg, 2005), there are indications that long-distance colonization by ferns is not necessarily a rare event (Shepherd et al., 2009). Moreover, most homosporous ferns are thought to be capable of establishment from a single spore (Peck et al., 1990; Wolf et al., 2001). Therefore, disjunct populations of ferns in particular yield excellent systems for studies on the genetic signature of long-distance colonization.
The capacity of homosporous ferns to establish from single spores results from the fact that spore germination gives rise to a (haploid) gametophyte that is potentially bisexual and capable of producing a sporophyte by intragametophytic selfing (i.e. self-fertilization on a single haploid gametophyte; Klekowski, 1979; Ranker and Geiger, 2008). Single-spore colonization via this extreme form of inbreeding may result in strong founder effects and effectively, in terms of genetics, inbreeding populations. Subsequent increases in genetic diversity and recombination through sexual reproduction are conditional on the immigration of additional genotypes (Pannell and Dorken, 2006). Although potential for intragametophytic selfing varies between species (Soltis and Soltis, 1992; Ranker and Geiger, 2008), recent breeding experiments by the authors for a number of temperate ferns indicated that genotypes capable of self-fertilization may be present in more species than is generally assumed (De Groot et al., 2012). Clear patterns of low population genetic variation and strong population differentiation have been shown for a number of temperate rock-dwelling ferns (e.g. Schneller and Holderegger, 1996; Vogel et al., 1999a; Suter et al., 2000). These patterns were attributed to a high incidence of intragametophytic selfing, as a result of their patchy habitats (Vogel et al., 1999b) and due to the fact that many of the species are polyploids, which may suffer less from inbreeding depression (Masuyama and Watano, 1990; Vogel et al., 1999a). Most populations of sexually reproducing (diploid) fern species seem to be mating randomly or primarily by outcrossing (Ranker and Geiger, 2008). However, in case some genotypes of these species are capable of self-fertilization, mate limitation in empty, disjunct habitats may result in strong selection for such genotypes.
Unfortunately, previous studies showing low variation in isolated fern populations may have been misled by two limitations in their experimental design which obscured evidence of the mechanisms behind this pattern. A first set of studies made use of allozymes as genetic markers (e.g. Schneller and Holderegger, 1996; Vogel et al., 1999a, b; Suter et al., 2000). As such markers typically show limited allelic diversity, their resolution may be too low for population genetic studies (Woodhead et al., 2005). Other studies, which overcame this problem by using high-resolution markers [amplified fragment length polymorphisms (AFLPs), random amplification of polymorphic DNAs (RAPDs) or microsatellites], focused on relatively old populations that experienced rather stable population sizes (Pryor et al., 2001), often located towards the centre of the species' distribution range (Jiménez et al., 2010). Their results were then discussed in relation to colonization histories over time scales of thousands of years (e.g. since the last ice age). This causes difficulties in disentangling the mechanisms that act at different moments after initial colonization. For instance, spatial structure may be caused either by a combination of founder effects and low variation among arriving colonizers, or by loss of variation at a later stage due to drift or non-random mating.
Very young populations established in a recent, disjunct habitat, form a useful study system to improve our understanding of the processes that shape patterns in genetic variation at different stages of colonization. In such populations, local equilibria between gene flow, mutation and genetic drift are of minor importance, and genetic structure is driven by a combination of the diversity and intensity of the spore rain and species-specific population dynamics. Here, we made use of an excellent example of such a young system (De Groot et al., 2008): isolated populations in a young approx. 1000 ha planted forest on Dutch polder land that was reclaimed from the sea in the 1940s. This woodland harbours the largest diversity of fern species in The Netherlands (Bremer, 2007), among which there are various rare calcicoles that must have reached the Dutch polders by LDD (Bremer, 1980). Most of these rare calcicoles established multiple spatially separated clusters in the forest, which is located towards the northern edge of the species' distribution ranges. From 1980 onwards, establishment of these clusters was carefully monitored (Bremer, 1980, 1994, 2007). This provides a unique opportunity to infer key mechanisms that shape the assembly of genetic diversity through time and in space subsequent to the initial long-distance colonization.
In this study, we used highly polymorphic microsatellite markers to infer the genetic structure of young disjunct populations in the Dutch polders of four rare calcicole fern species with varying mating strategies (as inferred from previous studies). This allowed us to gain evidence on two key premises with respect to fern colonization that were often described but rarely tested: disjunct habitats are frequently reached by fern spores through LDD, and such long-distance colonization commonly results in inbred populations with low genetic variability as population founders have to rely on self-fertilization. More specifically, we address the following four questions. (1) Are the observed spatial clusters of individuals the result of multiple, separate long-distance colonization events? (2) How much variation is present in the forest as a whole and within populations? (3) How much gene flow has already occurred between the local populations? (4) How do population genetic structure and observed levels of variation vary between species and how does this relate to their (local) ecology and reproductive biology?
MATERIALS AND METHODS
Study area and study species
We studied the genetic structure in populations of four fern species that are rare in The Netherlands, and which belong to two different genera and vary in ploidy level (Van der Meijden, 2005): Asplenium scolopendrium L., Asplenium trichomanes subsp. quadrivalens D.E.Meyer, Polystichum setiferum (Forssk.) Moore ex Woynar and Polystichum aculeatum (L.) Roth.
We studied young populations in the Kuinderbos forest, which is located in the Dutch Noordoostpolder. This polder was reclaimed from the sea in the 1940s. The new polder land was in fact a former sea bottom, most probably containing very limited numbers of viable diaspores. A few years after polder reclamation the soil was ploughed, moving any remaining spores in the top layer to a depth of at least 30 cm. Therefore, the area may in a way be seen as a huge ‘tabula rasa’-like system (Nordal, 1987): a large empty area that functioned as a huge diaspore trap and allowed for the development of new plant communities from scratch by means of immigration. The Kuinderbos forest was planted about 10 years later, after a period in which the area mainly consisted of brackish wetland (Feekes and Bakker, 1954). A mixture of tree species was used, but most ferns are found underneath stands of Picea sitchensis (Bong.) Carrière, Fraxinus excelsior L. and Quercus robur L. The forest's soil consists mainly of calcareous sand (due to the remains of sea shells), on top of the remnants of an old peat layer (Bremer, 1994). Throughout the forest, the top sand layer is intersected by a large number of drainage trenches. The steep slopes of the drainage trenches provide a substrate of moist calcareous sand, and form the main habitat of the study species (Bremer, 1980, 2007). All four are calcicoles with relatively high habitat specificities (Fitter and Peat, 1994; Page, 1997) and are mainly known from rocky, lime-rich substrates. In The Netherlands, P. aculeatum occurs almost exclusively on patches of lime-rich soil near the southern border (Maastricht). The other three species mainly occur on man-made walls (FLORON, 2010).
Asplenium scolopendrium is a diploid species. Gametophyte mating experiments showed a mixed mating system for at least the Kuinderbos genotypes of this species (Wubs et al., 2010). Its congener A. trichomanes subsp. quadrivalens is an allotetraploid taxonomic entity of the A. trichomanes complex and is known as a strong selfer (Suter et al., 2000). Both asplenioids are rare in The Netherlands, but are increasingly found in rural areas across the country (Bremer, 2007). The nearest source populations at the time of first establishment in the Kuinderbos were >30 km away (Bremer, 2007). The allotetraploid P. aculeatum and its diploid parent P. setiferum are rarer still, with the nearest populations at the southern Dutch border, in Germany and the UK. They must have dispersed over distances of at least 100 and 250 km, respectively (Bremer, 1980, 2007). Based on mating experiments, Pangua et al. (2003) reported evidence of inbreeding for the tetraploid P. aculeatum and obligate outcrossing for the diploid P. setiferum, at least for populations at the Iberian Peninsula.
In the drainage trenches of the Kuinderbos, the study species occur as spatially separated clusters of individuals, consistently called ‘populations’ throughout this text. Depending on the population, we estimate that there have been 2–4 generations since establishment for A. trichomanes subsp. quadrivalens and a maximum of seven generations for the other study species. Generations partly overlap, as sometimes even the founding individual is still present. Asplenium scolopendrium is by far the most abundant species in the forest, present with >13 000 individuals spread over numerous populations, mainly located in the north-western and central sections of the forest (Fig. 1; Bremer, 1994, 2007). Polystichum aculeatum is also abundant and present with 10–20 populations, again especially in the north-western corner (Bremer 1994). Asplenium trichomanes subsp. quadrivalens is present with only three populations of 10–11 individuals each, and P. setiferum has about ten populations of varying size, scattered throughout the forest.
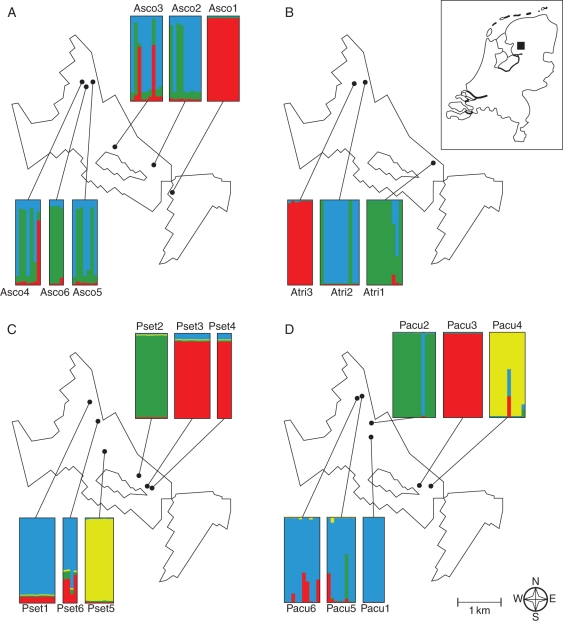
Fig. 1.
Maps of the Kuinderbos, showing the position of each sampled population, along with the assignment of individual samples to K different gene pools as inferred by Bayesian clustering analysis. Per population, each individual is represented by a vertical bar, in which different colours show the proportional membership in each gene pool. The inferred value of K varies per species. (A) Asplenium scolopendrium (K = 3), (B) Asplenium trichomanes (K = 3), (C) Polystichum setiferum (K = 4), (D) Polystichum aculeatum (K = 4). The inlay in the right upper corner shows the position of the Kuinderbos (black square) within The Netherlands.
Sampling strategy
Leaf fragments were sampled from six populations of A. scolopendrium, P. setiferum and P. aculeatum in the Kuinderbos, and all three known populations of A. trichomanes (Table 1). Each population was located in a different drainage trench and formed a spatially separated cluster of individuals (Fig. 1). Distances between populations ranged from 30 m (adjacent trenches) to 3·8 km. Population sizes differed within and between species (Table 2). At least ten sporophytes were sampled per population, except for two small populations of P. setiferum. Entire populations were sampled for A. trichomanes subsp. quadrivalens. Final sample sizes used in analyses are shown in Table 2.
Table 1.
Numbers of sampled populations (Npop) and total numbers of samples (Ns) per species in the Kuinderbos, in the rest of The Netherlands (excluding Kuinderbos) and throughout the rest of Europe (excluding The Netherlands)
| Kuinderbos |
The Netherlands |
Europe |
Acquired variation |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Npop | Ns | Npop | Ns | Npop | Ns | AKB/ANL | AKB/AEUR | ATOTAL |
| ASPS | 6 | 54 | 22 | 24 | 18 | 19 | 0·85 | 0·83 | 8·25 |
| ASPT | 3 | 31 | 11 | 11 | 29 | 29 | 0·70 | 0·54 | 7·50 |
| POLS | 6 | 51 | 10 | 21 | 12 | 12 | 0·59 | 0·55 | 11·00 |
| POLA | 6 | 60 | 3 | 9 | 17 | 18 | 0·91 | 0·71 | 7·50 |
Allelic variation is calculated as the mean number of different alleles per locus. The acquired genetic variation in the Kuinderbos is given both as the fraction of the total Dutch allelic variation that is present in the Kuinderbos (AKB/ANL) and as the fraction of the total European allelic variation that is present in the Kuinderbos (AKB/AEUR).
ATOTAL, total allelic variation across all samples per species; ASPS, Asplenium scolopendrium; ASPT, Asplenium trichomanes; POLS, Polystichum setiferum; POLA, Polystichum aculeatum.
Table 2.
Genetic characterization of all sampled populations in the Dutch Kuinderbos forest
| Species | Population | Pop. size | Yest | No. of samples | Ng | A | Ar | Ap/A | A/AKB | Ho | He | F | Founder |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASPS | Asco1 (KB1) | 339 (238) | 1980–1985 | 9 | 1 | 1·00 | 1·00 | 0·00 | 0·15 | 0·000 | 0·000 | NA* | ? |
| Asco2 (KB2) | 206 (145) | 1975–1980 | 10 | 7 | 2·50 | 2·02 | 0·07 | 0·38 | 0·081 | 0·258 | 0·684 | ? | |
| Asco3 (KB4) | 328 (259) | 1975–1980 | 10 | 8 | 3·25 | 2·65 | 0·03 | 0·50 | 0·103 | 0·391 | 0·737 | ? | |
| Asco4 (KB10) | 260 (139) | 1970–1975 | 9 | 9 | 3·75 | 3·13 | 0·12 | 0·58 | 0·238 | 0·431 | 0·448 | ? | |
| Asco5 (KB12) | 262 (95) | 1970–1980 | 9 | 9 | 3·25 | 2·57 | 0·00 | 0·50 | 0·119 | 0·458 | 0·740 | ? | |
| Asco6 (KB13) | 149 (81) | 1970–1980 | 7 | 6 | 2·75 | 2·52 | 0·10 | 0·42 | 0·207 | 0·474 | 0·563 | ? | |
| ASPT | Atri1 | 10 (10) | About 1975 | 10 | 4 | 2·50 | NA | 0·18 | 0·67 | NA | 0·270 | NA | ? |
| Atri2 | 11 (11) | 1985–1990 | 11 | 2 | 1·75 | NA | 0·25 | 0·47 | NA | 0·385 | NA | ? | |
| Atri3 | 10 (10) | About 1995 | 10 | 5 | 2·25 | NA | 0·38 | 0·60 | NA | 0·431 | NA | ? | |
| POLS | Pset1 | 28 (3) | 1970–1980 | 10 | 1 | 1·00 | 1·00 | 0·00 | 0·16 | 0·000 | 0·000 | NA* | Yes |
| Pset2 | 60 (36) | 1980–1990 | 12 | 5 | 1·50 | 1·59 | 0·38 | 0·24 | 0·083 | 0·239 | 0·651 | Yes | |
| Pset3 | 29 (20) | 1970–1980 | 10 | 1 | 1·00 | 1·00 | 0·00 | 0·16 | 0·000 | 0·000 | NA* | ? | |
| Pset4 | 6 (1) | 2005–2010 | 4 | 4 | 1·75 | 1·63 | 0·13 | 0·28 | 0·125 | 0·208 | 0·4 | Yes | |
| Pset5 | 12 (8) | 1970–1980 | 10 | 10 | 3·75 | 2·39 | 0·53 | 0·60 | 0·369 | 0·431 | 0·143 | ? | |
| Pset6 | 5 (1) | 1985–1990 | 5 | 4 | 1·67 | 1·40 | 0·25 | 0·27 | 0·400 | 0·175 | –0·600 | No | |
| POLA | Pacu1 | 10 (2) | 1965–1975 | 7 | 3 | 2·00 | NA | 0·06 | 0·38 | NA | 0·192 | NA | Yes† |
| Pacu2 | 138 (36) | 1986 | 12 | 5 | 2·25 | NA | 0·08 | 0·43 | NA | 0·281 | NA | Yes† | |
| Pacu3 | 55 (40) | 1965–1975 | 11 | 1 | 1·00 | NA | 0·00 | 0·19 | NA | 0·000 | NA | Yes | |
| Pacu4 | 50 (>25) | 1965–1975 | 10 | 3 | 2·75 | NA | 0·13 | 0·52 | NA | 0·299 | NA | Yes† | |
| Pacu5 | 147 (>100) | 1965–1975 | 10 | 8 | 3·75 | NA | 0·00 | 0·71 | NA | 0·542 | NA | ? | |
| Pacu6 | 31 (>10) | 1985–1990 | 10 | 10 | 4·25 | NA | 0·15 | 0·81 | NA | 0·569 | NA | ? |
Species acronyms are as in Table 1. Asplenium scolopendrium populations overlap with those in Wubs et al. (2010) (codes used in that article are given in parentheses).
Pop. size, population size at the time of sampling (number of spore-bearing plants in parentheses); Yest, estimated time of initial population establishment (P. Bremer, unpubl. res.); Founder, the population founder homozygous at the tested loci [yes, no or founder unknown (?)].
Measures of genetic variation: Ng, number of multilocus genotypes; A, allelic variability (mean number of alleles per locus); Ar, allelic richness (number of alleles per locus, corrected for sample size by means of rarefaction); Ap/A, mean fraction of private alleles (within the Kuinderbos) per locus; A/AKB, fraction of the total allelic variability in the Kuinderbos that is present per population.
Measures of heterozygosity and inbreeding: Ho, observed heterozygosity (undefined for polyploids); He, expected heterozygosity; F, inbreeding coefficient (undefined for polyploids).
* Only one multilocus genotype present. The inbreeding coefficient is therefore undefined, but effectively equals 2 (extreme inbreeding).
† Most probably homozygous, but heterozygosity cannot be excluded as this (tetraploid) individual showed two alleles at some loci.
For several populations, the founding individual was known from inventory data and sampled, and/or an estimate of the time of establishment of this founder was available (Table 2). Generally, observed levels of genetic diversity vary with both the study species and the markers applied. Therefore, to be able to compare the variation in Kuinderbos colonies between species, we additionally gathered samples from 3–22 populations per species in the rest of The Netherlands and 12–29 populations throughout Europe (‘context samples’, see Table 1), and then compared the genetic variation in the Kuinderbos with that in the entire data set. Dutch context populations were newly sampled (one sample per population), and European samples within an approx. 1000 km radius around the Kuinderbos (Supplementary Data Table S1) were either newly sampled or obtained from herbarium collections held at the Natural History Museum in London (BM, UK) and the University of Helsinki (H, Finland). Per individual, a few square centimetres of leaf tissue was sampled and put in a paper envelope. Per population, envelopes were put in a plastic bag with silica beads to rapidly dry and store the samples.
Microsatellite genotyping
DNA was extracted using the GenElute™ Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, St Louis, MO, USA) following the manufacturer's protocol. Genotyping was performed using microsatellite markers specifically developed for the study species (De Groot et al., 2011). Five polymorphic loci were available for A. scolopendrium (AS4N, AS7, AS9, AT5 and AT10) and four loci for each of the other species (A. trichomanes subsp. quadrivalens, AT1, AT3, AT8b and AT10; P. setiferum, PS3, PS5, PS9 and PA6; and P. aculeatum: PA4, PA5b, PA6 and PS5). Two loci, AT1 and AS9, are located in the chloroplast DNA (cpDNA). Amplification of all loci followed the standard protocol described in De Groot et al. (2011). Primers were labelled with fluorescent dyes (FAM or HEX), and fragment sizes were determined by automated detection using an ABI 3730 capillary sequencer. The GeneScan-500 ROX Size Standard (Life Technologies, Carlsbad, CA, USA) was used as an internal lane standard for all fragment size analyses (excluding the band at 250 bp that is known to be unstable). Allele scoring was performed using the ABI PeakScanner v.1·0 software (Life Technologies).
For two populations of P. setiferum (Pset3 and Pset6; Fig. 1), genotyping of marker PS9 failed for all individuals in the population. This result was interpreted as the presence of a null allele. For the characterization of genetic variation, these individuals were assumed to be homozygous at this locus for a single hypothetical allele not present in the other populations. The locus was excluded from tests of isolation by distance, population differentiation and in principal components analysis (PCA) and Bayesian cluster analyses.
Data analysis
Two of our four studied species have a tetraploid genome. This can be problematic in the analysis of microsatellite genotyping data, not only because part of the available software is only suited for diploid data sets, but also because banding patterns do not yield reliable information on allele dosages in heterozygous tetraploids (e.g. Van Puyvelde et al., 2010): the actual genotype of an individual with alleles A, B and C might for instance be AABC, ABBC or ABCC. As a result, exact allele frequencies cannot be estimated. To allow comparison between species, we aimed to use measures and statistical tests of genetic diversity and differentiation that could be calculated in a comparable way for both diploids and tetraploids. However, the use of alternative analytical procedures implemented in different software packages was unavoidable and a few heterozygosity-based tests (e.g. inbreeding rates and linkage disequilibrium) could only be performed for the two diploid species.
Linkage disequilibrium was tested for the two diploid species using Fisher's exact tests in Genepop (Raymond and Rousset, 1995) using both per population and across all populations estimations, for all possible loci combinations. For each population, the following measures of genetic diversity were calculated manually: mean number of different multilocus genotypes (Ng), mean number of alleles per locus (A), the number of private alleles (Npa) and the part of the total allelic diversity in the Kuinderbos (AKB) that was present in population i (Ai/AKB). For each species we estimated the percentage of the total Dutch and European genetic diversity that has arrived in the Kuinderbos since it was planted, by calculating the ratio between the number of alleles in the Kuinderbos and the number of alleles either in the total set of Dutch samples (AKB/ANL) or in the total data set (AKB/AEUR). As a measure of allelic diversity independent of sample size, we calculated the allelic richness Ar in Fstat (v 2·9·3; Goudet, 1995) by the method of rarefaction (El Mouradik and Petit, 1996). This was only possible for diploid species. Variation in sample size among populations was, however, very small for both tetraploid taxa (7–12 individuals), which justifies comparison of uncorrected values of A.
The markers AS9 and AT1 were excluded from all heterozygosity-based analyses, because they are located at the uniparentally inherited plastid genome. Expected heterozygosity (He) was calculated per population as the unbiased estimator described by Nei (1987), using FStat for diploids and ATetra (v 1·1; Van Puyvelde et al., 2010) for tetraploids. The program ATetra uses Monte Carlo simulations to estimate mean heterozygosity values based on a random sub-set of all possible combinations of allele configurations in tetraploid individuals. For the diploids, observed heterozygosities (Ho) and inbreeding coefficients (F) were calculated for all populations using FStat.
Genetic differentiation of populations was quantified using three different measures of partitioning of genetic variances: two recently developed measures, G'ST (Meirmans and Hedrick, 2011) and Jost's D (Jost, 2008), and the original estimator of GST as formulated by Nei (1987). Estimates of HS and HT unbiased for sample size differences (Nei and Chesser, 1983) were calculated per locus using SMOGD (v 1·2·5; Crawford, 2010) for diploids and manually for tetraploids based on expected heterozygosity values produced by ATetra. We then averaged HS and HT over loci before calculating GST and G'ST (Nei, 1973; Meirmans and Hedrick, 2011). For Jost's D, a multilocus value was obtained by taking the harmonic mean across loci (Crawford, 2010). For the diploid species, the significance of the population differentiation was tested using a Fisher's exact test in FStat, assessing whether the distribution of (multilocus) genotypes differed from that of a completely randomized data set (Goudet et al., 1996).
We performed two different types of cluster analyses to estimate more explicitly the minimum number of separate colonization events from which our sampled populations probably originated. As we studied the diversity of the spore rain and the colonization history, we were interested in the similarity in allelic variants among populations rather than in differences in dominance of alleles. We therefore first performed PCAs with PCord (v5·0; McCune and Mefford, 1999) based on a binary matrix of presence data per allele per individual. However, because each allele is interpreted as a separate variable, results may be dominated by variation at one highly polymorphic locus even when variation on average is relatively low. We therefore also applied Instruct (Gao et al., 2007) which, based on multilocus genotyping data, uses a Bayesian clustering algorithm to assign individuals to a pre-defined set of K different gene pools in such a way that each pool is characterized by its own set of allele frequencies. This program is an extension of the Structure software (Pritchard et al., 2000), but relaxes some of its key assumptions. Most importantly, it does not assume populations to be in Hardy–Weinberg equilibrium, but instead calculates expected genotype frequencies from estimated inbreeding rates. This approach suits our population data better, which are likely to be far from Hardy–Weinberg equilibrium, as only a few generations have passed, generations overlap and inbreeding rates are expected to be high. Unfortunately, Instruct does not accept polyploid genotype data (H. Gao, pers. comm.) and for the tetraploid taxa we applied Structure (v2·3·1), which has been adapted to handle polyploid genotypes with ambiguous allele copy numbers (Falush et al., 2007). Analyses were run for K-values of 1–8; the optimal value of K was determined from log likelihood values following the method of Evanno et al. (2005). Results from five Markov Chain Monte Carlo (MCMC) chains (burn-in: 100 000 iterations; run length: 200 000 iterations; admixture allowed) were matched using CLUMPP (Jakobsson and Rosenberg, 2007). As chains were very consistent (H' >0·995), they were combined to form a single output file, which was then visualized using DISTRUCT (Rosenberg, 2004).
To assess amounts of gene flow between our young populations, estimates based on the number of migrations per generation (such as Nm) are inappropriate as few generations have passed and populations thus probably have not reached an equilibrium between drift and migration (Whitlock and McCauley, 1999). We alternatively performed tests of isolation by distance using two different measures of pairwise genetic distance: one based on levels of heterozygosity (GST) and an alternative measure based on similarity in allelic variants (Sörensen's index of similarity, QS). Geographical distances between populations were calculated from GPS location data, and correlations between geographical and genetic distances were tested using Mantel tests in Genepop.
RESULTS
Marker performance
The mean number of alleles per locus was 7·5 in the tetraploids A. trichomanes and P. aculeatum, 8·3 in the diploid A. scolopendrium and 11·0 in the diploid P. setiferum. The total number of observed alleles in the data set ranged from 30 in A. trichomanes subsp. quadrivalens to 44 in P. setiferum. Tests of linkage disequilibrium per population for all combinations of loci were non-significant (tested for diploids only), indicating random associations between loci. Overall marker heterozygosities were low (mean heterozygosities of 0·13 for A. scolopendrium and 0·20 for P. setiferum), but higher when only European samples were included (0·15 and 0·31, respectively).
Overall genetic variation and population differentiation
Ratios between allelic variability within and outside the Kuinderbos (Table 1) for each species showed that well over half of the allelic variants sampled throughout The Netherlands, as well as those sampled throughout Europe, are already present in the Kuinderbos populations. The acquired variation was especially high in A. scolopendrium, for which the six populations harboured 83 % of the variation found in a total of 25 populations across Europe. The high value for AKB/ANL in P. aculeatum is probably an overestimation, as only three additional populations were sampled countrywide. Overall expected heterozygosities in the Kuinderbos (mean HT per locus) ranged from 0·43 in A. scolopendrium to 0·57 in P. setiferum.
Various measures of population differentiation consistently showed that, within the Kuinderbos, the variation in alleles was clearly partitioned over populations. This was especially the case in A. trichomanes, P. setiferum and P. aculeatum, as shown by the low ratios between allelic richness in the populations and overall allelic richness in the Kuinderbos (A/AKB; Table 2) and the relatively frequent occurrence of private alleles in populations where this ratio was higher (T1–3, S5). Given the use of highly polymorphic markers and the relatively low values of HS (Jost, 2008), values of G'ST and GST can be considered high and indicate strong population differentiation across species (Table 4). Values for Jost's D were lower, but still high for all species except A. scolopendrium, and showed almost the same interspecific pattern as the other two measures. Population differentiation was strongest in P. setiferum (G'ST = 0·906, D = 0·473); and weakest in A. scolopendrium, although still significant (Table 4).
Table 4.
Measures of population differentiation (GST, G'ST and Jost's D) and results of significance tests of population differentiation (P-values, diploids only) based on randomization tests
| Population differentiation |
Isolation by distance |
|||||
|---|---|---|---|---|---|---|
| GST | G'ST | Jost's D | P | P (GST) | P (QS) | |
| ASPS | 0·161 | 0·319 | 0·061 | < 0·001 | 0·224 | 0·079 |
| ASPT | 0·199 | 0·510 | 0·227 | n/a | 0·827 | 0·490 |
| POLS | 0·711 | 0·906 | 0·473 | < 0·001 | 0·544 | 0·853 |
| POLA | 0·268 | 0·531 | 0·198 | n/a | 0·276 | 0·003 |
Results of tests for isolation by distance, based on Mantel tests correlating pairwise geographic distances with either pairwise GST or pairwise Sörensen indices of similarity (QS). Significant P-values (<0·05) are given in bold. Species acronyms are as in Table 1.
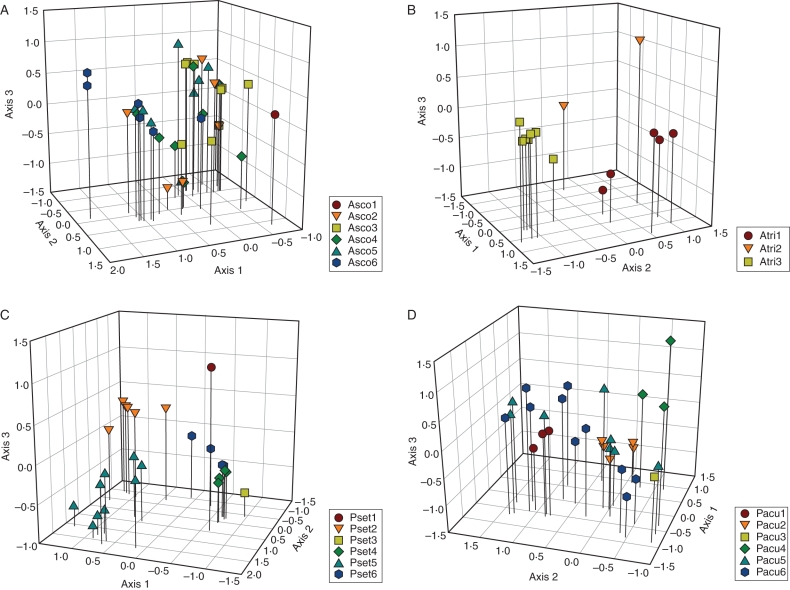
Results of PCAs showed a clear separation of individuals from different populations along the first three components in both A. trichomanes and P. setiferum (Fig. 2). The same was true for population Pacu1–Pacu4 in P. aculeatum, but populations Pacu5 and Pacu6 were more variable and contained several individuals that fell in between clusters of other populations. A more diffuse pattern is present in A. scolopendrium, although population Asco1 formed a separate and monomorphic group and the other individuals were loosely separated into two groups along the first axis, with individuals from all populations except Asco6 divided over the two groups. This pattern in A. scolopendrium was largely supported by the results of Bayesian cluster analysis (Fig. 1), which suggested K = 3 as the best-supported number of gene pools. Populations Asco1 and Asco6 were largely assigned to single gene pools, while the other populations contained a mixture of individuals assigned to different pools. Most individuals were assigned to a single gene pool (i.e. low admixture). In the other three species, cluster analysis assigned populations, with some exceptions, almost entirely to separate pools without admixture (Fig. 1). In A. trichomanes subsp. quadrivalens the number of pools equalled the number of populations. Two individuals of population Atri2 were assigned to the gene pool associated with population Atri1; two admixed individuals were found in Atri1. In both polystichoids the inferred number of pools was four. In P. setiferum, the small population Pset4 was assigned to the same gene pool as the nearby population Pset3. Population Pset6, the only population with a heterozygous founder (Table 1), showed differentiation between individuals and the presence of admixed genotypes. In accordance with PCA results, Bayesian inference showed various individuals with admixed ancestry from all four identified pools in population Pacu5 and Pacu6.
Fig. 2.
Principal components analysis (PCA) based on presence data per allele per locus. Position along the first three principle components is plotted for each genotyped individual. Note that individuals with the same multilocus genotype overlap each other in the plot. Different colours represent different populations. (A) Asplenium scolopendrium, (B) Asplenium trichomanes, (C) Polystichum setiferum, (D) Polystichum aculeatum. The cumulative percentage of variation explained by the first three components was 48·5, 83·5, 66·9 and 59·4 %, respectively, for the four species.
Within-population genetic variation and heterozygosity
Both allelic variability (A and Ar) and expected heterozygosities (He) were low in most populations in all four species (Table 2). In two populations of P. setiferum and one population of both P. aculeatum and A. scolopendrium, only a single multilocus genotype was found among the samples (Ng = 1). The remaining populations of all species, except those of A. scolopendrium and Pacu5 and Pacu6 of P. aculeatum, lacked variation at some loci (1–3). In all but one population of A. scolopendrium (Asco1), almost every sampled individual contained a unique multilocus genotype (Table 2). Observed heterozygosities were, however, low in both diploid species, resulting in high inbreeding coefficients (F). Considerable inbreeding was also detected in all but one population of P. setiferum. Strong outbreeding (F << 1) was detected in population Pset6. Due to unknown allele dosages, Ho and thereby F, remained ambiguous in the polyploid species.
Founding individuals
Based on inventory data, founder plants could be identified for four populations in both polystichoids. All founder plants of the diploid P. setiferum were completely homozygous at the tested loci, except for the founder of population Pset6. In one population of the tetraploid P. aculeatum (Pacu3), the founder plant showed only a single allele at all four loci. In three other populations (Pacu1, Pacu2 and Pacu4) the founder had two alleles at some loci and heterozygosity thus could not be excluded. However, in these populations the same combination of alleles found for the founder was also present in most other sampled individuals, suggesting that this individual still represents a homozygous genotype (with different alleles located at homoeologous chromosomes). In both species, the multilocus genotype of the founder plants was the most abundant haplotype in its population and a majority of the sampled individuals per population contained the full haplotype of the founder (Table 3).
Table 3.
The percentage of sampled individuals per population containing the full haplotype of the founder individual, for all populations of which the founder was known
| Species | Population | Percentage | N |
|---|---|---|---|
| POLS | Pset1 | 100 % | 10 |
| Pset2 | 67 % | 12 | |
| Pset4 | 50 % | 4 | |
| Pset6 | 80 % | 5 | |
| POLA | Pacu1 | 100 % | 7 |
| Pacu2 | 92 % | 12 | |
| Pacu3 | 100 % | 11 | |
| Pacu4 | 90 % | 10 |
N, total number of sampled individuals; POLS, Polystichum setiferum; POLA, Polystichum aculeatum. Population acronyms are as in Table 2.
Isolation by distance
Pairwise GST values were mostly high across species (Supplementary Data Table S2). In both polystichoids a much lower GST and higher QS was found for a single pair of populations located in adjacent trenches (Pset3–Pset4 and Pacu5–Pacu6, geographical distance ≤40 m). No significant isolation by distance (IBD) effects were found based on pairwise estimates of GST (Table 4). Mantel tests based on pairwise Sörensen similarity indices, however, did show strongly significant IBD in P. aculeatum (P = 0·003). This effect in P. aculeatum was mainly driven by a strong overlap in alleles between the nearby populations A5 and A6, but was still significant (P = 0·014) when these populations were merged.
DISCUSSION
Strong population differentiation due to multiple colonization events
While low immigration rates are generally expected to result in genetically differentiated populations with low levels of genetic variation within populations (Schaal and Leverich, 1996), the actual genetic signature of populations in isolated habitats will be very much dependent on their colonization history (Pannell and Dorken, 2006). The establishment of multiple spatially separated populations in the Kuinderbos could have resulted from two different scenarios. If arrival of spores by LDD was extremely rare, long-distance colonization might have occurred only once (or very rarely) and might have been followed by secondary founding via local dispersal. This would imply that local gene flow is possible and that local populations have a shared ancestry, resulting in limited spatial genetic structure. The fact that for our study species several populations established more or less simultaneously in different parts of the forest since its plantation in around 1950 (Table 2) suggests a different scenario, in which LDD was sufficiently common to allow multiple independent colonization events. As long as genotypic diversity among the arriving colonizers is sufficiently high, this scenario would initially result in a high overall genetic diversity and strong spatial genetic structure (strong population differentiation but no IBD). All our analyses consistently showed this pattern and indicated that all four studied species indeed colonized the Kuinderbos by means of multiple independent colonizations. However, the fact that such strong genetic evidence for this particular colonization history can still be shown in the current populations is surprising, even given their young age, as this particular signature of colonization is easily erased by subsequent gene flow between populations (Pannell and Dorken, 2006). Apparently, local (meta-)population dynamics (a combination of inbreeding and limited dispersal) have strongly restricted the possibilities for gene flow over the last decades. It should be noted, however, that only a limited number of generations has passed since colonization.
The observed pattern of low diversity per population and high overall genetic diversity and differentiation among populations in the Kuinderbos thus most probably was determined and maintained by a combination of factors, including a rich diversity among arriving colonizers, limited time since population establishment, and limited local gene flow due to high selfing and/or limited local dispersal. However, the exact importance of these factors varied both between populations and between species. Below, their separate and combined effects on fern population genetic patterns in isolated habitats will be discussed in more detail.
Regular arrival of diverse spores by long-distance dispersal
Given the large distances to possible spore sources, spore rains reaching the Kuinderbos can be expected to be sparse for all four study species, and especially for the polystichoids. However, the fact that all study species, except for A. trichomanes subsp. quadrivalens, established a large number of populations via independent dispersal events within a few decades shows that considerable numbers of spores reached the Kuinderbos within a period of 70 years maximum (starting when the sea bottom soil was drained). The same is suggested by a study on the young soil spore banks in the Dutch polders, which showed low dispersal limitation and the arrival of various very rare species (G. A. De Groot, unpubl. res.). This might mean that dispersal over large distances might be a less rare event than is generally appreciated, a suggestion that was also recently made by Shepherd et al. (2009) and Perrie et al. (2010) based on evidence for repeated dispersal of Asplenium hookerianum over hundreds of kilometres from New Zealand to two remote oceanic islands.
Interestingly, spores not only arrived in considerable numbers, but were also genetically diverse. This was shown by overall high expected heterozygosities (HT), the presence of a remarkably high part of the European allelic diversity in the Kuinderbos (high AKB/AEUR) and the fact that Bayesian clustering analysis indicated separate population founders to be of clearly distinct origin. The observed high allelic diversity could be thought of as resulting from the ‘inverse isolation effect’ (Sundberg, 2005), a hypothesis predicting an increase in spore rain diversity with increasing isolation, as a result of the fact that the majority of spores reaching isolated habitats originate from a large number of source populations, while less isolated habitats are swamped by spores from a few nearby sources. Species differences in the amount of European diversity present in the Kuinderbos are, however, in clear contrast to this theory. Values for AKB/AEUR were highest for A. scolopendrium, for which the nearest source populations are located approx. 30 km away, while being lower for P. setiferum and P. aculeatum, which must have dispersed over much larger distances (100–250 km; Bremer, 2007). This pattern can be explained by a lack of geographic structure in genetic variation at a European scale, as Sundberg's (2005) theory can only be valid if an increased source area results in a larger total diversity among source populations. In case large-scale phylogeographic structure is limited in A. scolopendrium, the ‘inverse isolation effect’ does not apply as a few nearby sources may show an equally high variation as a combination of many distant sources. Detailed studies on European fern phylogeography might help to resolve this issue.
Limited local dispersal
Our results indicate low genetic mixing between relatively nearby populations in the first decades after colonization. We found no indications for isolation by distance in three of the four species. This result is, at least for P. setiferum and A. trichomanes subsp. quadrivalens, most likely to be due to limited gene flow between populations with independent genetic histories. Fern dispersal kernels are known to be strongly leptokurtic (Penrod and McCormick, 1996), and a very low wind speed in the forest understorey and especially in the deep trenches inhabited by the study species will have hampered local dispersal even further (Bremer, 2007). Occasional dispersal seems to have occurred, but mainly to populations in the same trench or directly adjacent trenches. Both in P. setiferum and in P. aculeatum, we found some evidence of such short-distance dispersal, as in both species a strong genetic similarity was found between two populations located only tens of metres apart in adjacent trenches. In both cases, successful dispersal from one population might have resulted in the establishment of the other. While the absence of evidence for IBD is often interpreted as an indication for strong mixing between populations (e.g. Pálsson, 2004), here we show that such a relationship should not be drawn at face value. The age of a population and other ecological factors are of vital importance in interpreting this pattern.
Some indications for correlations between geographic and genetic distances were found for A. scolopendrium and P. aculeatum. Evidence of gene flow was especially strong for A. scolopendrium, which showed both a mixture of individuals of different genetic origin within populations and the occasional presence of recombinant individuals. These higher migration rates may result from the species' 10- to 500-fold larger local abundance compared with the other species. Populations are both larger and more abundant, which currently results in merging of populations within trenches and a high chance that adjacent trenches contain sporulating individuals. Stepwise migration via populations in between the studied populations may therefore explain the observed gene flow, especially in the north-western part of the forest, where the species is historically most abundant (Fig. 1A). Polystichum aculeatum, the second most abundant of these four species in the Kuinderbos, has a distribution similar to A. scolopendrium (Fig. 1D) and shows a comparable pattern of lower pairwise genetic distances between populations in the north-western part.
Bayesian analysis suggested a few individuals in two populations of A. trichomanes subsp. quadrivalens to result from dispersal between local populations. Their actual multilocus genotypes are, however, not a mixture of genotypes from any combination of the three populations present. Most probably, genetic variation in these populations must be attributed to the arrival of additional genotypes via LDD. A single, genetically distinct, LDD-based immigrant will, however, not easily be detected by clustering analysis, but rather will be assigned to one of the more common gene pools. Previous studies on A. trichomanes and other rock-dwelling asplenioids also suggested genetic variation within populations to be the result of LDD rather than local migration (Schneller and Holderegger, 1996; Suter et al., 2000; Pryor et al., 2001). The STRUCTURE algorithm is, however, unable to detect such sole representatives of additional gene pools, leading to an underestimation of K.
The detection of genetic variation in the Kuinderbos populations of A. trichomanes subsp. quadrivalens is, however, in marked contrast to previous results from a pilot study using allozymes, carried out at the Natural History Museum in London, which suggested all variation to be allocated among the three populations (G. A. de Groot and J. C. Vogel, unpubl. res). Our and other data (Pryor et al., 2001; Woodhead et al., 2005) show that previous studies using allozymes or AFLPs will have underestimated migration rates and levels of genetic variation.
Mating systems during and after colonization
Polyploid rock-dwelling ferns often show high levels of inbreeding and low genetic variation, a pattern attributed to their abilities for single-spore establishment by means of intragametophytic selfing (Schneller and Holderegger, 1996; Vogel et al., 1999a, b; Suter et al., 2000). In contrast, many diploids are outbreeders, harbouring significantly higher levels of genetic variation within populations (Soltis and Soltis, 1988; Hunt et al., 2009). Spatial genetic structure has been detected for some outcrossing rock-dwelling ferns, but this was attributed to either the patchiness of their habitat or a limited availability of safe sites for germination (Soltis et al., 1989). Both cause differentiation by non-random mating as a result of spatial aggregation of individuals over multiple separate patches. We found, however, strong population differentiation even in recently founded populations of two reportedly outcrossing diploids, a pattern most likely to be due to their colonization history rather than subsequent genetic drift. Especially in the diploid P. setiferum, most populations were (almost) completely monomorphic. Even though this species has previously been described as an obligate outcrosser (Pangua et al., 2003), this pattern is best explained by single-spore colonization via intragametophytic selfing. Genotyping of known founder plants supports this theory: all except one were homozygous at all loci scored. The only population of P. setiferum which contained a heterozygous founder also showed a low inbreeding coefficient and mostly heterozygous offspring. Apparently, even diploid ferns generally found to be outcrossing may show intraspecific variation in selfing potential, as previously observed in A. scolopendrium (Wubs et al., 2010), although obligate outcrossers may exist (Haufler and Soltis, 1984). Recent breeding experiments with gametophytes derived from Kuinderbos populations showed high capacities for intragametophytic selfing for all four species (De Groot et al., 2012). Mate limitation during long-distance colonization will have strongly limited establishment by means other than through intragametophytic selfing, leading to strong selection for any extant selfing genotypes and a frequent occurrence of inbreeding in populations (e.g. Flinn, 2006).
In the absence of genetic variation in a population, self- and cross-fertilization will both lead to inbred offspring (e.g. Keller and Waller, 2002). High inbreeding coefficients (F) can thus be the result of a mating system driven by intragametophytic selfing, as well as of a lack of genetic variation. Genetic isolation and small population size may lead to strong founder effects, and thereby a functionally inbreeding population. This effect is especially strong in fern populations founded by intragametophytic selfing, even if the founding genotype has a mixed mating system. In such cases, the founder plant is completely homozygous and outcrossing rates are thus initially dependent on the influx of new genotypes (Wubs et al., 2010). Inbreeding coefficients are high in most studied populations in the Kuinderbos, indicating low rates of outcrossing. Drawing conclusions about the ratios between intra- and intergametophyic selfing and thus the intrinsic mating strategy of the species and genotypes involved is, however, very difficult. As mate limitation is common in these ferns, rates of self-fertilization might remain high as long as the founding genotype, capable of intragametophytic selfing, remains dominant in the population.
Even in populations of A. scolopendrium, which showed higher levels of mixing and genetic variation, inbreeding coefficients were remarkably high (0·448–0·740). The lack of heterozygotes, combined with the high numbers of multilocus genotypes within populations, suggests that the variation in these populations is largely due to input of new genotypes, which subsequently reproduce by inbreeding instead of mixing (Pryor et al., 2001). Although Wubs et al. (2010) showed that many genotypes of A. scolopendrium have a mixed mating system and produce more sporophytes by outcrossing than by selfing, our results suggest that outcrossing has until now been very limited in the local populations. This pattern may partly be an effect of the limited number of generations that has passed, but can also be expected for ferns with a mixed mating system, as long as mate limitation is sufficiently high and/or local conditions are sufficiently benign to allow the survival of inbred individuals with a somewhat lower fitness (Wubs et al., 2010). Based on the low numbers of multilocus genotypes, populations of the other study species might experience even higher genetic isolation and inbreeding rates.
Interspecific variation in colonization success
The putative repeated population establishment through intragametophytic selfing indicates a regular arrival of genotypes with a low genetic load in all four studied species. This suggests that in many ferns, including outcrossing taxa, mating strategies do not limit the colonization of new habitat patches, something which even seems to be the case under the constraint of LDD. A filtering effect for selfing genotypes may reduce the incidence of successful colonization events, depending on the intraspecific variation in mating strategies and the abundance and genetic diversity of the arriving spores, and this may affect the speed of colonization and the local numbers of plants. However, while overall high capabilities for rapid establishment in temporarily available micro-sites suitable for establishment (rates of germination and juvenile growth, breeding system) are strong determinants of local success (Flinn, 2007), we hypothesize that a greater versatility in habitat preferences explains the relative success of some species in the Kuinderbos. Although still considered to be a strong habitat specialist, only colonizing habitats that provide shade, moisture and high pH, in north-western Europe A. scolopendrium is regularly found in deciduous woodlands, on stream banks and hedge banks, and along hollow/sunken roads (Hegi, 1965; Page, 1997). This may explain the relatively high speed at which this species has been able to colonize the sandy soils of the Kuinderbos trench banks. The same ability to colonize soil substrates is to some extent found in P. setiferum and P. aculeatum. Soil-dwelling individuals of these three species have been reported from various locations in The Netherlands. Apart from the Kuinderbos, we do know, however, of only one such record for A. trichomanes subsp. quadrivalens (Willems, 2005). All other reported examples of soil-dwelling individuals of A. trichomanes concerned a different subspecies, A. trichomanes subsp. trichomanes. Although A. trichomanes subsp. quadrivalens occurs on calcareous and siliceous substrates with a broad range in pH, it grows, unlike its relative, almost exclusively on steep or vertical rock surfaces (Bouharmont, 1968; Rasbach et al., 1991). In Europe, its occurrence outside the mountainous areas where such habitats occur is mainly due to its ability to colonize the mortar of old walls effectively (e.g. Lovis, 1955). Future experiments should clarify whether variation in habitat preferences among genotypes of A. trichomanes subsp. quadrivalens can explain the fact that a few genotypes were able to colonize the drainage trenches of the Kuinderbos. Such variation is not unlikely, as the allopolyploid subspecies quadrivalens probably originated multiple times, in different parts of Europe (Vogel, 1995). The various hybridization events of its progenitors (A. trichomanes subsp. trichomanes and A. trichomanes subsp. inexpectans) will probably have involved different genotypes of one or both species (Vogel, 1995), resulting in different inbred lineages of A. trichomanes subsp. quadrivalens with potentially slightly different habitat requirements.
Conclusions
The ability of ferns to disperse their spores over huge distances has long been known (Tryon, 1970), but the frequency of immigration by LDD, and how this varies between species and with distance from the spore source, has rarely been studied. Our results for four different species consistently show that dispersal over large distances is not a rare event. Isolated habitat patches may regularly receive spores from multiple distant sources, allowing the establishment of considerable genotypic diversity within a few decades after habitat creation. This pattern of long-distance colonization results, however, in a very patchy spatial distribution consisting of various small clusters of individuals, with a very distinct genetic signature in which all variation is partitioned among clusters. These results are consistent with general expectations for peripheral populations (Arnaud-Haond et al., 2006; Eckert et al., 2008) and the results of previous studies on rock-dwelling populations of A. trichomanes subsp. quadrivalens and other polyploid rock ferns (Schneller and Holderegger, 1996, Vogel et al., 1999b, Suter et al., 2000, Pryor et al., 2001) and attributed to multiple events of single-spore colonization via intragametophytic selfing. We show, however, that this scenario and the resulting genetic pattern can occur across species that differ in ploidy and dominant mating system, rather than being limited to polyploid selfers. Apparently, even some diploid species with a high proportion of outcrossing genotypes are capable of frequent single-spore colonization and show this capability in isolated habitats. Moreover, we show that the resulting genetic pattern not only occurs in very heterogeneous habitats (i.e. the patchy habitats normally inhabited by rock ferns), but can occur even in large uniform strips of suitable habitat as exist in the Kuinderbos. Thus, genetic differentiation among isolated populations may result from their effective dispersal and broad range of mating strategies alone, instead of necessarily being driven by their patchy habitats. Limited gene flow can conserve the genetic signature of multiple colonization events for decades. This is an important consideration for conservation of genetic diversity, as extinction of a single population (e.g. by habitat destruction) will result in a considerable loss of genetic diversity. In contrast to mountainous habitats, this pattern may, however, be easily erased over time in uniform habitats like the Kuinderbos, as ongoing establishment in between the original clusters increases the potential for (stepwise) gene flow. Increased opportunities for outcrossing and mixing between gene pools may primarily be a result of increasing local abundances, rather than being a main determinant of these abundances. Nevertheless, increased outcrossing rates will boost local abundances even further for species in which most genotypes have mixed or outcrossing mating systems.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We wish to thank Staatsbosbeheer for allowing us to work on their terrains. Sampling within The Netherlands was performed by G.A.G. under licence number FF/75A/2009/066a (Ministry of Economic Affairs, Agriculture and Innovation). This work was supported by: the Miquel Fund (Utrecht University, to G.A.G.); the Schure-Beijerinck-Popping Fund (KNAW, to G.A.G.); the Innovational Research Incentives Scheme (VENI grant number 863·09·017 to R.H.J.E.) of the Netherlands Organisation for Scientific Research (NWO).
LITERATURE CITED
- Arnaud-Haond S, Teixeira S, Massa SI, et al. Genetic structure at range edge: low diversity and high inbreeding in Southeast Asian Mangrove (Avicennia marina) populations. Molecular Ecology. 2006;15:3515–3525. doi: 10.1111/j.1365-294X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- Barton NH. Adaptation at the edge of a species' range. In: Silvertown J, Antonovics J, editors. Integrating ecology and evolution in a spatial context. Oxford: Blackwell Science; 2001. pp. 365–392. [Google Scholar]
- Bohrer G, Nathan R, Volis S. Effects of long-distance dispersal for metapopulation survival and genetic structure at ecological time and spatial scales. Journal of Ecology. 2005;93:1029–1040. [Google Scholar]
- Bouharmont J. Les forms chromosomiques d' Asplenium trichomanes L. Bulletin du Jardin botanique national de Belgique. 1968;38:103–114. [Google Scholar]
- Bremer P. The ferns of the Kuinderbos (The Netherlands), the establishment of 23 species in a planted forest. Acta Botanica Neerlandica. 1980;29:351–357. [Google Scholar]
- Bremer P. Flora, vegetatie en bosverjonging in het Kuinderbos. Driebergen: Staatsbosbeheer; 1994. [Google Scholar]
- Bremer P. The colonisation of a former sea-floor by ferns. The Netherlands: Wageningen University; 2007. PhD thesis. [Google Scholar]
- Broquet T, Yearsley J, Hirzel AH, Goudet J, Perrin N. Inferring recent migration rates from individual genotypes. Molecular Ecology. 2009;18:1048–1060. doi: 10.1111/j.1365-294X.2008.04058.x. [DOI] [PubMed] [Google Scholar]
- Cain ML, Milligan BG, Strand AE. Long-distance seed dispersal in plant populations. American Journal of Botany. 2000;87:1217–1227. [PubMed] [Google Scholar]
- Crawford NG. SMOGD: software for the measurement of genetic diversity. Molecular Ecology Resources. 2010;10:556–557. doi: 10.1111/j.1755-0998.2009.02801.x. [DOI] [PubMed] [Google Scholar]
- De Groot GA, Erkens RHJ, During HJ. WODAN: de invloed van verspreidings- en vestigingsmogelijkheden op het ontstaan van biodiversiteit. Gorteria. 2008;33:59–60. [Google Scholar]
- De Groot GA, Korpelainen H, Wubs ERJ, Erkens RHJ. Isolation of polymorphic microsatellite markers and tests of cross-amplification in four widespread European calcicole ferns. American Journal of Botany. 2011;98:e319–e322. doi: 10.3732/ajb.1100051. [DOI] [PubMed] [Google Scholar]
- De Groot GA, Verduyn B, Wubs ERJ, Erkens RHJ, During HJ. Inter- and intraspecific variation in fern mating systems after long-distance colonization: the importance of selfing. BMC Plant Biology. 2012;12 doi: 10.1186/1471-2229-12-3. doi:10.1186/1471-2229-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. Genetic variation across species' geographical ranges: the central–marginal hypothesis and beyond. Molecular Ecology. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- El Mousadik A, Petit RJ. High levels of genetic differentiation for allelic richness among populatoins of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theoretical and Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Eriksson O. Regional dynamics of plants: a review of evidence for remnant, source–sink and metapopulations. Oikos. 1996;77:248–258. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of cluster of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feekes W, Bakker D. De ontwikkeling van de natuurlijke vegetatie in de Noordoostpolder. Van Zee tot Land. 1954;6:1–92. [Google Scholar]
- Fitter AH, Peat HJ. The Ecological Flora Database. Journal of Ecology. 1994;84:415–425. [Google Scholar]
- Flinn KM. Reproductive biology of three fern species may contribute to differential colonization success in post-agricultural forests. American Journal of Botany. 2006;93:1289–1294. doi: 10.3732/ajb.93.9.1289. [DOI] [PubMed] [Google Scholar]
- Flinn KM. Microsite-limited recruitment controls fern colonization of post-agricultural forests. Ecology. 2007;88:3103–3114. doi: 10.1890/06-2124.1. [DOI] [PubMed] [Google Scholar]
- FLORON. FLORBASE and FLORIVON floral databases for the Netherlands. Nijmegen: FLORON and Leiden: Nationaal Herbarium Nederland (NHN); 2010. [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov Chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (vers. 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Goudet J, Raymond M, De Meeüs T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haufler CH, Soltis DE. Obligate outcrossing in a homosporous fern: field confirmation of a laboratory prediction. American Journal of Botany. 1984;71:878–881. [Google Scholar]
- Hegi G. Illustrierte Flora von Mittel-Europa. 1965 München: Carl Hanser/Verlag. [Google Scholar]
- Hunt HV, Ansell W, Russell SJ, Schneider H, Vogel JC. Genetic diversity and phylogeography in two diploid ferns, Asplenium fontanum subsp. fontanum and A. petrarchae subsp. bivalens, in the western Mediterranean. Molecular Ecology. 2009;18:4940–4954. doi: 10.1111/j.1365-294X.2009.04402.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jiménez A, Holderegger R, Csencsins D, Quintanilla LG. Microsatellites reveal substantial among-population genetic differentiation and strong inbreeding in the relict fern Dryopteris aemula. Annals of Botany. 2010;106:149–155. doi: 10.1093/aob/mcq094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology and Evolution. 2002;17:230–241. [Google Scholar]
- Klekowski EJ. The genetics and reproductive biology of ferns. In: Dyer AF, editor. The experimental biology of ferns. London: Academic Press; 1979. pp. 133–170. [Google Scholar]
- Lande R, Shannon S. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution. 1996;50:434–437. doi: 10.1111/j.1558-5646.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- Lovis JD. The problem of Asplenium trichomanes. In: Lousley JE, editor. Species studies in the British Flora. London: The Botanical Society of the British Isles; 1955. pp. 99–103. [Google Scholar]
- Maguire TL, Saenger P, Baverstock P, Henry R. Microsatellite analysis of genetic structure in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) Molecular Ecology. 2000;9:1853–1862. doi: 10.1046/j.1365-294x.2000.01089.x. [DOI] [PubMed] [Google Scholar]
- Masuyama S, Watano Y. Trends for inbreeding in polyploid pteridophytes. Plant Species Biology. 1990;5:13–17. [Google Scholar]
- McCune B, Mefford MJ. PC-ORD. Multivariate analysis of ecological data. Oregon: MjM Software; 1999. Version 5·0. [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Molecular Ecology Resources. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Annals of Human Genetics. 1983;47:253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x. [DOI] [PubMed] [Google Scholar]
- Nordal I. Tabula rasa after all? Botanical evidence for ice-free refugia in Scandinavia reviewed. Journal of Biogeography. 1987;14:377–388. [Google Scholar]
- Page CN. The ferns of Britain and Ireland. 2nd edn. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- Pálsson S. Isolation by distance, based on microsatellite data, tested with spatial autocorrelation (SPAIDA) and assignment test (SPASSIGN) Molecular Ecology Notes. 2004;4:143–145. [Google Scholar]
- Pangua E, Quintanilla LG, Sancho A, Pajarón S. A comparative study of the gametophytic generation in the Polystichum aculeatum group (Pteridophyta) International Journal of Plant Sciences. 2003;164:295–303. [Google Scholar]
- Pannell JR, Dorken ME. Colonization as a common denominator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landscape Ecology. 2006;21:837–848. [Google Scholar]
- Peck JH, Peck CJ, Farrar DR. Influences of life history attributes on formation of local and distant fern populations. American Fern Journal. 1990;80:126–142. [Google Scholar]
- Penrod KA, McCormick LH. Abundance of viable hay-scented fern spores germinated from hardwood forest soils at various distances from a source. American Fern Journal. 1996;86:69–79. [Google Scholar]
- Perrie LR, Ohlsen DJ, Shepherd LD, Garrett M, Brownsey PJ, Bayly MJ. Tasmanian and Victorian populations of the fern Asplenium hookerianum result from independent dispersal from New Zealand. Australian Systematic Botany. 2010;23:387–392. [Google Scholar]
- Pritchard JK, Stephens M, Donelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor KV, Young JE, Rumsey FJ, Edwards KJ, Bruford MW, Rogers HJ. Diversity, genetic structure and evidence of outcrossing in British populations of the rock fern Adiantum capillus-veneris using microsatellites. Molecular Ecology. 2001;10:1881–1894. doi: 10.1046/j.1365-294x.2001.01343.x. [DOI] [PubMed] [Google Scholar]
- Ranker TA, Geiger JMO. Population genetics. In: Ranker TA, Haufler CH, editors. Biology and evolution of ferns and lycophytes. Cambridge: Cambridge University Press; 2008. pp. 107–133. [Google Scholar]
- Rasbach H, Rasbach K, Reichstein T, Bennert HW. Asplenium trichomanes subsp. coriaceifolium, a new subspecies and two new intra-specific hybrids of the A. trichomanes complex (Aspleniaceae, Pteridophyta). II. Description and illustrations. With an appendix on pairing behaviour of chromosomes in fern hybrids. Wildenowia. 1991;21:239–261. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Schaal BA, Leverich WJ. Molecular variation in isolated plant populations. Plant Species Biology. 1996;11:33–40. [Google Scholar]
- Schneller JJ, Holderegger R. Genetic variation in small, isolated fern populations. Journal of Vegetation Science. 1996;7:113–120. [Google Scholar]
- Shepherd LD, De Lange PJ, Perrie LR. Multiple colonizations of a remote oceanic archipelago by one species: how common is long-distance dispersal? Journal of Biogeography. 2009;36:1972–1977. [Google Scholar]
- Soltis DE, Soltis PS. The distribution of selfing rates in homosporous ferns. American Journal of Botany. 1992;79:97–100. [Google Scholar]
- Soltis PS, Soltis DE. Genetic variation and population genetic structure in the fern Blechnum spicant (Blechnaceae) from western North America. American Journal of Botany. 1988;75:37–44. [Google Scholar]
- Soltis PS, Soltis DE, Ness BD. Population genetic structure in Cheilanthes gracillima. American Journal of Botany. 1989;76:1114–1118. [Google Scholar]
- Sundberg S. Larger capsules enhance short-range spore dispersal in Sphagnum, but what happens further away? Oikos. 2005;108:115–124. [Google Scholar]
- Suter M, Schneller JJ, Vogel JC. Investigations into the genetic variation, population structure, and breeding systems of the fern Asplenium trichomanes subsp. quadrivalens. International Journal of Plant Sciences. 2000;161:233–244. doi: 10.1086/314258. [DOI] [PubMed] [Google Scholar]
- Tryon AF. Origin of the fern flora of Tristan de Cunha. British Fern Gazette. 1966;9:269–276. [Google Scholar]
- Tryon RM. Development and evolution of fern floras of oceanic islands. Biotropica. 1970;2:76–84. [Google Scholar]
- Van der Meijden R. Heukels' Flora van Nederland. Groningen: Wolters-Noordhoff; 2005. [Google Scholar]
- Van Puyvelde K, Van Geert A, Triest L. ATETRA a new software program to analyse tetraploid microsatellite data: comparison with TETRA and TETRASAT. Molecular Ecology Resources. 2010;10:331–334. doi: 10.1111/j.1755-0998.2009.02748.x. [DOI] [PubMed] [Google Scholar]
- Vogel JC. Multiple origins of polyploids in European Asplenium (Pteridophyta) UK: Cambridge University; 1995. PhD thesis. [Google Scholar]
- Vogel JC, Rumsey FJ, Russell SJ, et al. Genetic structure, reproductive biology and ecology of isolated populations of Asplenium csikii (Aspleniaceae, pteridophyta) Heredity. 1999a;83:604–612. doi: 10.1038/sj.hdy.6886120. [DOI] [PubMed] [Google Scholar]
- Vogel JC, Rumsey FJ, Schneller JJ, Barrett JA, Gibby M. Where the glacial refugia in Europe? Evidence from pteridophytes. Biological Journal of the Linnean Society. 1999b;66:23–37. [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration. Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. FST doesn't equal 1/(4Nm+1) [DOI] [PubMed] [Google Scholar]
- Willems JH. Een groeiplaats van steenbreekvaren op de grond in Zuid-Limburg. Natuurhistorisch Maandblad. 2005;94:269–270. [Google Scholar]
- Wolf PG, Schneider H, Ranker TA. Geographic distributions of homosporous fern taxa: does dispersal obscure evidence of vicariance? Journal of Biogeography. 2001;18:263–270. [Google Scholar]
- Woodhead M, Russell J, Squirrell J, et al. Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Molecular Ecology. 2005;14:1681–1695. doi: 10.1111/j.1365-294X.2005.02543.x. [DOI] [PubMed] [Google Scholar]
- Wubs ERJ, De Groot GA, During HJ, et al. Mixed mating system in the fern Asplenium scolopendrium: implications for colonization potential. Annals of Botany. 2010;106:583–590. doi: 10.1093/aob/mcq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.