Abstract
Background and Aims
Nothoscordum gracile is an apomitic tetraploid widely distributed throughout the Americas and naturalized in many temperate regions of other continents. It has been suggested to form a species complex with sexual and apomictic N. nudicaule and N. macrostemon. Tetraploids of these species also share a structurally heterozygous chromosome complement 2n = 19 (13M + 6A). In this work, the origin of N. gracile and its relationships with its related species was investigated based on cytological and molecular data.
Methods
Cytogenetic analyses were based on meiotic behaviour, CMA bands, localization of 5S and 45S rDNA sites, and genomic in situ hybridization (GISH). Nuclear ITS and plastidial trnL-trnF sequences were also obtained for most individuals.
Key Results
Proximal CMA bands were observed in the long arms of all acrocentrics of 2x and 4x N. macrostemon but not in diploid and some tetraploid cytotypes of N. nudicaule. Samples of N. gracile showed a variable number of CMA bands in the long arms of acrocentrics. Analysis of ITS sequences, dot-blot, GISH, and 5S and 45S rDNA sites, revealed no differentiation among the three species. The trnL-trnF cpDNA fragment showed variation with a trend to geographical structuring irrespective of morphospecies and fully congruent with karyotype variation.
Conclusions
The 2n = 19 karyotype was probably formed by a centric fusion event occurring in N. nudicaule and later transmitted to tetraploid cytotypes of N. macrostemon. Diploids of N. nudicaule and N. macrostemon appeared as consistent recently diverged species, whereas tetraploid apomicts seem to constitute an assemblage of polyploid hybrids originating from multiple independent hybridization events between them, part of which are morphologically recognizable as N. gracile.
Keywords: Nothoscordum gracile, CMA bands, rDNA sites, haplotype network, Robertsonian translocations
INTRODUCTION
Taxonomical decisions in apomictic complexes may particularly affect the outcome of evolutionary and phylogenetic studies. The level of intraspecific variability is dependent on the circumscription of the entity under study which, in extreme cases, may constitute a monoclonal apomictic microspecies (Gornall, 1999). The broader distribution of some asexual taxa relative to their sexual relatives is referred to as ‘geographic parthenogenesis’ (Vandel, 1928). The success of an apomictic clone is attributed to its ability to colonize new environments where homogenous abiotic conditions limit the spread of sexual relatives. New populations may be efficiently established from a single propagule, or by the indefinite propagation of a particular genotype that is able to adapt to a wide range of environmental conditions, and which is maintained due to the absence of recombination (reviewed in Bierzychudek, 1985; Verduijn et al., 2004). Range expansions are also attributed to the effect of polyploidy irrespective of breeding systems (Lowry and Lester, 2006). In polyploid/agamic complexes, the effects of asexual reproduction, hybridity and polyploidy on range expansion are confounded, given that frequently the polyploid components are in turn of hybrid origin and reproduce by apomixis (Bierzychudek, 1985; Hörandl, 2006).
The genus Nothoscordum Kunth (Gilliesioideae, Alliaceae) is native to South America and comprises >20 described species (Guaglianone, 1972; Crosa, 1996). Nothoscordum gracile (Aiton) Stearn, also referred to as N. inodorum (Aiton) G.Nicholson or N. fragrans (Vent.) Kunth (Stearn, 1986), is the only Nothoscordum species that is widely distributed throughout the Americas and naturalized in many temperate regions of other continents. It belongs to the section Inodorum Guag. and it is closely related morphologically and cytologically to N. nudicaule (Lehm.) Guagl. and N. macrostemon Kunth (Guaglianone, 1972; Nuñez et al., 1974), which have previously been classified as varieties of N. gracile (Guaglianone, 1972). Nuñez et al. (1974), based mainly on mitotic and meiotic chromosome analyses, concluded that these three species constitute a closely related group forming the Inodorum complex.
The chromosomes of Nothoscordum, as well as those of other Alliaceae taxa, are easily amenable to detailed cytogenetic analysis and may provide a wealth of information in evolutionary studies. All representatives of the section Inodorum have large metacentric (M) or acrocentric (A) chromosomes, some >20 µm long (Nuñez et al., 1974). The basic number of the section is x = 5 (3M + 2A) and the number of chromosome arms, or fundamental number (FN), is always eight or a multiple of eight. Diploids have 2n = 10 (6M + 4A) and FN = 16, and tetraploids display 2n = 19 (13M + 6A) or rarely 2n = 18 (14M + 4A), both with FN = 32 (Crosa, 1972, 1996). Dyer (1967) and Nuñez et al. (1974) observed seedlings with 2n = 20 obtained from seeds of individuals of N. gracile with 2n = 19, suggesting that the perfectly doubled karyotype is occasionally formed. This variation in chromosome numbers among tetraploids (2n = 18, 19 and 20) seems to be due to centric fission/fusion events, which frequently occur in the genus Nothoscordum (Crosa, 1996; Jones, 1998; Souza et al., 2009).
Nothoscordum gracile, N. macrostemon and N. nudicaule were initially known as strictly polyploid (Nuñez et al., 1974), but the finding of diploid cytotypes of the two latter species provided some insight into the evolutionary history of the complex Inodorum (Crosa, 1996). Meiotic, morphological and reproductive biology studies have confirmed that the diploid cytotypes of both taxa are allogamous and self-incompatible (Crosa, 1972, 1996). They are easily distinguishable from one another due to their morphological and phenological differences. Both display a more restricted geographical distribution than their respective tetraploid cytotypes, diploid N. nudicaule having a relictual distribution along the river banks of eastern Uruguay. On the other hand, tetraploids of N. macrostemon and N. nudicaule are facultatively apomictic, self-pollinating and more widespread, the former being the dominant species in the south of Uruguay, colonizing roadside lands and cultivated areas (Crosa, 1996).
Nothoscordum gracile is an invasive species found exclusively in disturbed habitats. It is a strictly tetraploid, facultative apomictic, showing high variability in the shapes of its ovaries, staminate filaments and tepals (Guaglianone, 1972). Some of these variant forms are intermediate between N. nudicaule and N. macrostemon, which led Ravenna (1978) to propose a hybrid origin for this species. The species N. arenarium Herter, included in section Inodorum, is strictly diploid (Crosa, 1972; Souza et al., 2009).
Cytologically, N. gracile is the most studied species of the genus, although the origin of the number 2n = 19 is still controversial. Nuñez et al. (1974) suggested that 2n = 19 arose from the union of a non-reduced gamete of a species with 2n = 10 with a reduced gamete of a species with 2n = 18. Alternatively, 2n = 19 could have arisen from the fusion of two acrocentric chromosomes of a normal tetraploid with 2n = 20 (12M + 8A) (Crosa, 1996). Nuñez et al. (1974) and Jones (1998) suggested that N. gracile could be an allotetraploid involving the two basic numbers of the genus, x = 4 (4M) and x = 5 (3M + 2A), with three sets of n = 5 and one of n = 4. Whatever its origin, the maintenance of this singular chromosome number is ensured by adventitious nucellar embryony and bulb offsets, as well as by sexual reproduction (Dyer, 1967).
Karyotypic analyses of N. gracile using chromosome C-banding techniques have shown small bands on the short arms of the six acrocentric chromosomes and in the proximal regions of the long arms of five of them, one of the proximal bands being duplicated. This same heterozygotic banding pattern was observed in plants from Japan (Kurita and Kuroki, 1963; Sato et al., 1979) and from Argentina (Canelada and Fernandez, 1985), suggesting the occurrence of a single widespread apomictic clone. The heterochromatin on the short arms of acrocentric chromosomes is associated with nucleolus organizer regions (NORs), as first reported in N. gracile by Levan and Emsweller (1938) and later confirmed by a number of other authors (see also Kurita and Kuroki, 1963; Sato et al., 1982; Canelada and Fernandez, 1985). The presence of 45S rDNA sequences on these short arms was first demonstrated by Crosa (1996) in N. nudicaule using fluorescent in situ hybridization (FISH). 45S rDNA sites were also observed on the short arms of the acrocentric chromosomes of N. arenarium, N. hirtellum (Kunth) Herter, N. felipponei Beauverd and N. pulchellum Kunth (Guerra and Felix, 2000; Souza et al., 2009, 2010).
C-banding in chromosomes of N. arenarium and N. pulchellum revealed exactly the same banding pattern observed after double staining with the fluorochromes chromomycin A3 (CMA) and 4',6-diamidino-2-phenylindole (DAPI) (Guerra and Felix, 2000; Souza et al., 2009). The heterochromatic bands, including the 45S rDNA sites, were always positively stained with CMA and negatively stained with DAPI (CMA bands). As the CMA/DAPI technique is simpler than C-banding, does not alter the chromatin structure, and the best slides can be reutilized for FISH analyses, it became the most suitable method for sequential banding and FISH analyses in these species.
The relationships between N. gracile and the other species of the Inodorum complex may be further investigated by the comparison of DNA sequences. Analyses of the ITS (intergenic transcribed spacer) nuclear region of the 45S rDNA of Allium species revealed divergences among species greater than those detected with other DNA sequences, suggesting that this region could be adequate for the evolutionary studies of other members in the family Alliaceae (Friesen et al., 2006; Nguyen et al., 2008; Li et al., 2010). Sequences of plastid DNA (cpDNA) are also important in phylogenetic analyses as they show uniparental inheritance. As a consequence, taxa of different ploidy levels can be included in the analyses irrespectively of the auto- or allopolyploid nature of the species involved, and their phylogenies can be directly inferred by conventional methods (Rua et al., 2010). The combined use of nuclear ITS and plastid sequences has allowed us to analyse the relationships among some of the representatives of Gilliesioideae (Fay et al., 2006).
The present work investigates the origin of N. gracile and its relationships with N. macrostemon and N. nudicaule, based on cytological data including chromosome number variability, meiotic behaviour, CMA bands, 5S and 45S rDNA sites, and genomic in situ hybridization (GISH). Additionally, evolutionary relationships between species and cytotypes were compared using ITS and trnL-trnF sequences and the N. gracile intraspecific variability was investigated searching for clonal diversification.
MATERIALS AND METHODS
Plant material
Eighty-five plants of Nothoscordum gracile, N. macrostemon and N. nudicaule collected from natural populations were analysed. A sample of N. arenarium was also used for comparison in the sequence analysis. The collection sites and numbers of all examined individuals are shown in Table 1. Vouchers were deposited in the Bernardo Rosengurtt Herbarium at the Facultad de Agronomía, Universidad de la República, Montevideo, Uruguay (MVFA). Plants of Ipheion uniflorum (Lindl.) Raf. (2n = 12) and I. recurvifolium (C.H.Wright) Traub (2n = 20) were used as controls in GISH and slot-blot experiments, respectively (see Souza et al., 2010 for details of samples and karyotypes).
Table 1.
Species cytologically investigated with provenance, accession number in the living collection, number of individuals analysed, chromosome number (2n) and number of acrocentric chromosomes without CMA bands (A0), with one CMA band (A1) or two CMA bands (A2)
| CMA bands |
||||||||
|---|---|---|---|---|---|---|---|---|
| Species | Provenance (locality, state/department/province, country) | Accession no. | No. of individuals | 2n | Karyotype formulae | A0 | A1 | A2 |
| Nothoscordum gracile (Aiton) Stearn | Blumenau, Santa Catarina, Brazil | GRA1832 | 2 | 19 | 13M + 6A | 1 | 4 | 1 |
| Botucatu, São Paulo, Brazil | GRA1817 | 2 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Buenos Aires, Argentina | GRA1849 | 3 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Gramado, Rio Grande do Sul, Brazil | GRA1718 | 1 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Lima, Peru | GRA1813 | 3 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Londrina, Paraná, Brazil | GRA1846 | 3 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Montevideo, Montevideo, Uruguay | GRA1851 | 1 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Passo Fundo, Rio Grande do Sul, Brazil | GRA1843 | 5 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Santa Maria, Rio Grande do Sul, Brazil | GRA1845 | 6 | 19 | 13M + 6A | 1 | 4 | 1 | |
| São Paulo, São Paulo, Brazil | GRA1706 | 4 | 19 | 13M + 6A | 1 | 4 | 1 | |
| Lavras, Minas Gerais, Brazil | GRA1770 | 3 | 19 | 13M + 6A | 3 | 3 | – | |
| Londrina, Paraná, Brazil | GRA1716 | 10 | 19 | 13M + 6A | 3 | 3 | – | |
| Piracicaba, São Paulo, Brazil | GRA1848 | 2 | 19 | 13M + 6A | 3 | 3 | – | |
| Tucumán, Tucumán, Argentina | GRA1847 | 3 | 19 | 13M + 6A | 3 | 3 | – | |
| Maldonado, Uruguay | GRA1711 | 4 | 18 | 14M + 4A | – | 4 | – | |
| N. macrostemon Kunth | Ruta 5, Rivera, Uruguay | MAC1391 | 1 | 10 | 6M + 4A | – | 4 | – |
| Ruta 9, Arroyo Solís Grande, Canelones, Uruguay | MAC1358 | 4 | 10 | 6M + 4A | – | 4 | – | |
| Ruta 9, Arroyo Solís Grande, Canelones, Uruguay | MAC1712 | 2 | 19 | 13M + 6A | – | 6 | – | |
| N. nudicaule (Lehm.) Guagl. | Uruguay | NUD1082 | 1 | 10 | 6M + 4A | 4 | – | – |
| Ruta 7 Km 323, Durazno, Uruguay | NUD1392 | 1 | 10 | 6M + 4A | 4 | – | – | |
| Ruta 8, Arroyo Convoy, Treinta y Tres, Uruguay | NUD1363 | 1 | 10 | 6M + 4A | 4 | – | – | |
| Ruta 8, Arroyo Otazo, Treinta y Tres, Uruguay | NUD1361 | 1 | 10 | 6M + 4A | 4 | – | – | |
| Paso Aguiar, Rio Negro, Tacuarembó, Uruguay | NUD1386 | 1 | 10 | 6M + 4A | 4 | – | – | |
| Bella Unión, Artigas, Uruguay | NUD0104 | 2 | 10 | 6M + 4A | 4 | – | – | |
| Canelones, Uruguay | NUD1717 | 1 | 18 | 14M + 4A | 2 | 2 | – | |
| Ruta 9, Solís Grande, Canelones, Uruguay | NUD1714 | 5 | 18 | 14M + 4A | 2 | 2 | – | |
| Artigas, Uruguay | NUD1751 | 2 | 19 | 13M + 6A | 6 | – | – | |
| Bella Unión, Artigas, Uruguay | NUD1750 | 1 | 19 | 13M + 6A | 6 | – | – | |
| Ruta 3, Arroyo Malo, Flores, Uruguay | NUD1752 | 1 | 19 | 13M + 6A | 6 | – | – | |
| Canelones, Uruguay | NUD1717 | 1 | 19 | 13M + 6A | 4 | 2 | – | |
| Porto Alegre, Rio Grande do Sul, Brazil | NUD1842 | 4 | 19 | 13M + 6A | 4 | 2 | – | |
| Nothoscordum sp. 1 | Punta Ballena, Maldonado, Uruguay | 1707 | 3 | 15 | 9M + 6A | – | 6 | – |
| Nothoscordum sp. 2 | Punta Ballena, Maldonado, Uruguay | 1030 | 1 | 19 | 13M + 6A | – | 6 | – |
Diagnostic characters to identify species followed Guaglianone (1972). The principal traits defined by this author to separate these three species were as follows: N. gracile and N. macrostemon show a turbinate-evident perianth; stamens upright and closely spaced, sometimes forming a cylinder around the gynoecium during anthesis, and staminate filaments curved at their apices. Nothoscordum gracile differs from N. macrostemon by the former having an ellipsoidal to obovate sessile ovary, while the latter has an oblong and stipulate ovary. On the other hand, N. nudicaule has a cupuliform-subrotate perianth and lanceolate-subulate staminate filaments joined at their bases. In general, the flowers of N. macrostemon are nocturnal and those of N. nudicaule are diurnal, while those of N. gracile are morphologically and phenologically intermediate between these two types.
Meiotic analyses, pollen-grain mitosis and pollen-grain germination
For meiotic analysis, young inflorescences from N. gracile, N. macrostemon and N. nudicaule were directly fixed in ethanol : acetic acid (3 : 1, v/v) for 2–24 h at room temperature and stored at –20 °C. Prior to slide preparation, young flower buds were washed in distilled water, anthers were squashed in a drop of 45 % acetic acid, the coverslip was removed after freezing in liquid nitrogen, and the chromosomes were stained with CMA/DAPI as indicated below. For pollen-grain mitosis fixed anthers were directly squashed in 2 % acetocarmine. For pollen-tube germination, anthers of opened flowers were gently squeezed in a drop of 15 % sucrose solution and maintained during 12–24 h in a wet chamber.
CMA/DAPI banding and FISH
Root tips obtained from bulbs were pretreated with 0·05 % colchicine for 24 h at 10 °C, fixed and stored as described before. CMA/DAPI banding and FISH procedures were performed according to Souza et al. (2009). Fixed root tips were washed in distilled water and digested in a 2 % (w/v) celullase (Onozuka)/20 % (v/v) pectinase (Sigma) solution, at 37 °C, for 90 min. Meristem was macerated in a drop of 45 % acetic acid and the coverslip was later removed in liquid nitrogen.
The CMA/DAPI double-staining technique was used for fluorochrome banding. Slides were aged for 3 d, stained with CMA (0·1 mg mL−1) for 60 min and re-stained with DAPI (1 µg mL−1l) for 30 min. Slides were mounted in glycerol : McIlvaine buffer pH 7·0 (1 : 1) and aged for 3 d before analysis in an epifluorescence Leica DMLB microscope. Images were captured with a Cohu CCD video camera using the Leica QFISH software and later edited in Adobe Photoshop CS3 version 10·0.
To localize the rDNA sites, 5S rDNA from Lotus japonicus labelled with Cy3-dUTP (Amersham) and 45S rDNA from Arabidopsis thaliana labelled with digoxigenin-11-dUTP were used as probes (Souza et al., 2009, 2010). Both labellings were done by nick translation. The 45S rDNA probe was detected with sheep anti-digoxigenin FITC conjugate (Roche) and amplified with rabbit anti-sheep FITC conjugate (Dako). The hybridization mixture contained formamide 50 % (v/v), dextran sulfate 10 % (w/v), 2× SSC and 5 ng μL−1 of each probe. The slides were denaturated at 75 °C for 3 min. Stringent washes were performed reaching a final stringency of approx. 76 %. Images of the best cells were captured as previously described.
GISH and dot-blot hybridization
Total nuclear DNA was extracted following the CTAB method of Doyle and Doyle (1987). Genomic DNA utilized as probe or block was broken into fragments of approx. 500 bp by incubation at 100 °C for approx. 30 min. In the two first experiments, the genomic DNA of N. gracile labelled with Cy3-dUTP (Amersham) by nick translation was used as a probe, while non-labelled genomic DNA of N. nudicaule or N. macrostemon was separately utilized as blocking DNA in concentrations of 1 × , 25 × , 50× and 75×. The hybridization mixture containing the genomic probe and the blocking DNA was tested on metaphases of N. gracile or a control species, following the same protocol used for FISH.
In other experiments, the genomic DNA of N. macrostemon labelled with digoxigenin-11-dUTP was used as probe and non-labelled genomic DNA of N. nudicaule was used as blocking DNA. Inverse labelling was also tested using N. nudicaule DNA as probe and N. macrostemon as blocking DNA. As an internal control of hybridization efficiency, in some cases, root tips of Ipheion uniflorum, N. macrostemon (2x) or N. nudicaule (4x) (without CMA bands) were macerated together with root tips of N. gracile or N. nudicaule 4x on the same slide for in situ hybridization.
Dot-blot hybridization was performed using whole-genome DNA (500 ng) from N. gracile, N. macrostemon and N. nudicaule loaded manually onto a nylon membrane (Hybond N+; Amersham Biosciences, Amersham, Bucks, UK). Ipheion recurvifolium DNA was used as negative control. DNA of N. macrostemon and N. nudicaule used as probes was labelled with digoxigenin-11-dUTP as described above. The membranes were hybridized at 37 °C overnight. The genomic DNA was added to the hybridization buffer [1 % blocking reagent (Roche), 0·1 % sodium dodecyl sulfate (SDS), 5 % dextran sulfate, in 5× SSC, pH 7·0], according to Sambrook and Russell (2001). After hybridization, membranes were washed twice in 2× SSC, 0·1 % SDS for 5 min and in 0·5× SSC, 0·1 % SDS for 15 min, at 45 °C and 68 °C for low and high stringency, respectively. The genomic DNA probe was detected using anti-DIG alkaline phosphatase conjugate (Roche) and CDP-Star (Roche). The hybridization signals were captured on X-ray ECL film (Amersham Biosciences) and the intensity of probe signals was measured in the digital image using software QFISH version 2·1 (Leica). The highest relative signal intensity (100 %) was obtained when labelled genomic DNA of one species was hybridized with genomic DNA of this same species.
Sequence analyses
The non-coding DNA plastid region trnL-trnF was analysed in 33 samples, including representatives from N. macrostemon (2x), N. nudicaule (2x and 4x), N. gracile (4x) and N. arenarium (2x). Universal primers (C and F) described by Taberlet et al. (1991) were used to amplify trnL(UAA) and trnL(UAA)-trnF(GAA) regions. The nuclear ITS1-5·8S-ITS2 region of 16 samples of N. gracile, N. macrostemon, N. nudicaule and N. arenarium were amplified and sequenced using ITS4 and ITS5 universal primers (White et al., 1990).
All PCR amplifications were carried out in 50-μL reactions containing 5 U of Taq polymerase, 1·75 mm MgCl2, 0·5 µm of each primer and 0·1 mm of each dNTP in the manufacturer's buffer. The PCR programme for cpDNA amplification consisted of an initial step of 5 min at 95 °C, the first cycle consisted of 1 min at 94 °C, 1 min at 58 °C and 2 min 30 s at 72 °C, and then the annealing temperature was decreased by 1 °C for six cycles followed by 32 additional cycles with an annealing temperature of 52 °C and a final elongation step of 5 min at 72 °C. For ITS amplification, the PCR programme consisted of an initial denaturing step of 95 °C for 2 min, five cycles of 95 °C for 1 min, 53 °C for 1 min and 72 °C for 2 min, with a decrease of 1 °C per cycle in the annealing temperature, 35 cycles with an annealing temperature of 48 °C, and a final extension step of 72 °C for 12 min. All amplified fragments were sequenced in both directions at the Instituto Pasteur de Montevideo (Uruguay) or by Macrogen Inc. (Korea).
The sequences were edited manually using Sequencher™ (V4·1·4; Genecodes, AnnArbor, MI, USA) and all ambiguous end regions were removed. The resulting partial sequences were pre-aligned with the Clustal-W (Thompson et al., 1994) algorithm included in BioEdit version 5·0·6 (Hall, 2001) and manually adjusted. All sequences and alignments were submitted to Genbank for trnL-trnF (accession nos JN591627–JN591651) and ITS (accession nos JN591597–JN591618). A haplotype network was constructed based on this matrix using the software NETWORK version 4·5·1·6 based on the standard function of maximum parsimony (Fluxus Technology Ltd).
RESULTS
Mitotic analyses
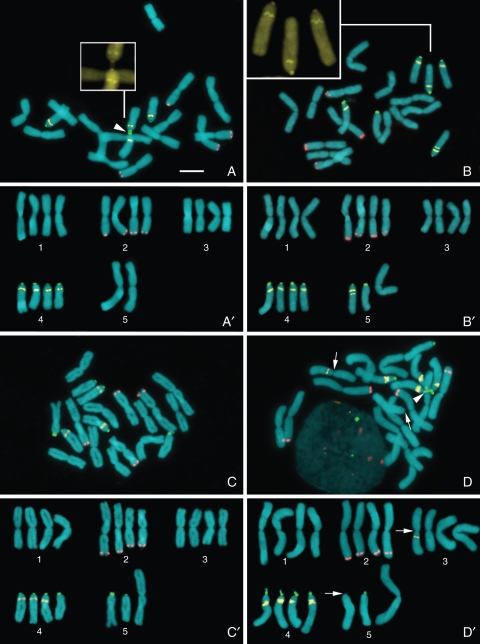
Chromosome counts of 85 individuals from different localities revealed the occurrence of diploid [2n = 10 (6M + 4A)] and tetraploid [2n = 18 (14M + 4A) or 2n = 19 (13M + 6A)] cytotypes of both N. macrostemon and N. nudicaule while N. gracile showed exclusively tetraploid cytotypes (2n = 18 or 2n = 19). Tetraploids with 2n = 18 were only rarely found in any of these species (13·0 %; Table 1). Diploid and tetraploid plants of the same species were very similar morphologically, and could be distinguished by cytological analyses only. Nothoscordum nudicaule was typically found in wet areas along river banks, while N. macrostemon and N. gracile were collected along roadsides. Plants with B chromosomes or mosaics with 2n = 18/19 were never observed. However, metaphases with two acrocentric chromosomes associated by the ends of their short arms, resembling a single metacentric chromosome, were often found (Fig. 1A). Multiple associations including three or more 45S rDNA sites were also common (Figs 1 and 2E) .
Fig. 1.
Karyotype variability in Nothoscordum gracile: (A) 2n = 18 (14M + 4A1); (B) 2n = 19 (13M + 1A0 + 4A1 + 1A2); (C) 2n = 19 (13M + 3A0 + 3A1); (D) 2n = 19 (13M + 2A0 + 4A1) with a reciprocal translocation between the long arms of chromosomes 3 and 5 (arrows). (A'–D') Karyograms based on metaphases in (A–D). Observe the variable number of CMA bands on the proximal region of the long arms of acrocentrics. The arrowhead in (A) indicates an association between 45S rDNA sites of two acrocentric chromosomes. Inserts in (A) and (B) show higher magnification of short arms of acrocentric chromosomes stained with CMA. CMA = yellow, DAPI = blue, 5S rDNA = red, 45S rDNA = green. Scale bar in (A) = 10 µm.
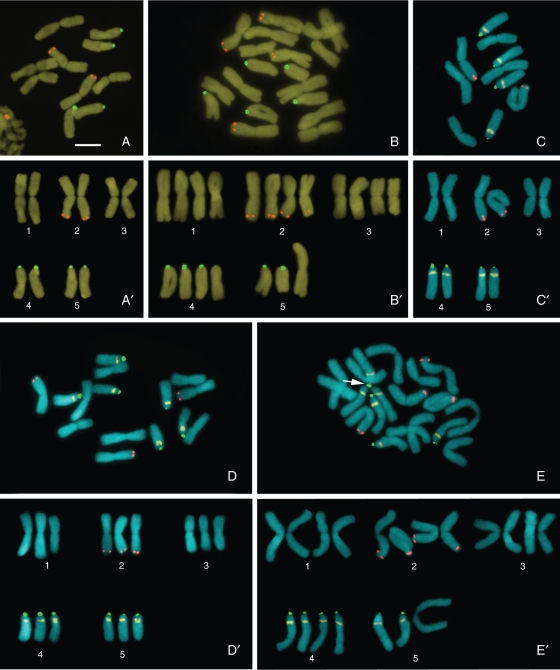
Fig. 2.
Cytotypes of Nothoscordum nudicaule, N. macrostemon and Nothoscordum sp. 1: (A, B) N. nudicaule with 2n = 10 (6M + 4A0) (A) and 2n = 19 (13M + 6A0) (B); (C, E) N. macrostemon with 2n = 10 (6M + 4A1) (C) and 2n = 19 (13M + 6A1) (E); (D) Nothoscordum sp. 1 2n = 15 (9M + 6A1). (A'–E') Karyograms based on metaphases in (A–E). The arrow in (E) indicates an association between three 45S rDNA sites. CMA = yellow, DAPI = blue, 5S rDNA = red, 45S rDNA = green. Scale bar in (A) = 10 µm.
CMA+/DAPI− banding (CMA bands) revealed the presence of heterochromatin on the short arms of the acrocentric chromosomes, and one or two CMA bands on the proximal region of the long arms of most acrocentrics (Figs 1 and 2). No DAPI+ heterochromatic blocks were observed. According to the number of CMA bands on the long arms, the acrocentric chromosomes were classified as A0 (without bands), A1 (with one band), and A2 (with two bands). Sequential analyses with CMA/DAPI and FISH using 5S and 45S rDNA probes revealed a single 5S rDNA site per monoploid complement, always located on the terminal region of a metacentric chromosome, and 45S rDNA sites on the short arms of all acrocentric chromosomes co-localized with CMA bands. No variation in the number or position of these sites was detected.
The most frequent karyotype of N. gracile was 2n = 19 (13M + 1A0 + 4A1 + 1A2), observed in 57·7 % of the individuals and considered the standard karyotype of this species. This karyotype was found in most Brazilian populations and in the samples from Lima (Peru), Buenos Aires (Argentina) and Montevideo (Uruguay) (Fig. 1B). Other populations from Brazil (Lavras, Londrina and Gramado) and Argentina (Tucumán), totalling 34·6 % of the individuals, showed 2n = 19 (13M + 3A0 + 3A1) (Fig. 1C), whereas a single sample from Uruguay (Canelones) had 2n = 18 (14M + 4A1) (Fig. 1A). Four out of six individuals from Santa Maria (Brazil) displayed an aberrant 2n = 19 karyotype (13M + 2A0 + 4A1) bearing a CMA band on the long arm of a metacentric chromosome, in the same position as that of the long arm of A1 acrocentrics. Noteworthy, this karyotype also had one A0 chromosome without a 45S rDNA site (Fig. 1D), suggesting a reciprocal translocation between an M and an A1 chromosome. The other two individuals from this locality showed the standard karyotype. For the distribution of the different cytotypes of N. gracile see below (Fig. 5).
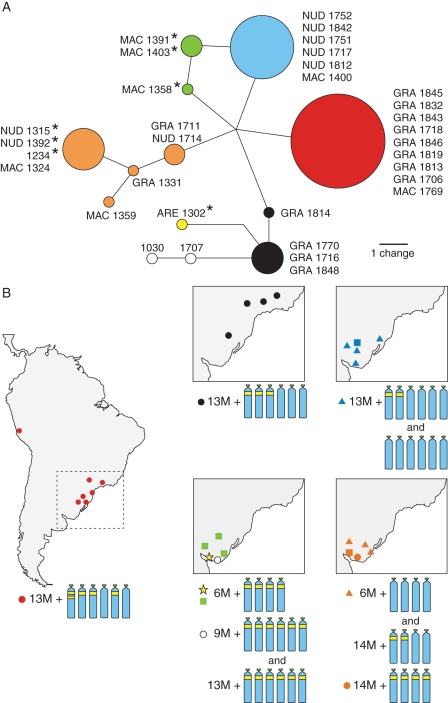
Fig. 5.
Network connecting trnL-trnF of section Inodorum (A) and their relationships with geographic and cytogenetic data (B). Each haplotype is indicated by a different colour in both (A) and (B). Circle sizes in the network are proportional to the number of individuals observed for each haplotype. Idiograms of acrocentric chromosomes represent the predominant cytotype for each haplotype, with the same colours as in (A). Diploid samples in (A) are indicated by asterisks. In (B) closed circles indicate N. gracile (GRA), squares N. macrostemon (MAC), triangles N. nudicaule (NUD), stars N. arenarium (ARE) and open circles Nothoscordum sp. 1 and sp. 2.
Diploid samples of N. nudicaule always showed acrocentric chromosomes without proximal CMA bands (6M + 4A0) (Fig. 2A). Likewise, tetraploid plants from northern (Departamentos de Artigas and Salto) and south-eastern (Departamento de Flores) Uruguay (Fig. 2B) always showed 2n = 19 (13M + 6A0). A mixed population with 2n = 19 (13M + 4A0 + 2A1) and 2n = 18 (14M + 2A0 + 2A1) was found in southern Uruguay (Departamento de Canelones). The diploid and tetraploid samples of N. macrostemon examined had exclusively A1 chromosomes, with karyotype formulae 2n = 10 (6M + 4A1) (Fig. 2C) or 2n = 19 (13M + 6A1) (Fig. 2E). Vigorous triploid individuals bearing many bulbils, referred to as Nothoscordum sp. 1 [2n = 15 (9M + 6A1); Fig. 2D], were only found in Punta Ballena, southern Uruguay, together with the closely related tetraploid Nothoscordum sp. 2 [2n = 19 (13M + 6A1)].
Meiotic analyses
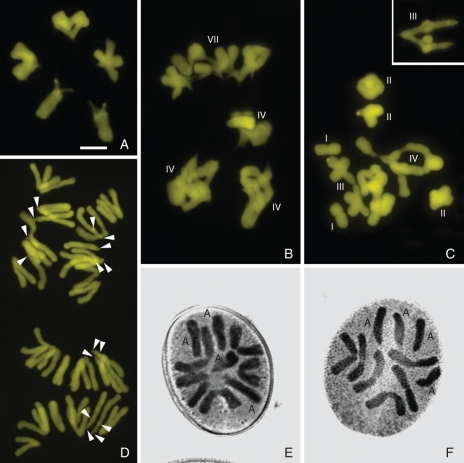
Meiosis was regular in the diploid samples of N. macrostemon and N. nudicaule, with five bivalents and a predominance of interstitial chiasmata (Fig. 3A). On the other hand, the tetraploid cytotypes showed irregular meiotic pairing, with univalents, bivalents and multivalents, but only rare anaphase bridges or lagging chromosomes. A predominance of tetravalents formed by metacentric chromosomes was observed in tetraploid N. macrostemon (2n = 19) (Fig. 3B). In addition, one heteromorphic trivalent and multivalents with up to six acrocentric and one metacentric chromosomes were often observed (Fig. 3B). The heteromorphic trivalent was formed by one metacentric paired with two acrocentric chromosomes by their long arms (Fig. 3C). In tetraploid N. nudicaule the most common meiotic arrangement was 2I + 3II + 1III + 2IV (Fig. 3C), with an identical heteromorphic trivalent. The meiotic pairing of N. gracile was similar to that of the other tetraploid species, with the heteromorphic trivalent being particularly noticeable (Fig. 3C, insert).
Fig. 3.
Meiosis in Nothoscordum macrostemon (A, D) and N. nudicaule (B, C) and first pollen-grain mitosis in N. gracile (E, F). (A–C) Metaphase I with five bivalents in N. macrostemon 2n = 10 (A) and with multivalents in N. macrostemon 2n = 19 (B) and N. nudicaule 2n = 19 (C). (D) Anaphase II in N. macrostemon 2n = 19 showing chromosome sets with 6M + 4A (above) and 7M + 2A (below). (E, F) First pollen-grain mitosis of N. gracile with n = 9 (5M + 4A) (E) and n = 10 (6M + 4A) (F). Inset in (C) shows a heteromorphic trivalent of another cell oriented to the poles. Univalent, bivalent, trivalent, quadrivalent and heptavalent are indicated by Roman numerals. Arrowheads in (D) point to CMA bands of some acrocentric short arms and proximal ones. Scale bar in (A) = 10 µm.
In spite of their meiotic irregularities, the tetraploids of the Inodorum complex had apparently normal and viable pollen grains. When pollen grains of any tetraploid sample were immersed in a drop of 15 % sucrose solution, all or almost all of the grains germinated within 24 h. Chromosome counts in 38 cells in anaphase II (Fig. 3D) and 96 first pollen-grain mitosis (Fig. 3E, F) revealed a predominance of haploid chromosome complements of n = 9 (46·8 %) or n = 10 (45·7 %) with different combinations of metacentric and acrocentric chromosomes. Cells with n = 8 and n = 11 were also observed (7·4 %).
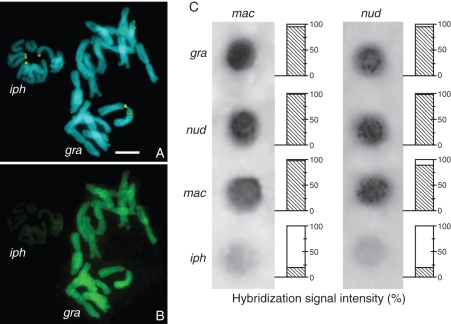
GISH and dot-blot experiments
None of the GISH experiments using N. gracile metaphases and total nuclear DNA of N. macrostemon or N. nudicaule as probe with or without blocking DNA could differentiate between the diploid genomes of N. gracile. All experiments showed uniform labelling of the chromosomes, whereas the chromosomes of the internal control (Ipheion uniflorum) were consistently weakly labelled (Fig. 4A, B). Moreover, after subsequent increments in the concentration of blocking DNA (25x, 50x and 75x) in the hybridization mixture, the labelling intensity of all chromosomes remained uniform. In the dot-blot analyses, the N. macrostemon probe hybridized with equal intensity on both N. gracile and N. nudicaule DNA, as well as on its own DNA. Similar results were observed when genomic DNA of N. nudicaule was used as probe (Fig. 4C). Only when DNA of N. macrostemon or N. nudicaule was hybridized with genomic DNA of an external group (Ipheion recurvifolium) was a much lower labelling intensity (approx. 17 %) obtained.
Fig. 4.
GISH (A, B) and dot-blot hybridization (C) among species of the complex Inodorum and the genus Ipheion. (A, B) Metaphase of N. gracile (gra) and prometaphase of I. uniflroum (iph) stained with CMA/DAPI (A) and in situ hybridized with N. nudicaule genomic probe (B). (C) Labelled genomic DNA of N. macrostemon (mac) and N. nudicaule (nud) hybridized in membrane with non-labelled genomic DNA of N. gracile, N. macrostemon and N. nudicaule. Relative hybridization intensity based on the 100 % value for homologous hybridization is indicated below. Scale bar in (A) = 10 µm.
Sequence analyses
The ITS1-5·8S-ITS2 nuclear region was highly conserved among the species analysed here, including N. arenarium. The aligned ITS matrix was 655 bp long, with only three parsimony-informative loci. All samples investigated were quite similar to each other, with the exception of two plants of N. nudicaule (JN591604 and JN591605) collected near Rio Quaraí, Bella Unión (Uruguay), which exhibited eight variable sites in relation to the remaining samples of the Inodorum complex. The samples of N. macrostemon JN591608 and JN591609, both from North Uruguay, shared the same A → G transition which distinguished them from the remaining N. macrostemon sample from South Uruguay.
The plastid DNA sequence trnL-trnF showed greater variability. When aligned, the trnL-trnF matrix was 849 bp long, with 831 constant and 17 parsimony-informative characters. Sequence variation included two substitutions (one autapomorphic), one single base-pair insertion, and sequence-length variations in repetitive regions of one AT microsatellite and in two poly-T regions. The analysis of haplotypes resulted in an incompletely resolved network. Plastid lineages did not appear clearly diverging and some haplotypes were shared by different morphospecies. The network separated most diploids of N. macrostemon (haplotypes in green; Fig. 5) from diploids of N. nudicaule (in orange) but the same did not occur with tetraploid accessions of those species or N. gracile. The triploid Nothoscordum sp. 1 and the tetraploid Nothoscordum sp. 2 (represented as empty circles) were more closely related to a less-common haplotype of N. gracile (black) and to N. arenarium (yellow). Three distinct haplotypes of N. gracile were detected: two widely distributed (red and black haplotypes) and one restricted to Uruguay (orange).The latter was also found in some diploid samples of N. macrostemon and N. nudicaule. The red and black haplotypes corresponded to the two most frequent cytotypes of N. gracile, 2n = 19 (13M + 1A0 + 4A1 + 1A2) and 2n = 19 (13M + 3A0 + 3A1), respectively (Fig. 5).
Tetraploid apomicts did not show a strict correspondence between morphospecies and cpDNA haplotypes. A geographical trend was apparent instead with tetraploid samples of both morphospecies possessing the same haplotype (orange in Fig. 5) towards south-western Uruguay and another haplotype (blue) towards the north-east. This trend was also apparent in Rio Grande do Sul (Brazil) where one haplotype (red) of N. gracile was found along the Atlantic coast and a different haplotype (black) was distributed further inland (Fig. 5).
DISCUSSION
Karyotype variability of the Inodorum complex
The chromosome numbers and morphologies of the species analysed here were similar to those previously described (Crosa, 1972, 1996; Nuñez et al., 1974). Intra-individual variation with 2n = 18 and 19 was not observed here, although several cells displayed two acrocentric chromosomes associated by the NORs on their short arms, resembling a metacentric chromosome. Associations between acrocentric chromosomes with terminal NORs have been described in a number of species, including Nothoscordum gracile (Nuñez et al., 1974) and Allium sativum (Sato and Kawamura, 1981) and might be the cause of the apparent mosaicism (2n =18/19) reported by Nassar and Aguiar (1978) for N. gracile.
The presence of CMA bands in the proximal region of the long arms of the acrocentric chromosomes of 2x and 4x N. macrostemon, and their absence in the diploids and most tetraploids of N. nudicaule, appears to be the most prominent cytological difference between these two species. The occurrence of acrocentric chromosomes with and without CMA bands in all of the samples of N. gracile constitutes a strong indication of their hybrid origin and the probable independent formation of different cytotypes. In spite of the high variability observed in the CMA banding pattern in the tetraploids of the Inodorum complex, little intrapopulational variation was observed. Moreover, the occurrence of karyotypic heterozigosity suggests that propagation of tetraploids is predominantly asexual.
The diversity of banding patterns and chromosome numbers was higher in localities in Uruguay where different cytotypes of the Inodorum complex occur. The largest variability occurred at the contact zone between tetraploid apomictics and related sexual diploids, whereas only some fixed genotypes may show considerably expanded geographic ranges (Daurelio et al., 2004; Hörandl, 2006). Consequently, our data confirm that the centre of diversity of the Inodorum complex is located in Uruguay (Crosa, 1996) because in that area the largest karyotypic and haplotypic variability occur and it is where only diploid cytotypes of N. macrostemon and N. nudicaule were found. Throughout its range, N. gracile was found in disturbed roadside environments, a pattern that has been found for apomictic hybrids in other complexes (Bierzychudek, 1985; Hörandl, 2006).
The production of viable pollen and effective pollination are necessary for endosperm formation in N. gracile because it is pseudogamous (Tandon and Kapoor, 1963). In the case of the tetraploids of the Inodorum complex, in spite of irregularities observed in their meiotic pairing the pollen grains were generally viable, with a high frequency of microspores with n = 10 and n = 9, thus indicating that sexual reproduction is also possible (see also Dyer, 1967; Nuñez et al., 1974). The occurrence of facultative sexually reproducing individuals in apomictic complexes is not rare (Akins and Dijk, 2007). Residual female sexuality or the production of functional male gametes seems to be sufficient to allow cross-fertilization between apomictic tetraploids as well as between tetraploids and diploids, which can account for the karyotype variability observed and the formation of triploids and individuals with rare translocations. The karyotype variability of the Inodorum complex, as in other apomictic complexes, may have been generated by multiple de novo origins of the apomictic polyploids, backcrossing to sexually related species or by somatic mutations (Hörandl et al., 2009). On the other hand, extensive asexual reproduction ensured by apomixis and the abundant production of bulbils may explain the stabilization and dissemination of a few cytotypes (Akins and Dijk, 2007).
Origin of the 2n = 19 karyotype
Considering the stability of the diploid chromosome number of the section Inodorum (2n = 10; 6M + 4A), the number 2n = 20 (12M + 8A) should be expected in the tetraploids. Indeed, tetraploids with 2n = 20 may have been formed a number of times in the wild, as pollen grains of N. gracile with n = 10 and n = 9 are very frequent. Nuñez et al. (1974) reported n = 10 in 22 % of the pollen grains and meiotic dyad cells of N. gracile, most of them having 6M + 4A, as in the present sample. Seedlings with 2n = 20 (14M + 6A) descendents of clones of N. gracile with 2n = 19 were reported by Dyer (1967) and Nuñez et al. (1974).
The 2n = 19 (13M + 6A) karyotype seems to be derived from a centric fusion in the ancestral karyotype with 2n = 20 (Crosa, 1996). This assumption is supported by the maintenance of the FN expected for 2n = 20 (FN = 16) and by the presence of a heteromorphic trivalent formed by a metacentric and two acrocentric chromosomes during meiosis (see also Levan and Emsweller, 1938; Dyer, 1967; Nuñez et al., 1974). This fusion must have occurred between the centromeres of two acrocentric chromosomes and resulted in the loss of their short arms, since no 45S rDNA signal were observed in the extra metacentric chromosomes. Fusion of acrocentric chromosomes bearing nucleolar organizer regions on their short arms, with the loss of ribosomal sites, has been reported in several other genera such as Lillium (Muratović et al., 2005) and Rumex (Koo et al., 2004). In Nothoscordum, Robertsonian translocations seems to be a common phenomenon and may also have occurred between acrocentric and metacentric chromosomes, as in N. arenarium (Souza et al., 2009) and in four individuals of N. gracile from Santa Maria (Brazil) which have a metacentric chromosome with a proximal CMA band on one of its arms.
The occurrence of 2n = 19 in tetraploid cytotypes of three closely related species suggests that the fusion probably occurred in a tetraploid plant with 2n = 20, and the extra metacentric chromosome was later transmitted to the other tetraploids of the complex Inodorum through interspecific hybridization. The absence of a CMA band in the proximal region of the extra metacentric chromosome suggests that this fusion may have initially occurred in N. nudicaule, as it is the only species in this complex without proximal CMA bands in their acrocentric chromosomes.
The union of a balanced gamete with n = 9 (7M + 2A0) from a 2n = 19 N. nudicaule and a non-reduced gamete of N. macrostemon 2x, with n = 10 (6M + 4A1), could have originated the 2n = 19 tetraploid hybrid N. nudicaule × N. macrostemon, which, by self-fertilization or further introgression, generated the diversity of banding patterns observed in the tetraploids of this complex. Analysis of trnL-trnF sequences showed that each tetraploid cytotype exhibited a single and exclusive haplotype, with a few exceptions, suggesting that each tetraploid lineage had an independent origin. A non-reduced gamete of diploid N. macrostemon or N. arenarium may also have contributed to form the triploid plants (Nothoscordum sp. 1) with 2n = 15 (9M + 6A1) without the extra metacentric. Dyer (1967) proposed that the maintenance of this fusion in heterozygosis was due to the presence of a balanced lethal system, as has been observed in other species with permanent heterozygotic translocations (reviewed by Crosa, 1996; Levin, 2002). However, some homozygous samples of N. gracile [2n = 18 (14M + 4A1)] and N. nudicaule [2n = 18 (14M + 2A0 + 2A1)] were observed here, suggesting that they did not have a lethal allele system. It is interesting to observe that the combination 7M + 2A is by far the most frequent one both in dyad cells of meiosis and in pollen-grain mitosis (Levan and Emsweller, 1938; Dyer, 1967; Nuñez et al., 1974), and it could have allowed the formation of eventual tetraploids (14M + 4A) bearing two copies of the extra metacentric chromosome.
The origin of Nothoscordum gracile and its relationships with N. macrostemon and N. nudicaule
The ability of GISH to differentiate among parental genomes in a hybrid depends mainly on the size of the genomes, on the degree of divergence of the principal families of repetitive DNA sequences, and on the phylogenetic distance between the ancestral species (Markova and Vyskot, 2009). The fact that GISH and dot-blot could not differentiate between genomes of the species of the complex Inodorum and that artificial hybrids between diploids N. nudicaule and N. macrostemon are viable (O. Crosa, unpubl. res.) suggest that this complex is very young. A recent origin is also compatible with the lack of differentiation of the ITS sequences among members of this complex and the morphological and cytogenetic similarities between their tetraploids.
The variation among trnL-trnF haplotypes was consistent with the CMA banding patterns. Most populations with the same haplotype share the same cytotype and all N. gracile samples with the dominant or standard cytotype had the same haplotype. Despite the low level of variation among plastid sequences, the strict correspondence between them and the cytological complements supports the interpretation that haplotype similarity is not due to homoplasy but rather inherited from the progenitors of each lineage together with karyotypic features.
At least three distinct lineages can be recognized in N. gracile both by sequence and karyotypic data. The plastid sequences of the polyploids are related to different degrees to distinct putative sexual sources, and their cytological constitution can be explained by different contributions from the different diploid sources. In addition, the presence of shared plastid sequences among different morphospecies and the lack of distinct lineages suggest both recent diversification and ongoing gene flow. The karyotypes of asexual polyploids can be explained by a combination, but not the simple addition, of those of the diploid sources. Mechanisms explaining such combinations and variability may include recombination at the polyploid level following multiple origins of the tetraploids (Soltis et al., 2003) and/or repeated introgression from sexuals in a reticulate pattern (Lo et al., 2010) including unilateral sexual polyploidization involving triploid bridges (Ramsey and Schemske, 1998).
In the Inodorum complex, polyploid apomicts appear more widespread than their sexual progenitors, which is congruent with the concept of geographic parthenogenesis. Each of the lineages identified by an individual cytotype/haplotype may represent a single apomictic clone. When individual clones can be identified, their geographic distributions can be asymmetrical, some clones being far more widespread than others (Richards, 2003). The most widespread cytotype/haplotype of N. gracile in South America is a structural heterozygote [2n = 19 (13M + 1A0 + 4A1 + 1A2)], also found in Japan (Sato et al., 1979). The maintenance of structural heterozygosity and the occurrence of this lineage in areas far removed from the distributional ranges of either diploid or tetraploid sexual partners, support the interpretation that it possibly represents the most vigorous and successful apomictic clone generated within the Inodorum complex. Because other apomictic lineages of the tetraploid N. gracile are reported here, its comparatively greater success cannot be attributed to the advantages of apomixis or polyploidy but must be explained by some inherent property of this individual clone. It has been proposed that some clones represent general-purpose genotypes (Lynch, 1984) that possess wide adaptation capabilities to a variety of environmental conditions. Further evidence of the exclusively clonal nature of this karyotype/cytotype may be needed to support this interpretation.
The diploid cytotypes of N. nudicaule and N. macrostemon represent the extremes of morphological and cytogenetic (CMA bands) variability within this complex and appear to constitute closely related true biological species in spite of the putative occurrence of chloroplast capture, causing the incongruent distribution of cpDNA haplotypes. At the tetraploid level, a greater trend towards geographical structuring of chloroplast haplotypes and a close correspondence between chloroplast sequences and karyotypes, irrespective of morphological variation, was found. Cytological evidence is highly congruent with a hybrid origin of not only N. gracile but also polyploid cytotypes assigned to N. macrostemon and N. nudicaule. This pattern strongly suggests that the sexual diploid components of the Inodorum complex hybridize frequently, producing apomictic clones spanning the morphological extremes represented by their parental species. Some of those clones are morphologically recognizable as N. gracile.
In this work we have shown that cytological and sequence information is congruent with the pattern suggested by morphological evidence that the Inodorum section is a species complex and that N. gracile is likely to be an assemblage of hybrids involving at least N. macrostemon and N. nudicaule. We have also shown that differentiation at the sequence level is incipient, suggesting a recent divergence among the sexual species of the complex. Analysis of ITS sequences points to possible gene flow constraints between the northern and southern populations of Uruguay, although no differences were observed in karyotype or trnL-trnF plastidial sequences between these populations. Further support for the evolutionary hypotheses and processes in this complex are likely to emerge from more intensive sampling and sensitive molecular markers.
ACKNOWLEDGEMENTS
We are grateful to Drs M. E. L. Canelada, L. C. Davide, A. C. Brasileiro Vidal, L. P. Felix, A. P. Moraes and D. C. Cabral-de-Mello who provided N. gracile samples for this study, the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE) for financial support, and Coordenacão de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for a grant to L. G. R. Souza.
LITERATURE CITED
- Akins PO, Dijk PJ. Mendelian genetics of apomixis in plant. Annual Review of Genetics. 2007;41:509–537. doi: 10.1146/annurev.genet.40.110405.090511. [DOI] [PubMed] [Google Scholar]
- Bierzychudek P. Patterns in plant parthenogenesis. Experientia. 1985;41:1255–1264. doi: 10.1007/978-3-0348-6273-8_9. [DOI] [PubMed] [Google Scholar]
- Canelada MEL, Fernandez AMF. Bandeo cromosomico en Nothoscordum inodorum (Soland. Ex Aiton) Nich. var. inodorum. Lilloa. 1985;36:181–186. [Google Scholar]
- Crosa O. Estudios cariología en el género Nothoscordum (Liliaceae) Boletin de la Facultad de Agronomia de Uruguay. 1972;122:3–8. [Google Scholar]
- Crosa O. 1996 Sistemática e evolução das espécies da seção Inodorum Guag. do gênero Nothoscordum Kunth (Allieae, Alliaceae). PhD Thesis Departamento de Genética, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil. [Google Scholar]
- Daurelio LD, Espinoza F, Quarin CL, Pessino SC. Genetic diversity in sexual diploid and apomictic tetraploid populations of Paspalum notatum situated in sympatry or allopatry. Plant Systematics and Evolution. 2004;244:189–199. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Dyer AE. The maintenance of structural heterozygosity in Nothoscordum fragrans Kunth. Carylogia. 1967;20:287–308. [Google Scholar]
- Fay MF, Rudall PJ, Chase M. Molecular studies of subfamily Gilliesioideae (Alliaceae) Aliso. 2006;22:367–371. [Google Scholar]
- Friesen N, Fritsch RM, Blattner FR. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso. 2006;22:372–395. [Google Scholar]
- Gornall RJ. Population genetic structure in agamospermous plants. In: Hollingsworth PM, Bateman RM, Gornall RJ, editors. Molecular systematics and plant evolution. London: Taylor & Francis; 1999. pp. 118–138. [Google Scholar]
- Guaglianone EA. Sinopsis de las especies de Ipheion Raf. y Nothoscordum Kunth (Liliáceae) de Entre Ríos y regiones vecinas. Darwiniana. 1972;17:159–240. [Google Scholar]
- Guerra M, Felix LP. O cariótipo de Nothoscordum pulchellum (Alliaceae) com ênfase na heterocromatina e sítios de DNAr. Boletín de la Sociedad Argentina de Botánica. 2000;35:283–289. [Google Scholar]
- Hall T. 2001 BioEdit, version 5·0·6. Department of Microbiology, North Carolina State University. http://www.mbio.ncsu.edu/BioEdit/bioedit.html . [Google Scholar]
- Hörandl E. The complex causality of geographical parthenogenesis. New Phytologist. 2006;171:525–538. doi: 10.1111/j.1469-8137.2006.01769.x. [DOI] [PubMed] [Google Scholar]
- Hörandl E, Greilhuber J, Klimova K, et al. Reticulate evolution and taxonomic concepts in the Ranunculus auricomus complex (Ranunculaceae): insights from morphological, karyological and molecular data. Taxon. 2009;58:1194–1215. [PMC free article] [PubMed] [Google Scholar]
- Jones K. Robertsonian fusion and centric fission in karyotype evolution of higher plants. Botanical Review. 1998;64:273–289. [Google Scholar]
- Koo DH, Yoonkang H, Bang JW. Variability of rDNA loci in dioecious Rumex acetosa L. detected by fluorescence in situ hybridization. Korean Journal of Genetics. 2004;26:9–13. [Google Scholar]
- Kurita M, Kuroki Y. Heterochromaty in Nothoscordum chromosomes. Memoirs of Ehime University Section II. 1963;4:493–500. [Google Scholar]
- Levan A, Emsweller SL. sStructural hibridity in Nothoscordum fragrans. Journal of Heredity. 1938;29:291–294. [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. Oxford: Oxford University Press; 2002. [Google Scholar]
- Li QQ, Zhou SD, He XJ, Yu Y, Zhang YC, Wei XQ. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps 16 sequences, focusing on the inclusion of species endemic to China. Annals of Botany. 2010;106:709–733. doi: 10.1093/aob/mcq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo EYY, Stefanović S, Dickinson TA. Reconstructing reticulation history in a phylogenetic framework and the potential of allopatric speciation driven by polyploidy in an agamic complex in Crataegus (Rosaceae) Evolution. 2010;64:3593–608. doi: 10.1111/j.1558-5646.2010.01063.x. [DOI] [PubMed] [Google Scholar]
- Lowry E, Lester SE. The biogeography of plant reproduction: potential determinants of species' range sizes. Journal of Biogeography. 2006;33:1975–1982. [Google Scholar]
- Lynch M. Destabilizing hybridization, general-purpose genotypes and geographic parthenogenesis. Quarterly Review of Biology. 1984;59:257–290. [Google Scholar]
- Markova M, Vyskot B. New horizons of genomic in situ hybridization. Cytogenetic and Genome Research. 2009;126:368–75. doi: 10.1159/000275796. [DOI] [PubMed] [Google Scholar]
- Muratović E, Bogunié F, Šoljan D, Siljak-Yakovlev Does Lilium bosniacum merit species rank? A classical and molecular-cytogenetic analysis. Plant Systematics and Evolution. 2005;252:97–109. [Google Scholar]
- Nassar NMA, Aguiar MLR. Multiple karyotypes in individuals of Nothoscordum fragrans (Liliaceae) Caryologia. 1978;31:7–14. [Google Scholar]
- Nguyen NH, Driscoll HE, Specht CD. A molecular phylogeny of the wild onions (Allium; Alliaceae) with a focus on the western North American center of diversity. Molecular Phylogenetics and Evolution. 2008;47:1157–1172. doi: 10.1016/j.ympev.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Nuñez O, Frayssinet N, Rodriguez RH, Jones K. Cytogenetic studies in the genus Nothoscordum Kunt. I. The N. inodorum polyploid complex. Caryologia. 1974;27:403–441. [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Ravenna PF. Studies in the Genus Nothoscordum. Plant Life. 1978;34:136–145. [Google Scholar]
- Richards AJ. Apomixis in flowering plants: an overview. Philosophical Transactions of the Royal Society of London. 2003;358:1085–1093. doi: 10.1098/rstb.2003.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua GH, Speranza PR, Vaio M, Arakaki M. A phylogenetic analysis of the genus Paspalum (Poaceae) based on cpDNA and morphology. Plant Systematic and Evolution. 2010;288:227–243. [Google Scholar]
- Sambrook J, Russell DW. In: Molecular cloning: a laboratory manual. 3rd edn., editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sato M, Sato S, Matsumoto E. Chromosome banding produced by UV-light exposure in the presence of Hoechst 33258. Caryologia. 1982;35:405–409. [Google Scholar]
- Sato S, Kawamura S. Cytological studies on the nucleolus and the NOR-carrying segments of Allium sativum. Cytologia. 1981;46:781–790. [Google Scholar]
- Sato S, Kuroki Y, Ohta S. Two types of color-diferentiated C-banding positive segments in chromosomes of Nothoscordum fragrans, Liliaceae. Cytologia. 1979;44:715–725. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytologist. 2003;161:173–91. [Google Scholar]
- Souza LGR, Crosa O, Guerra M. The karyotype of Nothoscordum arenarium Herter (Gilliesioideae, Alliaceae): a populational and cytomolecular analysis. Genetics and Molecular Biology. 2009;32:111–116. doi: 10.1590/S1415-47572009005000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza LGR, Crosa O, Guerra M. Karyological circumscription of Ipheion Rafinesque (Gilliesioideae, Alliaceae) Plant Systematics and Evolution. 2010;287:119–127. [Google Scholar]
- Stearn WT. Nothoscordum gracile, the correct name of N. fragrans and the N. inodorum of authors (Alliaceae) Taxon. 1986;35:335–338. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tandon SL, Kapoor BM. Contributions to the cytology of endosperm in some Angiosperms II. Nothoscordum fragrans Kunth. Caryologia. 1963;16:377–395. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandel A. La parthénogenese geographique Contribution a l'étude biologique et cytologique de la parthénogenese naturelle. Bulletin Biologique de la France et de la Belgique. 1928;62:164–182. [Google Scholar]
- Verduijn MH, Van Dijk PJ, Van Damme JMM. Distribuition, phenology and demography of sympatric sexual and asexual dandelions (Taraxacum officinale s.l.): geographic parthenogenesis on a small scale. Biological Journal of the Linnean Society. 2004;82:205–218. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand D, Sminsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. pp. 315–322. [Google Scholar]