Structures of IPMDH from the obligate piezophile S. benthica DB21MT-2 (SbIPMDH) and the nonpiezophile S. oneidensis MR-1 (SoIPMDH) were determined at atmospheric pressure. Comparison of the structures revealed that SbIPMDH is in a more open conformation and has a larger internal cavity volume than SoIPMDH.
Keywords: 3-isopropylmalate dehydrogenase, pressure tolerance, obligate piezophiles
Abstract
Organisms living in deep seas such as the Mariana Trench must be adapted to the extremely high pressure environment. For example, the 3-isopropylmalate dehydrogenase from the obligate piezophile Shewanella benthica DB21MT-2 (SbIPMDH) remains active in extreme conditions under which that from the land bacterium S. oneidensis MR-1 (SoIPMDH) becomes inactivated. In order to unravel the differences between these two IPMDHs, their structures were determined at ∼1.5 Å resolution. Comparison of the structures of the two enzymes shows that SbIPMDH is in a more open form and has a larger internal cavity volume than SoIPMDH at atmospheric pressure. This loosely packed structure of SbIPMDH could help it to avoid pressure-induced distortion of the native structure and to remain active at higher pressures than SoIPMDH.
1. Introduction
Organisms living in deep seas such as the Mariana Trench are exposed to an extremely high pressure environment of up to 100 MPa and their survival depends on pressure adaptation of their constituent macromolecules. For example, the activity of dihydrofolate reductase (DHFR) from the nonpiezophile Escherichia coli decreases as pressure increases, whereas the DHFR from the piezophile Shewanella violacea strain DSS12, which was isolated from the Ryukyu Trench at a depth of 5110 m, has optimal activity at approximately 100 MPa (Ohmae et al., 2004 ▶). This fact suggests that DHFRs from deep-sea bacteria have pressure adaptations that those from organisms living at atmospheric pressure do not possess.
3-Isopropylmalate dehydrogenase (IPMDH) has been used as a model enzyme to investigate pressure adaptation of proteins. IPMDH catalyzes the reduction of 3-isopropylmalate (IPM) to 2-isopropyl-3-oxosuccinate in the presence of divalent metal cations such as magnesium or manganese ion and nicotinamide adenine dinucleotide. This pathway is part of leucine biosynthesis (Hayashi-Iwasaki & Oshima, 2000 ▶). Crystal structures of IPMDH from various organisms and in several enzymatic states have been determined and described (Imada et al., 1991 ▶, 1998 ▶; Hurley & Dean, 1994 ▶; Kadono et al., 1995 ▶). IPMDHs have also been investigated in order to understand the thermostability of proteins. For example, the thermostability of IMPDHs from mesophiles was improved by random and site-directed mutagenesis (Akanuma et al., 1999 ▶; Tamakoshi et al., 2001 ▶; Ohkuri & Yamagishi, 2007 ▶).
In addition, previously published work shows that IPMDH from the obligate piezophile S. benthica DB21MT-2, originally isolated from the Mariana Trench (Kato et al., 1998 ▶; Nogi & Kato, 1999 ▶), is more tolerant towards high-pressure stress than the same enzyme originating from the nonpiezophile S. oneidensis MR-1. The k cat/K m values of SbIPMDH at 150–200 MPa are 50–70% of those at atmospheric pressure. In contrast, those of SoIPMDH are less than 20% of the values at atmospheric pressure (Kasahara et al., 2009 ▶), even though these two enzymes share 85% amino-acid identity. However, the mechanism of pressure adaptation possessed by proteins from piezophiles is not well understood. In this study, we have determined the structures of IPMDHs from the obligate piezophile S. benthica DB21MT-2 (SbIPMDH) and the nonpiezophile S. oneidensis MR-1 (SoIPMDH) at ∼1.5 Å resolution. We describe the relationship between structural features and pressure tolerance in SbIPMDH.
2. Materials and methods
2.1. Expression and purification
SoIPMDH and SbIPMDH were overexpressed in Escherichia coli BL21-CodonPlus-(DE3)-RIL cells (Agilent Technologies, Santa Clara, California, USA) transformed with pQE MR1-leuB and pQE DB21MT-2-leuB vectors, respectively. The pQE MR1-leuB and pQE DB21MT-2-leuB vectors carry the subcloned leuB genes of S. oneidensis MR-1 and S. benthica DB21MT-2 inserted into the BamHI/HindIII restriction sites of pQE-80L vector (Qiagen, Hilden, Germany), respectively. These vectors allow the expression of N-terminally His6-tagged SoIPMDH and SbIPMDH fusion proteins (Kasahara et al., 2009 ▶). Bacterial cultures were grown to an OD600 of 0.6 at 310 K in medium containing 100 µg ml-1 ampicillin. Expression of the proteins was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.5 mM. The cells were cultivated for 6 h after induction and then harvested by centrifugation at 4000g for 15 min. To remove residual broth, cell pellets were washed with cell-lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol). The cells were thawed on ice, suspended in cell-lysis buffer supplemented with lysozyme (0.1 mg ml−1 final concentration) and incubated for 30 min before being lysed by sonication. Cell debris was removed by centrifugation (15 000g, 277 K, 30 min). The proteins were purified by Ni2+–NTA affinity chromatography using 250 mM imidazole for elution in 50 mM Tris–HCl pH 8.0 containing 300 mM NaCl and 10 mM β-mercaptoethanol. Fractions containing the enzymes were dialyzed against gel-filtration buffer (25 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM β-mercaptoethanol) and loaded onto a HiLoad Superdex 200 26/60 prep-grade column (GE Healthcare, Freiburg, Germany) equilibrated with gel-filtration buffer. The collected protein fractions were dialyzed against 10 mM Tris–HCl buffer pH 8.0 and concentrated to 60 mg ml−1.
2.2. Crystallization
Crystals of the SoIPMDH–IPM–Mg2+ and SbIPMDH–IPM–Mg2+ complexes were grown in a hanging-drop vapour-diffusion setup using the microseeding method. The seed crystals were obtained via the hanging-drop vapour-diffusion method by mixing the protein solution (15 mg ml−1 in 10 mM magnesium chloride, 10 mM IPM, 10 mM Tris–HCl pH 8.0) with reservoir solution in a 1:1 ratio. The reservoir solution for crystallization of SoIPMDH consisted of 17% PEG 3350, 100 mM Na HEPES pH 7.0, 100 mM sodium chloride, while that for SbIPMDH consisted of 15% PEG 3350, 100 mM Na HEPES pH 6.7. Crystals of SoIPMDH and SbIPMDH grew to typical dimensions of 0.2 × 0.2 × 0.05 mm within 4–5 d. The crystals belonged to space groups C2 and P21, respectively. All crystallization experiments were carried out at 293 K.
2.3. Diffraction data collection and processing
Prior to low-temperature data collection at 100 K, an SoIPMDH crystal was soaked in cryoprotectant solution consisting of 25% PEG 3350, 100 mM Na HEPES pH 7.0, 100 mM sodium chloride, 10 mM magnesium chloride, 10 mM IPM. An SbIPMDH crystal was soaked in solution consisting of 32% PEG 3350, 100 mM Na HEPES pH 6.7, 10 mM magnesium chloride, 10 mM IPM. Data were collected from the SoIPMDH and SbIPMDH crystals on beamlines BL5A and NE3A at Photon Factory, Tsukuba, Japan, respectively. The diffraction data were processed using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). The data-collection parameters and processing statistics are summarized in Table 1 ▶.
Table 1. Data-collection parameters and refinement statistics for SoIPMDH and SbIPMDH crystals.
Values in parentheses are for the highest resolution shell.
| SoIPMDH–IPM–Mg2+ | SbIPMDH–IPM–Mg2+ | |
|---|---|---|
| Data collection | ||
| Beamline | BL5A | NE3A |
| Detector | ADSC Quantum 210r | ADSC Quantum 270 |
| Temperature (K) | 100 | |
| Wavelength (Å) | 1.000 | |
| Oscillation angle per frame (°) | 1 | |
| Exposure time per frame (s) | 1 | 3 |
| Crystal-to-detector distance (mm) | 142 | 167 |
| No. of crystals used | 1 | |
| Space group | C2 | P21 |
| Unit-cell parameters | ||
| a (Å) | 102.68 | 51.97 |
| b (Å) | 57.50 | 59.21 |
| c (Å) | 76.33 | 119.78 |
| β (°) | 119.03 | 95.12 |
| Resolution range (Å) | 50.00–1.56 (1.59–1.56) | 50.00–1.48 (1.51–1.48) |
| No. of reflections | 55280 (2621) | 118167 (5710) |
| Rmerge† (%) | 3.8 (25.5) | 4.0 (32.2) |
| Completeness (%) | 99.2 (93.9) | 98.1 (96.1) |
| 〈I/σ(I)〉 | 40.06 (4.66) | 38.34 (4.60) |
| Multiplicity | 3.6 (2.8) | 3.6 (3.2) |
| Refinement | ||
| Rwork‡ (%) | 17.80 | 19.34 |
| Rfree§ (%) | 21.61 | 23.19 |
| No. of atoms | ||
| Protein | 2826 | 5654 |
| Ligand/ion | 14 | 28 |
| Water | 368 | 709 |
| B factor (Å2) | ||
| Protein | 19.32 | 18.96 |
| Ligand/ion | 15.37 | 11.84 |
| Water | 30.72 | 27.26 |
| R.m.s.d. from ideality | ||
| Bond lengths (Å) | 0.029 | 0.029 |
| Bond angles (°) | 2.415 | 2.446 |
R
merge is defined as 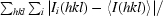
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted mean of all observations (after rejection of outliers).
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted mean of all observations (after rejection of outliers).
R
work is defined as 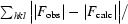
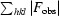 .
.
R free is calculated using 5% of the data randomly chosen and excluded from the refinement.
2.4. Structure determination
The initial crystal structures of SoIPMDH and SbIPMDH were solved with the program MOLREP (Vagin & Teplyakov, 2010 ▶) from the CCP4 suite (Winn et al., 2011 ▶) using an SoIPMDH monomer structure obtained at room temperature (PDB entry 3vkz; Nagae et al., 2012 ▶) as the search model. The structures of each IPMDH were then refined with the program REFMAC5 (Murshudov et al., 2011 ▶) and manually fitted using the program Coot (Emsley & Cowtan, 2004 ▶). Atomic coordinates and structure factors for SoIPMDH and SbIPMDH have been deposited in the Protein Data Bank with accession codes 3vmj and 3vmk, respectively.
The volumes of all of the cavities in the IPMDH dimer were first calculated with the program CASTp using a probe radius of 1.4 Å (Dundas et al., 2006 ▶). The net volumes of the internal cavities were then estimated by subtracting the volume of the water molecules and chloride anions in the cavities from the volume calculated by CASTp, where the volume used for the water molecule was 24.5 Å3 (Perkins, 2001 ▶) and that for the chloride anion was 24.4 Å3, corresponding to the volume of a sphere with a radius of 1.8 Å. The root-mean-square deviation (r.m.s.d.) between equivalent Cα atoms in the structures was calculated with SSM (Schneider, 2002 ▶). Schematic figures of the structures were drawn with the PyMOL visualization software (DeLano, 2002 ▶). The vectors in Fig. 2 were calculated using the modevector.py script from the PyMOL Wiki website (http://www.pymolwiki.org). The internal cavities in Figs. 3(a) and 3(b) were calculated with the program HOLLOW (Ho & Gruswitz, 2008 ▶).
3. Results and discussion
3.1. Overall structure of SoIPMDH and SbIPMDH
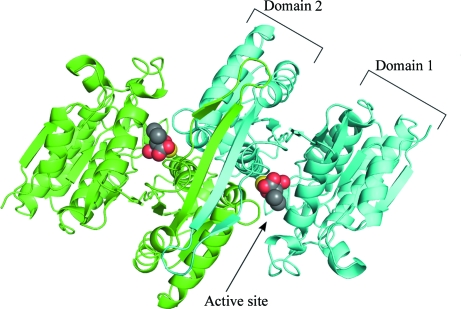
In common with other IPMDHs, SoIPMDH and SbIPMDH have a dimer in the biological unit. An overview of the SoIPMDH dimer is shown in Fig. 1 ▶. The crystal of SoIPMDH contains one subunit and that of SbIPMDH contains two subunits per asymmetric unit. Therefore, the SoIPMDH dimer is composed of two subunits related by the crystallographic twofold axis, while the SbIPMDH subunits are related by a noncrystallographic twofold axis. Each IPMDH monomer consists of two domains, referred to as domain 1 and domain 2, connected by a hinge region (Fig. 2 ▶). The relative orientation of the two domains defines whether the enzyme is in the open form or the closed form. In this study, the enzymes adopt a closed conformation owing to the binding of IPM.
Figure 1.
Cartoon representation of the overall structure of the SoIPMDH dimer (green, subunit A; cyan, subunit B). The IPM substrate molecules and the magnesium cations are represented as spheres (grey, carbon; red, oxygen; yellow, magnesium).
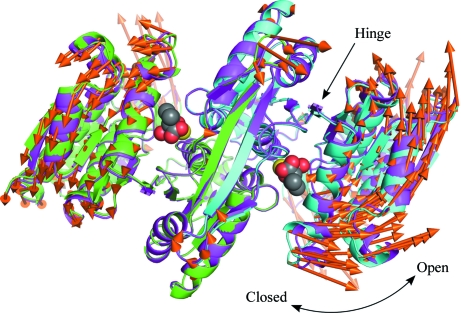
Figure 2.
Superposition of the overall structures of SoIPMDH and SbIPMDH. Domain 2 of the IPMDHs are superposed. Cartoon representation of the overall structure of the SoIPMDH and SbIPMDH dimers (green and cyan, subunit A and B of SoIPMDH, respectively; magenta, SbIPMDH). The IPM substrate molecules and the magnesium cations are represented as spheres. The vectors between the corresponding Cα positions of SoIPMDH and SbIPMDH are represented as orange arrows. For easier visualization, the lengths of the vectors are magnified eight times.
The two enzymes have highly similar structures as expected from their sequence similarity, although they show quite different pressure tolerances (Kasahara et al., 2009 ▶). The r.m.s.d. between equivalent Cα atoms in the SoIPMDH and SbIPMDH dimers is 0.322 Å. The vectors between the corresponding Cα positions of SoIPMDH and SbIPMDH are represented as orange arrows in Fig. 2 ▶. The vectors show that the SbIPMDH dimer is in a more open form than the SoIPMDH dimer.
3.2. Cavities in the protein interior and the dimer interface
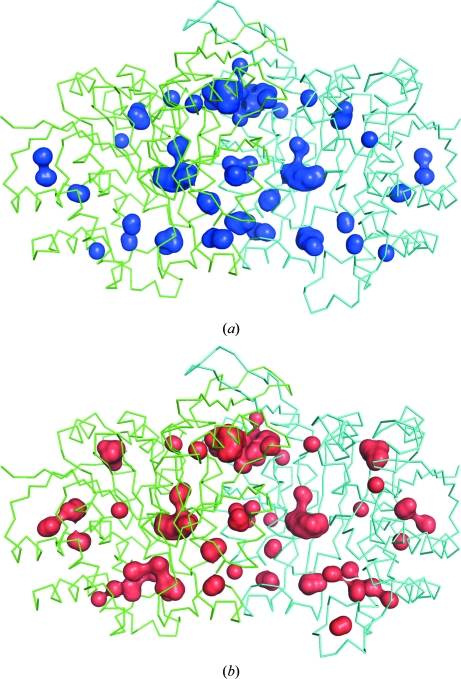
Fig. 3 ▶ shows the internal cavities in the SoIPMDH and SbIPMDH dimers. The program HOLLOW found 32 cavities in the subunits and ten cavities at the subunit interface of the SoIPMDH dimer. In the case of SbIPMDH the program found 39 and ten cavities in the subunits and the subunit interface, respectively. The total volumes of the internal cavities of SoIPMDH and SbIPMDH were estimated to be 1426 and 1537 Å3, respectively. Their net volumes, from which the volume of waters and chloride ions inside them were excluded, were estimated to be 592 and 655 Å3, respectively (Table 2 ▶). The cavity volume in the SbIPMDH dimer is larger than that in the SoIPMDH dimer at atmospheric pressure.
Figure 3.
The internal cavities of (a) SoIPMDH and (b) SbIPMDH dimers. The wire representation shows the overall structure of the IPMDHs (each subunit is drawn in green and cyan) and the surface representation in blue and red shows the internal cavities of SoIPMDH and SbIPMDH, respectively.
Table 2. Cavity volume of the protein interior and the dimer interface.
| SoIPMDH | SbIPMDH | |
|---|---|---|
| Total volume of the cavities (Å3) | 1426 | 1537 |
| No. of water molecules in the cavities | 32 | 34 |
| No. of chloride anions in the cavities | 2 | 2 |
| Net volume of the cavities (Å3) | 592 | 655 |
3.3. Protein structure and pressure tolerance
Under high pressure, proteins adapt a state of lower partial molar volume (PMV) according to Le Chatelier’s principle. The PMV of a macromolecule is expressed as the summation of the van der Waals volume, the void volume (internal cavities) and the volume change caused by hydration. A protein molecule should have a larger compressibility if it has larger void volume, or larger internal cavities, in its interior. The total PMV reduction of the protein ubiquitin was found to primarily be caused by a decrease in the void volume (Imai et al., 2007 ▶). At moderate pressures of up to a few hundred megapascals, the SoIPMDH dimer becomes more closed and the internal cavities of the dimer are compressed monotonically (Nagae et al., 2012 ▶). In this study, it is revealed that the SbIPMDH dimer has a more open conformation and also has a larger internal cavity volume than that of SoIPMDH. These results seem to show that SbIPMDH is more compressible than SoIPMDH under pressure. This might be one of the reasons for the pressure tolerance of SbIPMDH. It could be said that the SbIPMDH is able to avoid pressure-induced distortion of its native structure because its large voids play a role as a kind of damper and enable it to remain active at high pressure, whereas SoIPMDH becomes inactivated.
4. Conclusions
The structures of SoIPMDH and SbIPMDH were determined at a high resolution of ∼1.5 Å. The SbIPMDH dimer is in a more open form and has a larger cavity volume than the SoIPMDH dimer at atmospheric pressure. These features could help SbIPMDH from the obligate piezophile S. benthica DB21MT-2 to avoid pressure-induced distortion of the native structure and remain active at higher pressure than SoIPMDH from the nonpiezophile S. oneidensis MR-1.
A more detailed investigation should be performed using high-pressure protein crystallography; the crystals presented in this report are suitable for application to this method because of their high resolution. Work in this direction is in progress.
Supplementary Material
PDB reference: SoIPMDH, 3vmj
PDB reference: SbIPMDH, 3vmk
Acknowledgments
The authors acknowledge the technical staff of beamlines BL5A and NE3A of the Photon Factory for assistance during experiments. This work was partially supported by a KAKENHI Grant-in-Aid for Challenging Exploratory Research (21657027).
References
- Akanuma, S., Yamagishi, A., Tanaka, N. & Oshima, T. (1999). Eur. J. Biochem. 260, 499–504. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Dundas, J., Ouyang, Z., Tseng, J., Binkowski, A., Turpaz, Y. & Liang, J. (2006). Nucleic Acids Res. 34, W116–W118. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hayashi-Iwasaki, Y. & Oshima, T. (2000). Methods Enzymol. 324, 301–322. [DOI] [PubMed]
- Ho, B. K. & Gruswitz, F. (2008). BMC Struct. Biol. 8, 49. [DOI] [PMC free article] [PubMed]
- Hurley, J. H. & Dean, A. M. (1994). Structure, 2, 1007–1016. [DOI] [PubMed]
- Imada, K., Inagaki, K., Matsunami, H., Kawaguchi, H., Tanaka, H., Tanaka, N. & Namba, K. (1998). Structure, 6, 971–982. [DOI] [PubMed]
- Imada, K., Sato, M., Tanaka, N., Katsube, Y., Matsuura, Y. & Oshima, T. (1991). J. Mol. Biol. 222, 725–738. [DOI] [PubMed]
- Imai, T., Ohyama, S., Kovalenko, A. & Hirata, F. (2007). Protein Sci. 16, 1927–1933. [DOI] [PMC free article] [PubMed]
- Kadono, S., Sakurai, M., Moriyama, H., Sato, M., Hayashi, Y., Oshima, T. & Tanaka, N. (1995). J. Biochem. 118, 745–752. [DOI] [PubMed]
- Kasahara, R., Sato, T., Tamegai, H. & Kato, C. (2009). Biosci. Biotechnol. Biochem. 73, 2541–2543. [DOI] [PubMed]
- Kato, C., Li, L., Nogi, Y., Nakamura, Y., Tamaoka, J. & Horikoshi, K. (1998). Appl. Environ. Microbiol. 64, 1510–1513. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nagae, T., Kawamura, T., Chavas, L. M. G., Niwa, K., Hasegawa, M., Kato, C. & Watanabe, N. (2012). Acta Cryst. D68, 300–309. [DOI] [PMC free article] [PubMed]
- Nogi, Y. & Kato, C. (1999). Extremophiles, 3, 71–77. [DOI] [PubMed]
- Ohkuri, T. & Yamagishi, A. (2007). J. Biochem. 141, 791–797. [DOI] [PubMed]
- Ohmae, E., Kubota, K., Nakasone, K., Kato, C. & Gekko, K. (2004). Chem. Lett. 33, 798–799.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Perkins, S. J. (2001). Proc. Natl Acad. Sci. USA, 93, 129–139.
- Schneider, T. R. (2002). Acta Cryst. D58, 195–208. [DOI] [PubMed]
- Tamakoshi, M., Nakano, Y., Kakizawa, S., Yamagishi, A. & Oshima, T. (2001). Extremophiles, 5, 17–22. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: SoIPMDH, 3vmj
PDB reference: SbIPMDH, 3vmk