In order to characterize the type IV pili of nontypeable Haemophilus influenzae, an attempt to solve the atomic structure of the major pilin subunit PilA was initiated. A 1.73 Å resolution X-ray diffraction data set was collected from native N-terminally truncated PilA (ΔN-PilA).
Keywords: nontypeable Haemophilus influenzae, type IV pili, otitis media
Abstract
The type IV pili of nontypeable Haemophilus influenzae (NTHi) are involved in twitching motility, adherence, competence and biofilm formation. They are potential virulence factors for this important human pathogen and are thus considered to be vaccine targets. To characterize these pili, an attempt to solve the atomic structure of the major pilin subunit PilA was initiated. A 1.73 Å resolution X-ray diffraction data set was collected from native N-terminally truncated PilA (ΔN-PilA). Data processing indicated a hexagonal crystal system, which was determined to belong to space group P61 or P65 based on the systematic absences and near-perfect twinning of the crystal. The unit-cell parameters were a = b = 68.08, c = 197.03 Å with four molecules in the asymmetric unit, giving a solvent content of 50%. Attempts to solve the ΔN-PilA structure by molecular replacement with existing type IV pilin and type II secretion pseudopilin structures are in progress.
1. Introduction
Nontypeable Haemophilus influenzae (NTHi) are opportunistic Gram-negative bacterial pathogens that infect both the upper and the lower respiratory tracts (Murphy, 2003 ▶). These unencapsulated strains of H. influenzae cause numerous illnesses, including sinusitis, recurrent otitis media and exacerbations of chronic obstructive pulmonary disease and bronchitis. NTHi express type IV pili (T4P), hairlike filaments with diverse functions critical to pathogenesis in many Gram-negative bacteria (Ayers et al., 2010 ▶; Craig & Li, 2008 ▶; Pelicic, 2008 ▶). In NTHi, T4P are involved in adherence, twitching motility and biofilm formation (Bakaletz et al., 2005 ▶; Jurcisek & Bakaletz, 2007 ▶; Jurcisek et al., 2007 ▶).
Type IV pili are only 60–90 Å in diameter and 1 µm or more in length and are comprised of thousands of copies of the major pilin protein. Crystal structures of full-length type IV pilins revealed an extended N-terminal α-helix and a globular C-terminal domain with a central β-sheet and a C-terminal disulfide bridge delineating the D-region (Craig et al., 2003 ▶, 2006 ▶; Parge et al., 1995 ▶; Hartung et al., 2011 ▶). The highly conserved N-terminal α-helix serves as the polymerization domain and the variable C-terminal globular domain forms the pilus surface and defines many of its diverse functions (Craig & Li, 2008 ▶). The NTHi pilin subunit PilA is a member of the type IVa pilin subclass, which differ from the type IVb pilins in the lengths of the signal peptide, D-region and mature protein as well as in the topology of the globular domain (Craig & Li, 2008 ▶). In addition to the conserved C-terminal cysteine pair, NTHi PilA has two central cysteines, similar to PilA from Pseudomonas aeruginosa strain K122-4 and FimA from Dichelobacter nodosus. The PilA sequence is relatively conserved in NTHi disease isolates, with the exception of a short segment flanking one of the central cysteines and another segment towards the C-terminus of the protein. Thus, these pili are attractive targets for broad-specificity NTHi vaccines (Novotny et al., 2009 ▶, 2011 ▶). The atomic structure of the NTHi type IV pilin would be of great value in the design of vaccine candidates and may provide insights into the role of these pili in pathogenesis. Here, we describe our progress in determining the X-ray crystal structure of NTHi PilA.
2. Materials and methods
2.1. Expression and purification of ΔN-PilA
The pilA gene fragment encoding residues 29–137 (ΔN-PilA) was PCR-amplified from NTHi strain 86-028NP genomic DNA (Bakaletz et al., 2005 ▶; Jurcisek & Bakaletz, 2007 ▶; Jurcisek et al., 2007 ▶) using the primer GCGCATATGACTAAAAAAGCAGCGGTATCTG containing an NdeI site and the primer GCCAGATCTGCCATTTTGAGCGGTTACAC containing a BglII site. The pilA gene fragment was cloned into the pET15b vector (Novagen), which encodes an N-terminal hexahistidine tag (His tag) and linker, as described by Carruthers et al. (work submitted). A plasmid with the correct sequence was designated pRSPILA. Escherichia coli Origami (DE3) cells (Novagen) transformed with pRSPILA were grown in 6 l Luria broth containing ampicillin (100 µg ml−1) and kanamycin (30 µg ml−1) with shaking at 310 K until they reached an OD600 of ∼0.4. At this point, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.4 mM to induce ΔN-PilA expression and the cells were grown for a further 18 h at 292 K. All purification steps were carried out at 277 K. The cells were harvested by centrifugation at 5000g for 30 min and then flash-cooled in liquid nitrogen and thawed to disrupt the cell membranes. The cell pellets were resuspended in lysis buffer (50 mM Na2HPO4/NaH2PO4 pH 7.4, 500 mM NaCl) with 1 mg ml−1 lysozyme and protease-inhibitor cocktail (EDTA-free, Roche Pharmaceuticals) and incubated for 1 h at room temperature. The cells were lysed by sonication and centrifuged at 40 000g for 1 h to remove cell debris. The supernatant was loaded onto an Ni–NTA column (GE Healthcare) pre-equilibrated with lysis buffer and 40 mM imidazole pH 7.4. The column was washed with the same buffer and the protein was eluted using 300 mM imidazole. The protein was concentrated using an Amicon stirred-cell concentrator (Millipore) and loaded onto a Sephacryl S-100 size-exclusion column pre-equilibrated with 20 mM Tris–HCl pH 7.4, 50 mM NaCl, 1 mM EDTA. Peak fractions were pooled together and concentrated to 20 mg ml−1 using a stirred-cell concentrator. The ΔN-PilA protein was greater than 95% pure as assessed by a Coomassie-stained SDS–PAGE gel. Its identity was confirmed by mass spectrometry (data not shown).
2.2. Crystallization
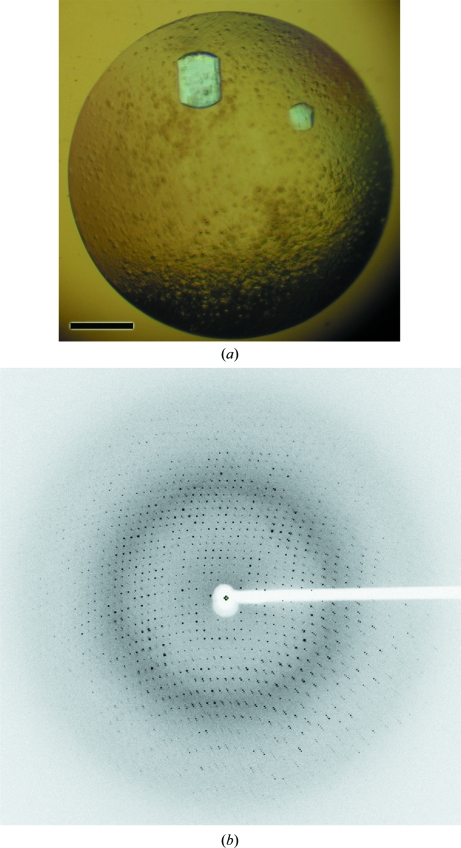
ΔN-PilA crystals were obtained using the hanging-drop vapour-diffusion method with 2 µl protein solution (20 mg ml−1) and 2 µl reservoir solution. Diffraction-quality crystals of ΔN-PilA grew in 20 mM Tris–HCl pH 7.4, 200 mM LiSO4, 1 M potassium/sodium tartrate (Fig. 1 ▶ a) after approximately seven months at 293 K. The crystals were cryoprotected using glycerol by first transferring them into mother liquor with 5%(v/v) glycerol and then increasing the glycerol content incrementally by 5% to a final concentration of 25% prior to freezing and storage in liquid nitrogen.
Figure 1.
ΔN-PilA crystals and X-ray diffraction. (a) H. influenzae ΔN-PilA crystal after seven months of growth in 20 mM Tris–HCl pH 7.4, 200 mM LiSO4, 1 M potassium/sodium tartrate. (b) Representative diffraction image for the ΔN-PilA crystal displayed in the program MOSFLM (Leslie, 1992 ▶). The resolution at the edge of the image is 1.7 Å.
2.3. Diffraction data collection and processing
A native ΔN-PilA data set was collected on beamline 08ID-1 (CMCF-ID) at the Canadian Light Source (CLS; Fig. 1 ▶ b). Raw diffraction data were reduced using XDS (Kabsch, 2010 ▶) and scaled to 1.73 Å resolution using XSCALE. The data quality was assessed using SFCHECK (Vaguine et al., 1999 ▶) and the solvent content was calculated using MATTHEWS_COEF from CCP4 (Winn et al., 2011 ▶). The programs Phaser (McCoy et al., 2007 ▶), MOLREP (Vagin & Teplyakov, 2010 ▶), EPMR (Kissinger et al., 1999 ▶; Long et al., 2008 ▶) and BALBES (Long et al., 2008 ▶) were used for molecular-replacement trials, with the type IVa pilins from Neisseria gonorrhoeae (PilE; PDB entry 2hi2; Craig et al., 2006 ▶), Pseudomonas aeruginosa (PilA, PDB entry 1oqw, Craig et al., 2003 ▶; K122-4 pilin, PDB entry 1qve, Audette et al., 2004 ▶; Pa110594 pilin, PDB entry 3jyz, Nguyen et al., 2010 ▶), Dichelobacter nodosus (FimA; PDB entry 3sok; Hartung et al., 2011 ▶) and Francisella tularensis (PilA; PDB entry 3soj; Hartung et al., 2011 ▶) as well as the type II secretion pseudopilin PulG from Klebsiella oxytoca (PDB entry 1t92; Köhler et al., 2004 ▶) as search models.
3. Results and discussion
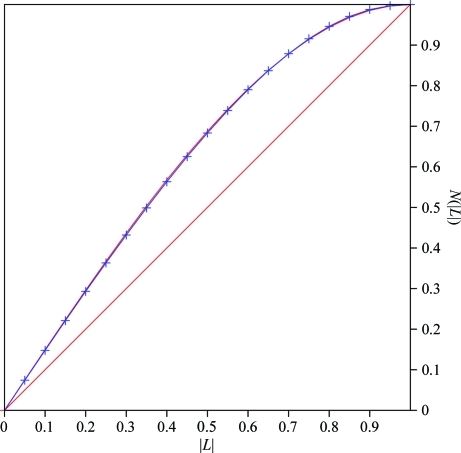
Recombinant ΔN-PilA was expressed in E. coli Origami cells at high levels, resulting in a yield of 5 mg purified protein per litre of cell culture at ∼95% purity. The crystals took many months to grow and resulted in only a few diffraction-quality crystals, from which a single 1.73 Å resolution data set was collected. The native ΔN-PilA data set was initially processed using XDS, which suggested a hexagonal crystal system (quality of fit 2.2 for primitive hexagonal). The unit-cell parameters were a = b = 68.08, c = 197.03 Å, α = β = 90, γ = 120° (Table 1 ▶). The processed data scaled well in 622, the highest symmetry in the hexagonal system, and XSCALE gave an R merge of 5.9% overall (41.6% for the highest resolution shell, 1.90–1.73 Å) and an 〈I/σ(I)〉 of 10.2 (2.2 for the highest resolution shell). Upon manual inspection of the reflection file, only the l = 6n reflections were present for 00l, thereby identifying a 61/65 screw and suggesting that the ΔN-PilA crystal belonged to either space group P6122 or P6522. A twin test was performed on the data set using the Merohedral Twin Detector: Padilla–Yeates Algorithm (http://nihserver.mbi.ucla.edu/pystats/; Padilla & Yeates, 2003 ▶), which gave 〈|L|〉 and 〈L 2〉 of 0.344 and 0.171 (Fig. 2 ▶), respectively, consistent with close-to-perfect twinning (〈|L|〉 = 0.500 and 〈L 2〉 = 0.333 for untwinned crystals and 〈|L|〉 = 0.375 and 〈L 2〉 = 0.200 for perfectly twinned crystals). The data set was further evaluated with CTRUNCATE in CCP4 (Winn et al., 2011 ▶), which confirmed near-perfect twinning. Since twinning is not possible for the 622 point group (Yeates, 1997 ▶) and the systematic absences ruled out all space groups in the P312 and P321 crystal systems, the space group was assigned as P61 or its enantiomorph P65. For a solvent content of ∼50%, there are four ΔN-PilA molecules in the asymmetric unit based on the Matthews coefficient calculation (Matthews, 1968 ▶).
Table 1. Crystallographic data-collection statistics for ΔN-PilA.
Values in parentheses are for the highest resolution shell.
| Beamline | CLS 08ID-1 |
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Space group | P61/P65 |
| Unit-cell parameters (Å, °) | a = b = 68.08, c = 197.03, α = β = 90.0, γ = 120.0 |
| Resolution (Å) | 20.0–1.73 |
| Completeness (%) | 92.2 (66.2) |
| Observed reflections | 488919 |
| Unique reflections | 52990 |
| Rmerge† (%) | 6.4 (48.4) |
| 〈I/σ(I)〉 | 22.0 (4.4) |
| Multiplicity | 9.2 (8.5) |
| Wilson B value (Å2) | 29.8 |
| Mosaicity (°) | 0.2 |
R
merge = 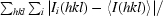
 , where I
i(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity of that reflection.
, where I
i(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity of that reflection.
Figure 2.
Twinning analysis. Results of analysis of ΔN-PilA (P61/P65) crystallographic data using the Merohedral Twin Detector: Padilla–Yeates Algorithm (http://nihserver.mbi.ucla.edu/pystats/). The red line represents theoretical untwinned data and the red curve represents theoretical perfectly twinned data. The blue curve is the observed data.
We are currently attempting to solve the ΔN-PilA structure by molecular replacement with type IV pilins of known structure using data scaled in P61/P65 as well as other possible trigonal and hexagonal space groups. NTHi is most similar in sequence to the type IV pilin PilE from N. gonorrhoeae (46% amino-acid sequence identity for the first 86 residues) and PAK pilin (44% identity) and K122-4 pilin (41% identity) from P. aeruginosa. However, sequence similarity is strongest in the N-terminal ∼25 amino acids of these pilins, which are absent in ΔN-PilA. All available type IVa pilin structures and also the type II secretion pseudopilin PulG were used as search models. Side chains were removed and replaced by alanines (polyalanine models) or by glycines (polyglycine models) in Coot (Emsley & Cowtan, 2004 ▶). Polyalanine/polyserine models were also generated in which a hydroxyl was added to Cβ of residues with large side chains. In addition, a composite model generated in Coot from an overlay of all available type IVa pilin structures was used in molecular-replacement trials. Loops were removed from the models and the protruding segment of the N-terminal α-helix α1N was removed from full-length pilin structures. Thus far, none of the searches have yielded a structure solution. We are also attempting ab initio structure determination using the ARCIMBOLDO set of programs (Rodríguez et al., 2009 ▶).
4. Conclusions
We have purified and crystallized N-terminally truncated NTHi PilA and obtained a high-resolution data set. However, structure determination by molecular replacement was impeded because of crystal twinning and the large number of molecules in the asymmetric unit. Furthermore, the existing pilin structures may not be sufficiently similar to PilA to provide a molecular-replacement solution. The native data set is likely to be of sufficient quality to determine the ΔN-PilA crystal structure if phases can be obtained by ab initio or MAD methods. Though challenging, the PilA structure is sure to reveal unique structural features that may provide insights into its functions in NTHi colonization and pathogenesis as well as aid in the design of vaccine candidates based on this important virulence determinant.
Acknowledgments
We thank Benjamin Hon for protein expression and purification and the beamline staff at the Canadian Light Source. This work was supported by operating grant R01 DC007464 from the National Institutes of Health to RSM. LC was supported by a New Investigator Award from the Canadian Institutes of Health Research and a Scholar Award from the Michael Smith Foundation for Health Research.
References
- Audette, G. F., Irvin, R. T. & Hazes, B. (2004). Biochemistry, 43, 11427–11435. [DOI] [PubMed]
- Ayers, M., Howell, P. L. & Burrows, L. L. (2010). Future Microbiol. 5, 1203–1218. [DOI] [PubMed]
- Bakaletz, L. O., Baker, B. D., Jurcisek, J. A., Harrison, A., Novotny, L. A., Bookwalter, J. E., Mungur, R. & Munson, R. S. (2005). Infect. Immun. 73, 1635–1643. [DOI] [PMC free article] [PubMed]
- Craig, L. & Li, J. (2008). Curr. Opin. Struct. Biol. 18, 267–277. [DOI] [PMC free article] [PubMed]
- Craig, L., Taylor, R. K., Pique, M. E., Adair, B. D., Arvai, A. S., Singh, M., Lloyd, S. J., Shin, D. S., Getzoff, E. D., Yeager, M., Forest, K. T. & Tainer, J. A. (2003). Mol. Cell, 11, 1139–1150. [DOI] [PubMed]
- Craig, L., Volkmann, N., Arvai, A. S., Pique, M. E., Yeager, M., Egelman, E. H. & Tainer, J. A. (2006). Mol. Cell, 23, 651–662. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hartung, S., Arvai, A. S., Wood, T., Kolappan, S., Shin, D. S., Craig, L. & Tainer, J. A. (2011). J. Biol. Chem., doi:10.1074/jbc.M111.297242. [DOI] [PMC free article] [PubMed]
- Jurcisek, J. A. & Bakaletz, L. O. (2007). J. Bacteriol. 189, 3868–3875. [DOI] [PMC free article] [PubMed]
- Jurcisek, J. A., Bookwalter, J. E., Baker, B. D., Fernandez, S., Novotny, L. A., Munson, R. S. & Bakaletz, L. O. (2007). Mol. Microbiol. 65, 1288–1299. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999). Acta Cryst. D55, 484–491. [DOI] [PubMed]
- Köhler, R., Schäfer, K., Müller, S., Vignon, G., Diederichs, K., Philippsen, A., Ringler, P., Pugsley, A. P., Engel, A. & Welte, W. (2004). Mol. Microbiol. 54, 647–664. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murphy, T. F. (2003). Curr. Opin. Infect. Dis. 16, 129–134. [DOI] [PubMed]
- Nguyen, Y., Jackson, S. G., Aidoo, F., Junop, M. & Burrows, L. L. (2010). J. Mol. Biol. 395, 491–503. [DOI] [PubMed]
- Novotny, L. A., Adams, L. D., Kang, D. R., Wiet, G. J., Cai, X., Sethi, S., Murphy, T. F. & Bakaletz, L. O. (2009). Vaccine, 28, 279–289. [DOI] [PMC free article] [PubMed]
- Novotny, L. A., Clements, J. D. & Bakaletz, L. O. (2011). Mucosal Immunol. 4, 456–467. [DOI] [PMC free article] [PubMed]
- Padilla, J. E. & Yeates, T. O. (2003). Acta Cryst. D59, 1124–1130. [DOI] [PubMed]
- Parge, H. E., Forest, K. T., Hickey, M. J., Christensen, D. A., Getzoff, E. D. & Tainer, J. A. (1995). Nature (London), 378, 32–38. [DOI] [PubMed]
- Pelicic, V. (2008). Mol. Microbiol. 68, 827–837. [DOI] [PubMed]
- Rodríguez, D. D., Grosse, C., Himmel, S., González, C., de Ilarduya, I. M., Becker, S., Sheldrick, G. M. & Usón, I. (2009). Nature Methods, 6, 651–653. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yeates, T. O. (1997). Methods Enzymol. 276, 344–358. [PubMed]