Enoyl-acyl carrier protein reductase (FabK) from S. mutans strain UA159 was cloned, overexpressed, purified and crystallized. X-ray diffraction data were collected to a resolution of 2.40 Å.
Keywords: enoyl-acyl carrier protein reductases, FabK, Streptococcus mutans
Abstract
A triclosan-resistant flavoprotein termed FabK is the sole enoyl-acyl carrier protein reductase in Streptococcus pneumoniae and Streptococcus mutans. In this study, FabK from S. mutans strain UA159 was overexpressed in Escherichia coli, purified and crystallized. Diffraction data were collected to 2.40 Å resolution using a synchrotron-radiation source. The crystal belonged to space group P62, with unit-cell parameters a = b = 105.79, c = 44.15 Å. The asymmetric unit contained one molecule, with a corresponding V M of 2.05 Å3 Da−1 and a solvent content of 39.9%.
1. Introduction
Bacterial type II fatty-acid biosynthesis provides essential fatty acids for use in the assembly of key cellular components, including the cell envelope, phospholipids, lipoproteins and lipopolysaccharides (Campbell & Cronan, 2001 ▶). The bacterial type II fatty-acid synthase (FAS) complex is composed of different monofunctional enzymes which catalyse each of the reactions, and the reaction intermediates in bacterial FAS are covalently linked to acyl carrier proteins (ACPs) and carried through the cytosol (Rock & Cronan, 1996 ▶). In contrast, mammals use a single large multifunctional protein, type I FAS, in which the growing chain is covalently attached to the protein to make fatty acids. Owing to this difference between bacteria and mammals, bacterial FAS enzymes are attractive targets for the development of novel antibiotics (Heath et al., 2000 ▶). Enoyl-ACP reductase catalyzes the final step in each elongation cycle of bacterial FAS by a conjugation reduction using NADPH or NADH as a cofactor (Bergler et al., 1996 ▶).
In Escherichia coli and Staphylococcus aureus FabI is the sole enoyl-ACP reductase, and triclosan is known to inhibit FabI (Heath et al., 1998 ▶). The antituberculosis agent isoniazid also targets the FabI homologue (InhA) in Mycobacterium tuberculosis (Quémard et al., 1995 ▶). Other compounds, including diazaborines, indole naphthyridinones and thiopyridines, have been reported to be FabI inhibitors (Levy et al., 2001 ▶; Seefeld et al., 2003 ▶; Ling et al., 2004 ▶). However, recent genomic studies have shown that another bacterial enoyl-ACP reductase, FabK, is present in several pathogens and that this protein is resistant to triclosan. FabK is the sole enoyl-ACP reductase in Streptococcus pneumoniae and S. mutans, while both FabI and FabK exist in key pathogens such as Enterococcus faecalis and Pseudomonas aeruginosa (Heath & Rock, 2000 ▶). Unlike FabI, FabK contains a prosthetic group, flavin monophosphate (FMN), for enzymatic activity (Marrakchi et al., 2003 ▶). FabI catalyses the reduction of trans-2-enoyl-ACP using the cofactor NAD(P)H as a hydride source. In contrast, FabK shows a two-step ping-pong catalytic mechanism that couples the reduction of FMN to the oxidation of NADH to NAD+, and the product, FMNH2 −, acts as the final hydride source for the reduction of the substrate. To date, only one crystal structure of FabK from S. pneumoniae in complex with FMN has been determined (Saito et al., 2008 ▶). This structure showed a fold that is unrelated to those of mammalian enoyl-ACP reductases (Maier et al., 2006 ▶) and the bacterial counterparts FabI and InhA, which contain a Rossmann fold that supports a binding site for NAD(P)H but not for FMN (Baldock et al., 1996 ▶; Dessen et al., 1995 ▶). A sequence-homology search revealed two S. mutans proteins that were closely homologous to S. pneumoniae FabK, with identities of 80% and 48% (accession Nos. NP_722069 and NP_721702).
To provide further structural data with regard to the catalytic mechanism of FabK, we have crystallized and performed X-ray crystallographic experiments on the enzyme from the Gram-positive bacterium S. mutans, which is most strongly associated with dental caries.
2. Materials and methods
2.1. Cloning, protein expression and purification
The gene encoding full-length (321 amino acids) FabK (accession No. NP_722069) was amplified from genomic DNA of S. mutans strain UA159 by the polymerase chain reaction (PCR) using specific primers. The forward primer contained an NdeI restriction site (bold) and had the sequence 5′-GGG CCC CAT ATG AAA ACG CGT ATT ACA GAA T-3′, while the reverse primer contained a XhoI site (bold) and had the sequence 5′-GGG CCC CTC GAG TTT CTC CAC GTC TGC CC-3′. The PCR product was then subcloned between the NdeI and XhoI sites of pET-22b vector (Novagen, USA). This construct contains an additional hexahistidine tag (LEHHHHHH) at the C-terminus for purification purposes. E. coli BL21 (DE3) strain (Novagen) was transformed with the recombinant plasmid and the cells were grown in a shaking incubator at 310 K in LB broth medium supplemented with 50 µg ml−1 ampicillin. Protein expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the cells reached an OD600 of 0.6 and the cells were cultured at 293 K for ∼16 h. Cultured cells were harvested by centrifugation at 3000g for 30 min at 277 K. The cell pellet was resuspended in binding buffer (20 mM Tris pH 8.0, 300 mM NaCl) and disrupted by sonication at 277 K. The crude lysate was centrifuged at 25 000g for 1 h at 277 K. The supernatant was then loaded onto an Ni2+-chelated HisTrap HP column (GE Healthcare, USA) which had been pre-equilibrated with binding buffer. After washing with wash buffer (20 mM Tris pH 8.0, 300 mM NaCl, 50 mM imidazole), the bound protein was eluted with elution buffer (20 mM Tris pH 8.0, 300 mM NaCl, 400 mM imidazole). The eluted protein was dialyzed for 6 h at 277 K in buffer A (20 mM Tris pH 7.0, 50 mM NaCl) and loaded onto a HiTrap SP HP column (GE Healthcare, USA) which had been pre-equilibrated with buffer A. After washing with ten volumes of buffer A, a linear gradient of NaCl (50–1000 mM) was applied to elute the protein. The eluted protein was then further purified by gel-filtration chromatography on a Superdex 200 column (GE Healthcare, USA) equilibrated with 20 mM Tris pH 7.0, 0.25 M NaCl. The purified protein was concentrated to 50 mg ml−1 using a Centriprep-10 (Amicon) and the purity of the protein was examined by 12% SDS–PAGE; the protein was determined to be >95% pure.
2.2. Crystallization and data collection
Crystallization of the protein was initiated by crystal screening at 293 K using the hanging-drop vapour-diffusion method in 24-well VDX plates (Hampton Research, USA) with a ratio of 1 µl protein solution concentrated in the gel-filtration buffer to 1 µl well solution over 500 µl well solution. Commercial screening kits from Hampton Research and Emerald BioSystems (Crystal Screen, Crystal Screen 2, Index, SaltRx, Natrix, MembFac and Wizard I and II) were used in preliminary screening. Suitable-sized crystals were obtained within a week under the following condition: 0.05 M Na HEPES pH 7.0, 0.02 M magnesium chloride, 0.01 M ammonium acetate, 5% PEG 8000 (Fig. 1 ▶). The crystals were cryoprotected by soaking them for 3 s in a cryoprotectant solution containing an additional 30%(v/v) glycerol and were flash-cooled in liquid nitrogen. Cooled crystals were mounted on the goniometer in a stream of cold nitrogen at 100 K. X-ray diffraction data were collected from a cooled crystal using an ADSC Quantum 210r CCD detector on beamline BL-5A at the Photon Factory, Japan. A total rotation range of 180° was covered with 1.0° oscillation and 5 s exposure per frame. The wavelength of the synchrotron X-ray beam was 1.00 Å and the crystal-to-detector distance was set to 180 mm. X-ray diffraction data were collected to 2.40 Å resolution. Data were indexed, integrated, scaled and merged using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶).
Figure 1.
A crystal of FabK from S. mutans strain UA159 grown using 0.05 M Na HEPES pH 7.0, 0.02 M magnesium chloride, 0.01 M ammonium acetate, 5% PEG 8000. The crystal dimensions are approximately 0.1 × 0.05 × 0.5 mm.
3. Results and discussion
FabK from S. mutans strain UA159 was cloned, overexpressed, purified and crystallized for structural studies. X-ray diffraction data from the crystal indicated that the crystal belonged to space group P62 or P64 on the basis of systematic absences, with unit-cell parameters a = b = 105.79, c = 44.15 Å. Data-collection statistics are provided in Table 1 ▶. The Matthews coefficient suggested the presence of one molecule in the crystallographic asymmetric unit, with a V M of 2.05 Å3 Da−1 and a solvent content of 39.9% (Matthews, 1968 ▶). The molecular-replacement (MR) method was used in an attempt to solve the structure using the structure of FabK from S. pneumoniae (PDB entry 2z6i; Saito et al., 2008 ▶) as a model. The MR solution obtained using the CNS package (Brünger et al., 1998 ▶) made it clear that the space group of the crystal was P62 and that the asymmetric unit of the crystal contained one protein molecule. The best MR solution gave an R factor of 41.9% for data in the resolution range 15–3.5 Å. The other solutions showed R factors of over 53%. The results of gel-filtration chromatography implied that the protein eluted as dimer and examination of the best MR solution showed a similar dimeric interface as in the dimeric structure of S. pneumoniae FabK between symmetry-related molecules in the crystal packing. The final model is currently being refined.
Table 1. Data-collection statistics.
Values in parentheses are for the last resolution shell.
| X-ray source | PF beamline BL-5A |
| Wavelength (Å) | 1.00 |
| Resolution range (Å) | 50.0–2.40 (2.44–2.40) |
| Space group | P62 |
| Unit-cell parameters (Å) | a = b = 105.79, c = 44.15 |
| No. of unique reflections | 11005 |
| Multiplicity | 10.4 (10.8) |
| Completeness (%) | 98.4 (100.0) |
| Molecules per asymmetric unit | 1 |
| VM (Å3 Da−1) | 2.05 |
| Solvent content (%) | 39.9 |
| Average I/σ(I) | 58.9 (5.7) |
| Rmerge† (%) | 5.8 (38.1) |
R
merge = 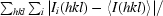
 , where Ii(hkl) is the intensity of an individual reflection hkl and 〈I(hkl)〉 is the average intensity of reflection hkl.
, where Ii(hkl) is the intensity of an individual reflection hkl and 〈I(hkl)〉 is the average intensity of reflection hkl.
Acknowledgments
We thank the manager of beamline BL-5A at Photon Factory for his assistance. This work was supported by Konkuk University in 2011.
References
- Baldock, C., Rafferty, J. B., Sedelnikova, S. E., Baker, P. J., Stuitje, A. R., Slabas, A. R., Hawkes, T. R. & Rice, D. W. (1996). Science, 274, 2107–2110. [DOI] [PubMed]
- Bergler, H., Fuchsbichler, S., Högenauer, G. & Turnowsky, F. (1996). Eur. J. Biochem. 242, 689–694. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Campbell, J. W. & Cronan, J. E. (2001). Annu. Rev. Microbiol. 55, 305–332. [DOI] [PubMed]
- Dessen, A., Quémard, A., Blanchard, J. S., Jacobs, W. R. & Sacchettini, J. C. (1995). Science, 267, 1638–1641. [DOI] [PubMed]
- Heath, R. J., Li, J., Roland, G. E. & Rock, C. O. (2000). J. Biol. Chem. 275, 4654–4659. [DOI] [PubMed]
- Heath, R. J. & Rock, C. O. (2000). Nature (London), 406, 145–146. [DOI] [PubMed]
- Heath, R. J., Yu, Y. T., Shapiro, M. A., Olson, E. & Rock, C. O. (1998). J. Biol. Chem. 273, 30316–30320. [DOI] [PubMed]
- Levy, C. W., Baldock, C., Wallace, A. J., Sedelnikova, S., Viner, R. C., Clough, J. M., Stuitje, A. R., Slabas, A. R., Rice, D. W. & Rafferty, J. B. (2001). J. Mol. Biol. 309, 171–180. [DOI] [PubMed]
- Ling, L. L., Xian, J., Ali, S., Geng, B., Fan, J., Mills, D. M., Arvanites, A. C., Orgueira, H., Ashwell, M. A., Carmel, G., Xiang, Y. & Moir, D. T. (2004). Antimicrob. Agents Chemother. 48, 1541–1547. [DOI] [PMC free article] [PubMed]
- Maier, T., Jenni, S. & Ban, N. (2006). Science, 311, 1258–1262. [DOI] [PubMed]
- Marrakchi, H., Dewolf, W. E., Quinn, C., West, J., Polizzi, B. J., So, C. Y., Holmes, D. J., Reed, S. L., Heath, R. J., Payne, D. J., Rock, C. O. & Wallis, N. G. (2003). Biochem. J. 370, 1055–1062. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 277, 307–326. [DOI] [PubMed]
- Quémard, A., Sacchettini, J. C., Dessen, A., Vilcheze, C., Bittman, R., Jacobs, W. R. & Blanchard, J. S. (1995). Biochemistry, 34, 8235–8241. [DOI] [PubMed]
- Rock, C. O. & Cronan, J. E. (1996). Biochim. Biophys. Acta, 1302, 1–16. [DOI] [PubMed]
- Saito, J., Yamada, M., Watanabe, T., Iida, M., Kitagawa, H., Takahata, S., Ozawa, T., Takeuchi, Y. & Ohsawa, F. (2008). Protein Sci. 17, 691–699. [DOI] [PMC free article] [PubMed]
- Seefeld, M. A. et al. (2003). J. Med. Chem. 46, 1627–1635. [DOI] [PubMed]



