Translation elongation factor eEF1A2 was purified to homogeneity from rabbit muscles. Obtained crystals of eEF1A2 diffracted to 2.5 Å resolution and belonged to space group P6122 or P6322.
Keywords: eEF1A2, translation elongation factors
Abstract
Translation elongation factor eEF1A2 was purified to homogeneity from rabbit muscle by two consecutive ion-exchange column-chromatography steps and this mammalian eEF1A2 was successfully crystallized for the first time. Protein crystals obtained using ammonium sulfate as precipitant diffracted to 2.5 Å resolution and belonged to space group P6122 or P6322 (unit-cell parameters a = b = 135.4, c = 304.6 Å). A complete native data set was collected to 2.7 Å resolution.
1. Introduction
Efficient translation of eukaryotic mRNAs is realised by the coordinated action of aminoacyl-tRNA synthetases, ribosomes and three groups of translation-promoting proteins called initiation, elongation and termination factors. Elongation factors ensure effective polypeptide-chain synthesis on 80S ribosomes. Translation elongation factor 1A (eEF1A) is one of the main participants in this process (Andersen et al., 2003 ▶; Negrutskii & El’skaya, 1998 ▶). The GTP-bound form of eEF1A generates a ternary complex with aminoacyl-tRNA regardless of the tRNA species. This complex delivers charged tRNA to the A site of the translating ribosome. In the case of correct codon/anticodon base pairing, the ribosome triggers GTP hydrolysis and inactive eEF1A–GDP leaves the A site. eEF1A–GDP then associates with the eFF1B complex, catalyzing GDP/GTP exchange and thus recycling the active GTP-bound form of eEF1A. It has been suggested (Petrushenko et al., 2002 ▶) that eEF1A–GDP may chaperone a deacylated tRNA molecule towards the cognate aminoacyl-tRNA synthetase. Considering the variety of essential functions performed by eEF1A, this factor is clearly a key component of the translation elongation cycle (Negrutskii et al., 1994 ▶; Negrutskii & El’skaya, 1998 ▶).
It has been found that eEF1A is not only involved in protein synthesis but also in other cellular processes. Thus, eEF1A interacts with actin and tubulin and participates in cytoskeleton rearrangement (Gillardon, 2009 ▶; Pittman et al., 2009 ▶). It is engaged in apoptosis (Chang & Wang, 2007 ▶) and lipotoxic cell death (Borradaile et al., 2006 ▶) and can also bind and regulate the expression of several cellular and viral mRNAs (Davis et al., 2007 ▶; Li et al., 2009 ▶; Miura et al., 2010 ▶; Fan et al., 2009 ▶). This is a far from complete list of the multiple so-called moonlighting functions of eEF1A.
In mammals, two isoforms of eEF1A, eEF1A1 and eEF1A2, exist; they contain 462 and 463 amino-acid residues, respectively, and are 92% identical. The isoforms are encoded by different genes (Bischoff et al., 2000 ▶) and display very different patterns of expression that are mutually exclusive: eEF1A2 is expressed only in myocytes and neurons, whereas the first isoform is expressed in all other cell types (Khalyfa et al., 2001 ▶). Importantly, eEF1A2 has been shown to be a potential oncogene that is overexpressed in cells where it is not normally present; its appearance has been correlated with cancer development (Anand et al., 2002 ▶; Tomlinson et al., 2005 ▶).
X-ray structures of a prokaryotic analogue of eEF1A, EF-Tu, in GDP-bound and GTP-bound conformations were solved some time ago (Kjeldgaard et al., 1993 ▶; Song et al., 1999 ▶). Subsequently, structures of ternary complexes of EF-Tu–GMPPNP with two different aminoacyl-tRNAs were published (Nissen et al., 1995 ▶, 1999 ▶). The prokaryotic EF-Tu and the eukaryotic eEF1A are more than 30% homologous and perform the same function in translation. In view of this, a great deal of similarity between prokaryotic and eukaryotic translation elongation factors at the level of their structural organization might be expected. However, the X-ray structure of yeast eEF1A in complex with a truncated eEF1Bα subunit revealed some unique features compared with the prokaryotic EF-Tu–EF-Ts complex (Andersen et al., 2000 ▶). Therefore, a crystal structure of mammalian eEF1A is of great interest and importance. However, despite extensive efforts, no crystals of eEF1A1 from higher eukaryotes have been obtained to date. We reported previously that the eEF1A1 isoform might possess a nonglobular extended conformation in solution that changes to an essentially more compact conformation upon interaction with tRNAs (Budkevich et al., 2002 ▶). This fact may to some extent explain the reluctance of eEF1A1 to crystallize. On the other hand, we have recently shown that the eEF1A2 isoform is more thermodynamically stable (Novosylna et al., 2007 ▶) and probably more compact than eEF1A1. Consequently, we came to the decision that the eEF1A2 molecule would be more appropriate for crystallization trials.
As the first step to resolving the three-dimensional structure of a mammalian eEF1A, we report here the purification, crystallization and preliminary crystallographic studies of native intact eEF1A2 protein from rabbit muscle.
2. Methods and results
2.1. Purification of translation elongation factor eEF1A2 from rabbit muscle
Rabbit muscle (200 g) was homogenized in 400 ml buffer A (30 mM potassium phosphate pH 7.5, 1 mM MgCl2, 15% glycerol, 6 mM β-mercaptoethanol supplemented with 1 mM PMSF) using a tissue blender at 8000 rev min−1 for 1 min. After 40 min centrifugation at 12 000g (277 K), the post-mitochondrial supernatant (about 500 ml) was recovered and filtered through four layers of sterile gauze to remove fat. The clear supernatant was immediately loaded onto a DEAE-cellulose column (4 × 14 cm, 175 ml; Whatman) previously equilibrated with buffer A. Unbound proteins were collected and loaded directly onto an SP-Sepharose column (1.1 × 10 cm, 9.5 ml; GE Healthcare) equilibrated with buffer A. The column was extensively washed with buffer A containing 100 mM KCl until the OD at 280 nm reached the baseline. eEF1A2 was eluted from the column with a 20 column volume linear gradient of 100–350 mM KCl in buffer A. The presence of eEF1A2 in the fractions was monitored using a [3H]GDP-binding assay (Shalak et al., 1997 ▶) and SDS–PAGE. The fractions containing the elongation factor with more than 90% purity (according to SDS–PAGE) were pooled, dialyzed against buffer A and concentrated using a small SP-Sepharose column (1 ml; GE Healthcare). Finally, eEF1A2 was dialyzed against storage buffer (25 mM potassium phosphate pH 7.5, 1 mM MgCl2, 25% glycerol, 2 mM DTT) and stored in liquid nitrogen. The purity of the eEF1A2 sample used for crystallization is shown in Fig. 1 ▶. The functional activity of purified eEF1A2 was demonstrated by two control tests: a [3H]GDP/GDP-exchange assay and stimulation of poly[14C]phenylalanine synthesis on poly(U)-programmed 80S ribosomes (data not shown).
Figure 1.
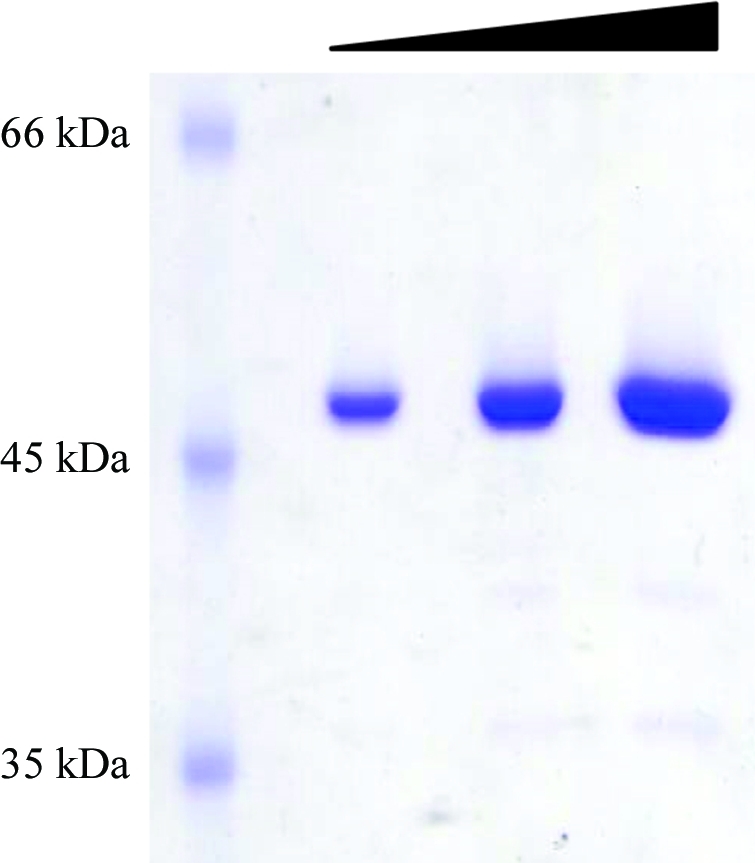
eEF1A2 sample prior to crystallization. The protein collected from the final purification step (SP-Sepharose column) was analyzed on 10% SDS–PAGE and stained with Coomassie blue. Several amounts of the same sample were loaded: from left to right, 0.5, 1 and 2 µg.
2.2. Crystallization and preliminary X-ray analysis
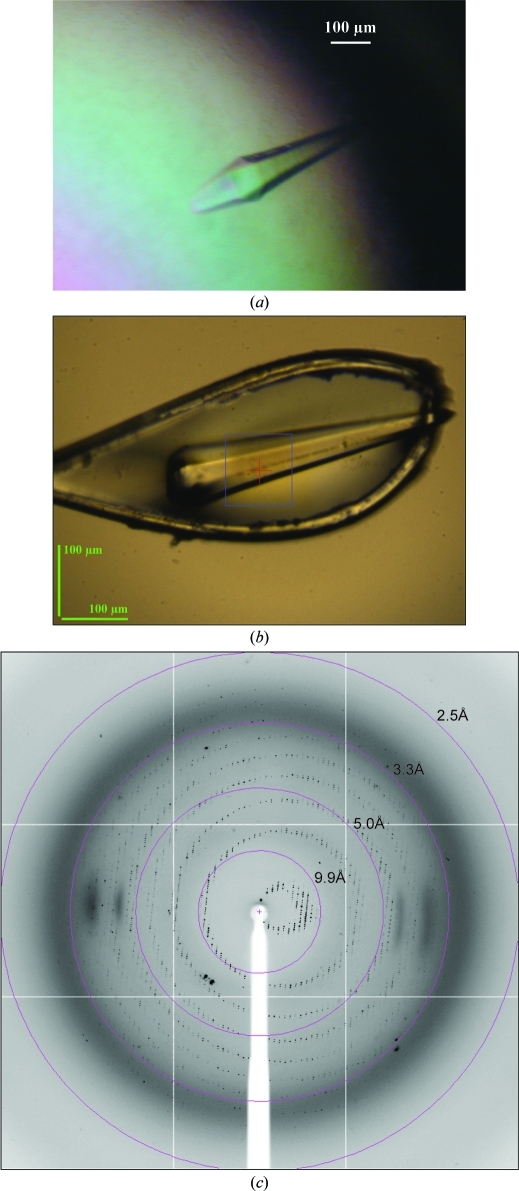
Prior to crystallization, the purified protein was dialyzed against 5 mM Na HEPES pH 7.5, 5 mM MgCl2, 2 mM DTT, 15% glycerol, 0.005% NaN3 and concentrated to about 10 mg ml−1 by filtration through a YM30 membrane (Amicon, UK). Protein samples were mixed with GDP to give a final GDP concentration of 100 µM prior to setting up crystallization experiments. Initial crystallization experiments were carried out at 293 K using the sitting-drop vapour-diffusion method with various sparse-matrix screens from Qiagen and Hampton Research available at the high-throughput crystallization platform at EMBL, Grenoble. Drops were prepared by mixing 0.15 µl protein solution with an equal volume of reservoir solution and were equilibrated against 120 µl reservoir solution. Only one condition, 2.5 M ammonium sulfate containing 50 mM MES pH 6.5 and 10 mM magnesium acetate, gave single crystals from over 600 screened conditions. These crystals were too small for X-ray diffraction studies. Optimization was performed by fine-tuning the protein, precipitant and glycerol concentrations using the hanging-drop vapour-diffusion technique and microseeding experiments. The best crystals were obtained from the following microseeding experiment: droplets consisting of 5 µl protein (3.5 mg ml−1 in 25 mM MES pH 5.6, 5 mM MgCl2, 100 µM GDP, 2 mM DTT, 5% glycerol, 0.005% NaN3) and 5 µl reservoir solution (50 mM MES pH 5.6, 2.4 M ammonium sulfate, 10 mM magnesium acetate) were equilibrated for 4–5 h against 0.8 ml reservoir solution. Crystallization was initiated by mixing the droplets with 0.2 µl seed stock. The seed stock was produced by washing and crushing previously prepared small crystals in 100 µl reservoir solution. After adding the seed stock, the concentration of ammonium sulfate in the reservoir was increased to 2.9 M. Under these crystallization conditions, eEF1A2 crystals typically grew within two weeks to approximate dimensions of 0.6 × 0.1 × 0.1 mm (Fig. 2 ▶). Crystals were mounted in cryoloops and flash-cooled by direct immersion into liquid nitrogen prior to X-ray diffraction analysis. 88%(v/v) saturated Li2SO4 in reservoir solution was used as a cryoprotectant. Crystals were soaked for 1 min in the cryoprotectant before cooling.
Figure 2.
eEF1A2 crystals and their diffraction pattern. (a) A single crystal in the mother-liquor solution. (b) A single crystal mounted in a cryoloop just before X-ray exposure. (c) Diffraction image of a native eEF1A2 crystal diffracting to 2.4 Å resolution. A native data set was collected to 2.7 Å resolution (Table 1 ▶)
The crystals were analysed on beamline ID14-EH4 at the ESRF (Grenoble, France). MOSFLM (Leslie, 2006 ▶) was used for indexing and the data-collection strategy. The crystals of eEF1A2 belonged to space group P6122 or P6322, with unit-cell parameters a = b = 135.4, c = 304.6 Å. A complete native data set was collected to 2.7 Å resolution (Table 1 ▶) and integrated with XDS (Kabsch, 2010 ▶). Crystals were soaked in a variety of heavy-atom solutions in order to obtain useful derivatives and solution of the structure is in progress.
Table 1. Data-collection statistics for native eEF1A2.
Values in parentheses are for the highest resolution shell.
| X-ray source | ID14-EH4, ESRF |
| Wavelength (Å) | 0.979 |
| Space group | P6122/P6322 |
| Unit-cell parameters | |
| a (Å) | 135.4 |
| b (Å) | 135.4 |
| c (Å) | 304.6 |
| Resolution range (Å) | 40–2.7 (2.8–2.7) |
| Completeness (%) | 98.4 (95.6) |
| Rmerge(I)† (%) | 8.8 (86.6) |
| 〈I/σ(I)〉 | 18.52 (2.76) |
| Total reflections | 341059 (34459) |
| Unique reflections | 45528 (4452) |
| Multiplicity | 7.5 (7.7) |
R
merge(I) = 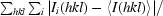
 .
.
Acknowledgments
The authors would like to thank the members of the HTX Lab (EMBL, Grenoble, France) and the ESRF–EMBL Joint Structural Biology Group for access to the ESRF synchrotron facilities (Grenoble, France). This work was supported by the NASU–CNRS collaboration program PICS.
References
- Anand, N., Murthy, S., Amann, G., Wernick, M., Porter, L. A., Cukier, I. H., Collins, C., Gray, J. W., Diebold, J., Demetrick, D. J. & Lee, J. M. (2002). Nature Genet. 31, 301–305. [DOI] [PubMed]
- Andersen, G. R., Nissen, P. & Nyborg, J. (2003). Trends Biochem. Sci. 28, 434–441. [DOI] [PubMed]
- Andersen, G. R., Pedersen, L., Valente, L., Chatterjee, I., Kinzy, T. G., Kjeldgaard, M. & Nyborg, J. (2000). Mol. Cell, 6, 1261–1266. [DOI] [PubMed]
- Borradaile, N. M., Buhman, K. K., Listenberger, L. L., Magee, C. J., Morimoto, E. T., Ory, D. S. & Schaffer, J. E. (2006). Mol. Biol. Cell, 17, 770–778. [DOI] [PMC free article] [PubMed]
- Bischoff, C., Kahns, S., Lund, A., Jørgensen, H. F., Praestegaard, M., Clark, B. F. & Leffers, H. (2000). Genomics, 68, 63–70. [DOI] [PubMed]
- Budkevich, T. V., Timchenko, A. A., Tiktopulo, E. I., Negrutskii, B. S., Shalak, V. F., Petrushenko, Z. M., Aksenov, V. L., Willumeit, R., Kohlbrecher, J., Serdyuk, I. N. & El’skaya, A. V. (2002). Biochemistry, 41, 15342–15349. [DOI] [PubMed]
- Chang, R. & Wang, E. (2007). J. Cell. Biochem. 100, 267–278. [DOI] [PubMed]
- Davis, W. G., Blackwell, J. L., Shi, P.-Y. & Brinton, M. A. (2007). J. Virol. 81, 10172–10187. [DOI] [PMC free article] [PubMed]
- Fan, K., Chrzanowska-Lightowlers, Z. M. & Hesketh, J. E. (2009). Biochem. Biophys. Res. Commun. 386, 82–88. [DOI] [PMC free article] [PubMed]
- Gillardon, F. (2009). Neuroscience, 163, 533–539. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Khalyfa, A., Bourbeau, D., Chen, E., Petroulakis, E., Pan, J., Xu, S. & Wang, E. (2001). J. Biol. Chem. 276, 22915–22922. [DOI] [PubMed]
- Kjeldgaard, M., Nissen, P., Thirup, S. & Nyborg, J. (1993). Structure, 1, 35–50. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Li, Z., Pogany, J., Panavas, T., Xu, K., Esposito, A. M., Kinzy, T. G. & Nagy, P. D. (2009). Virology, 385, 245–260. [DOI] [PMC free article] [PubMed]
- Miura, P., Coriati, A., Bélanger, G., De Repentigny, Y., Lee, J., Kothary, R., Holcik, M. & Jasmin, B. J. (2010). Hum. Mol. Genet. 19, 1211–1220. [DOI] [PubMed]
- Negrutskii, B. S. & El’skaya, A. V. (1998). Prog. Nucleic Acid Res. Mol. Biol. 60, 47–78. [DOI] [PubMed]
- Negrutskii, B. S., Stapulionis, R. & Deutscher, M. P. (1994). Proc. Natl Acad. Sci. USA, 91, 964–968. [DOI] [PMC free article] [PubMed]
- Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B. F. & Nyborg, J. (1995). Science, 270, 1464–1472. [DOI] [PubMed]
- Nissen, P., Thirup, S., Kjeldgaard, M. & Nyborg, J. (1999). Structure, 7, 143–156. [DOI] [PubMed]
- Novosylna, O. V., Timchenko, A. A., Tiktopulo, E. I., Serdyuk, I. N., Negrutskii, B. S. & El’skaya, A. V. (2007). Biopolym. Cell, 23, 386–390.
- Petrushenko, Z. M., Budkevich, T. V., Shalak, V. F., Negrutskii, B. S. & El’skaya, A. V. (2002). Eur. J. Biochem. 269, 4811–4818. [DOI] [PubMed]
- Pittman, Y. R., Kandl, K., Lewis, M., Valente, L. & Kinzy, T. G. (2009). J. Biol. Chem. 284, 4739–4747. [DOI] [PMC free article] [PubMed]
- Shalak, V. F., Budkevich, T. V., Negrutskii, B. S. & El’skaia, A. V. (1997). Ukr. Biokhim. Zh. 69, 104–109. [PubMed]
- Song, H., Parsons, M. R., Rowsell, S., Leonard, G. & Phillips, S. E. (1999). J. Mol. Biol. 285, 1245–1256. [DOI] [PubMed]
- Tomlinson, V. A., Newbery, H. J., Wray, N. R., Jackson, J., Larionov, A., Miller, W. R., Dixon, J. M. & Abbott, C. M. (2005). BMC Cancer, 5, 113. [DOI] [PMC free article] [PubMed]