Crystals of the XccFimXEAL–c-di-GMP and SeMet-XccFimXEAL–c-di-GMP–XccPilZ complexes from X. campestris diffracted to resolutions of 2.5 and 2.7 Å, respectively.
Keywords: FimXEAL–c-di-GMP–PilZ complex, type IV pili, Xanthomonas campestris
Abstract
c-di-GMP is a major secondary-messenger molecule in regulation of bacterial pathogenesis. Therefore, the c-di-GMP-mediated signal transduction network is of considerable interest. The PilZ domain was the first c-di-GMP receptor to be predicted and identified. However, every PilZ domain binds c-di-GMP with a different binding affinity. Intriguingly, a noncanonical PilZ domain has recently been found to serve as a mediator to link FimXEAL to the PilB or PilT ATPase to control the function of type IV pili (T4P). It is thus essential to determine the structure of the FimXEAL–PilZ complex in order to determine how the binding of c-di-GMP to the FimXEAL domain induces conformational change of the adjoining noncanonical PilZ domain, which may transmit information to PilB or PilT to control T4P function. Here, the preparation and preliminary X-ray diffraction studies of the XccFimXEAL–c-di-GMP and XccFimXEAL–c-di-GMP–XccPilZ complexes from Xcc (Xanthomonas campestris pv. campesteris) are reported. Detailed studies of these complexes may allow a more thorough understanding of how c-di-GMP transmits its effects through the degenerate EAL domain and the noncanonical PilZ domain.
1. Introduction
Cyclic di-GMP (c-di-GMP) was first identified as a positive allosteric effector of cellulose synthase in the bacterium Acetobacter xylinum more than 20 years ago (Ross et al., 1987 ▶, 1990 ▶), but has recently emerged as an important secondary messenger that controls a variety of cellular activities, such as the biogenesis of biofilms, flagella and pili in diverse bacteria. These activities have been correlated with bacterial pathogenicity (Römling et al., 2005 ▶; Jenal & Malone, 2006 ▶; Römling & Amikam, 2006 ▶). Diguanylate cyclases (DGCs) containing the GGDEF domain and phosphodiesterases (PDEs) containing the EAL domain (Tal et al., 1988 ▶; Simm et al., 2004 ▶; Tischler & Camilli, 2004 ▶; Römling et al., 2005 ▶) or the HD-GYP domain (Slater et al., 2000 ▶; Ryan et al., 2006 ▶) are responsible for the synthesis and degradation of c-di-GMP, respectively. However, it is still unclear how many targets of c-di-GMP are available and how this important secondary messenger mediates signal transduction in the cell. The components and responses of c-di-GMP signalling pathways are hot topics that are still being actively pursued (Römling, 2011 ▶).
PilZ-domain-containing proteins were suggested to be c-di-GMP receptors by a bioinformatics study (Amikam & Galperin, 2006 ▶) and this was subsequently demonstrated to be the case by several biochemical and structural studies (Ryjenkov et al., 2006 ▶; Merighi et al., 2007 ▶; Pratt et al., 2007 ▶; Ramelot et al., 2007 ▶). However, two types of PilZ domains were soon discovered: a type I PilZ domain that contains conserved RXXXR and D/NXSXXG signature motifs in the N-terminal region and experiences considerable conformational changes upon c-di-GMP binding (Benach et al., 2007 ▶), and a type II PilZ domain that lacks such signature motifs and is unable to bind c-di-GMP directly. PA2960 from Pseudomonas aeruginosa is possibly the best known type II protein and is the first PilZ domain (Alm et al., 1996 ▶) known to be required for T4P-mediated twitching mobility (Mattick, 2002 ▶). In Xanthomonas campestris pv. campesteris (Xcc), four PilZ-domain proteins were discovered and were found to be essential for its pathogenicity (McCarthy et al., 2008 ▶); two of them contain a regular type I sequence and the other two contain a type II noncanonical sequence. XCC1028 is one of the type II domain-containing proteins; it adopts a similar five-stranded β-barrel structure, yet exhibits considerable differences at the N-terminal end owing to a lack of the characteristic N-terminal c-di-GMP binding signature motifs (Li, Chin, Liu et al., 2009 ▶). XCC6012 is another example; it adopts a monomer structure similar to that of XCC1028, yet is interrupted in the middle by two extra long helices between the β1 and β2 strands and self-assembles into a tetramer via the extra α3 heptad-repeat helix (Li et al., 2011 ▶). How type II PilZ domains respond to the c-di-GMP signal remains unclear to date.
FimX is a large multi-domain protein containing a tandem of REC, PAS, GGDEF and EAL domains that governs bacterial twitching motility (Huang et al., 2003 ▶; Kazmierczak et al., 2006 ▶). Interestingly, the REC, GGDEF and EAL domains in FimX are all degenerate: the REC domain lacks the crucial Asp essential for phosphotransfer, the GGDEF domain contains a rather unusual GDSIF motif and the EAL domain contains a modified EVL motif at the active site. Several biochemical and structural analyses have revealed the role of the degenerate EAL domain as the high-affinity binding receptor of c-di-GMP (Navarro et al., 2009 ▶; Qi et al., 2011 ▶). The crystal structure of a degenerate EAL domain from P. aeruginosa has also been solved (Navarro et al., 2009 ▶).
Recently, a detailed study of the interaction between the FimXEAL and PilZ domains from Xac (X. axonopodis pv. citri) has been carried out using a variety of techniques such as NMR, thermal melting, far-Western blotting and in vivo motility assay methods (Guzzo et al., 2009 ▶). From these studies, it was concluded that XacPilZ binds to the XacPilB ATPase required for T4P polymerization and to the XacFimXEAL domain required for binding c-di-GMP to regulate T4P biogenesis. However, the crucial FimXEAL−PilZ complex structure is not available to date. Since the sequence and structure of XacPilZ were found to be identical to those of XccPilZ1028 (Guzzo et al., 2009 ▶; Li, Chin, Liu et al., 2009 ▶), and the FimXEAL sequences from both Xcc and Xac align very well with an identity of 90.9% and a similarity of 93.3% (data not shown), a similar interaction between the FimXEAL and PilZ domains is very likely to exist in Xcc. Here, we report the expression, purification, crystallization and preliminary X-ray diffraction studies of the XccFimXEAL–c-di-GMP and XccFimXEAL–c-di-GMP–XccPilZ1028 complexes from Xcc. This phytopathogen is ideal for studying c-di-GMP-related issues since it contains a considerable number of GGDEF-domain, EAL-domain, HD-GYP-domain and PilZ-domain proteins (Ryan et al., 2007 ▶; McCarthy et al., 2008 ▶). Detailed studies of the XccFimXEAL–c-di-GMP and XccFimXEAL–c-di-GMP–XccPilZ complexes may allow a more thorough understanding of how c-di-GMP transmits its effects through noncanonical PilZ-domain proteins.
2. Materials and methods
2.1. Reagents
c-di-GMP was produced by an enzymatic method using an altered thermophilic DGC enzyme as described previously (Rao et al., 2009 ▶).
2.2. Cloning and purification
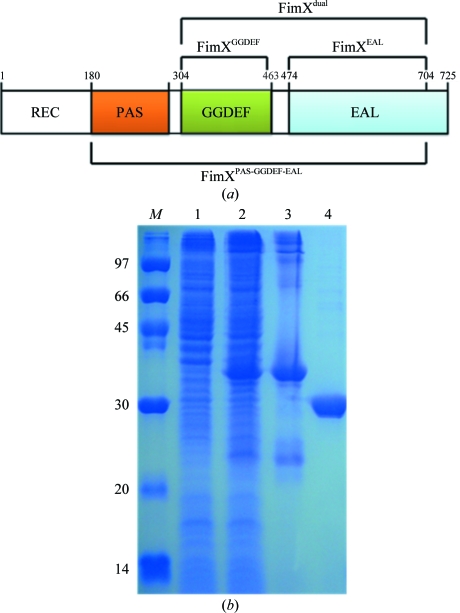
XccFimXEAL was PCR-amplified directly from the plant pathogen X. campestris pv. campestris strain 17 (Xcc) using the forward primer 5′-TACTTCCAATCCAATGCTGAGGAAGAACGCATCGAGCGC-3′ and the reverse primer 5′-TTATCCACTTCCAATGCTAGTAGTCGCCCGGCCACCCGCG-3′. The PCR fragment exhibited the correct size in an agarose-gel electrophoresis experiment and was confirmed by DNA sequencing. A ligation-independent cloning (LIC) approach (Aslanidis & de Jong, 1990 ▶; Stols et al., 2001 ▶; Wu et al., 2005 ▶) was used to obtain the desired constructs. The final construct codes for an N-terminal His6 tag, a 17-amino-acid linker and the XccFimXEAL target under the control of a T7 promoter. Overexpression of the His6-tagged target protein was induced by the addition of 800 µl 500 mM IPTG to the medium solution (to give a final IPTG concentration of 0.5 mM) at 293 K for 18 h. The target protein was purified by immobilized metal-affinity chromatography (IMAC) on a nickel column (Sigma) equilibrated with a buffer consisting of 20 mM Tris–HCl pH 8.0, 80 mM NaCl. The target protein was eluted with a gradient of 50–300 mM imidazole in the same buffer. The fractions containing XccFimXEAL were monitored using 13% SDS–PAGE and recombined. The His6 tag and linker were then cleaved from the XccFimXEAL target using TEV (tobacco etch virus) protease at 289 K for 10 h. For crystallization, the XccFimXEAL protein was further purified on a Sephadex gel-filtration column (ÄKTA, Pharmacia Inc.). The final target-protein sample exhibited a purity greater than 99% as revealed by SDS–PAGE gel analysis (Fig. 1 ▶). It contained only an extra tripeptide (SNA) from the vector at the N-terminal end. SeMet-labelled XccFimXEAL was prepared in a similar way, except that it was produced using a non-auxotroph Escherichia coli strain BL21 (DE3) as host in the absence of methionine but with ample amounts of SeMet (100 mg l−1). The M9 medium consisted of 1 g ammonium chloride, 3 g KH2PO4, 6 g Na2HPO4 supplemented with 20%(w/v) glucose, 0.3%(w/v) MgSO4 and 10 mg FeSO4 in 1 l double-distilled water. Induction was sustained at 293 K for 18 h by the addition of 0.45 ml 0.5 mM IPTG. The purification of the SeMet-labelled XccFimXEAL protein was performed using the same procedure as for the native protein.
Figure 1.
(a) The domain architecture and constructs used in these studies. (b) SDS–PAGE (13%) monitoring of the overexpression and purification of XccFimXEAL. Lane 1, protein markers (labelled in kDa); lane 2, whole cell lysate before IPTG induction; lane 3, whole cell lysate after IPTG induction; lane 4, supernatant of His6-tagged XccFimXEAL; lane 5, gel-purified XccFimXEAL after TEV cleavage.
The XccPilZ1028 sample was obtained using a similar protocol to that previously published (Li, Chin, Shih et al., 2009 ▶).
2.3. Crystallization
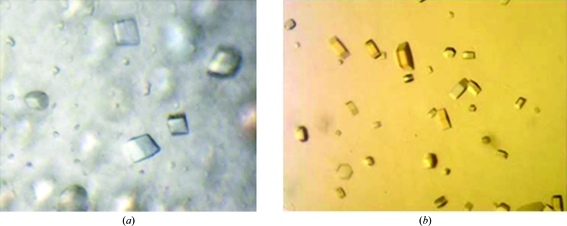
For crystallization, the native XccFimXEAL protein was concentrated to 0.56 mM in 20 mM Tris–HCl pH 8.0, 80 mM sodium chloride using an Amicon Ultra-10 (Millipore). The SeMet-XccFimXEAL–XccPilZ1028 complex (1:1 ratio) was concentrated to 0.075 mM in a similar way. Appropriate volumes of 25.6 mM c-di-GMP were added to the XccFimXEAL and SeMet-XccFimXEAL–XccPilZ1028 complex solutions to prepare samples for cocrystallization with a 2:1 ligand:protein ratio. Screening for crystallization conditions for each protein was performed using sitting-drop vapour diffusion in 96-well plates (Hampton Research) at 277 K by mixing 0.5 µl protein solution with 0.5 µl reservoir solution and equilibrating against 50 µl reservoir solution. Initial screens including the sparse-matrix Crystal Screen and Crystal Screen 2 (Hampton Research), a systematic PEG–pH screen and the PEG/Ion screen (Hampton Research) were performed using a Gilson C240 crystallization workstation. Cubic crystals of the XccFimXEAL–c-di-GMP complex appeared in 7 d from drops equilibrated against 50 µl reservoir solution comprising 20% PEG 3350, 0.2 M sodium formate pH 7.2, while hexagonal crystals of the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complex appeared in 21 d from drops equilibrated against 50 µl reservoir solution comprising 0.2 M NaCl, 0.1 M HEPES pH 7.5, 20% PEG 3K (Fig. 2 ▶). Crystals of both complexes suitable for diffraction experiments were grown from drops by mixing 1.5 µl protein solution with 1.5 µl reservoir solution and equilibrating against 500 µl reservoir solution at 277 K. Crystals of the XccFimXEAL–c-di-GMP complex reached dimensions of 0.2 × 0.2 × 0.2 mm after one week, while those of the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complex reached dimensions of 0.01 × 0.01 × 0.01 mm after three weeks.
Figure 2.
Crystals of the XccFimXEAL–c-di-GMP and SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complexes. (a) XccFimXEAL–c-di-GMP crystals grown in 0.2 M sodium formate pH 7.2, 20% PEG 3350 using the hanging-drop vapour-diffusion method at 277 K. These crystals reached average dimensions of 0.2 × 0.2 × 0.2 mm after one week. (b) SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 crystals grown in 0.1 M HEPES pH 7.5, 0.2 M NaCl, 20% PEG 3K using the hanging-drop vapour-diffusion method at 277 K. These crystals reached average dimensions of 0.01 × 0.01 × 0.01 mm after three weeks.
2.4. Data collection and processing
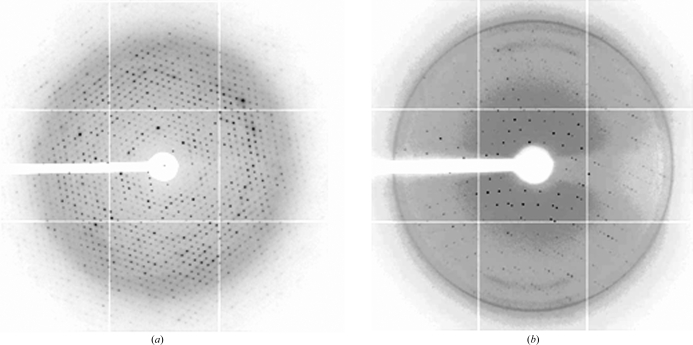
Crystals of both complexes were flash-cooled at 100 K under a stream of cold nitrogen gas using the reservoir solution as cryoprotectant. Before data collection, the crystals were scanned for Se absorption and 0.97934 Å was found to be the peak wavelength of the anomalous signal. X-ray diffraction data for the native XccFimXEAL–c-di-GMP complex were obtained to 2.5 Å resolution on beamline 13B1 at the National Synchrotron Radiation Research Center (NSRRC), Taiwan, while those for the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complex were collected to 2.7 Å resolution on beamline 12B2 at SPring-8, Japan (Fig. 3 ▶). The data were indexed and integrated using the HKL-2000 processing software (Otwinowski & Minor, 1997 ▶), generating data sets that were 97.1 and 99.4% complete with an overall R merge of 3.6 and 13.8% on intensities, respectively. Determination and refinement of the selenium positions, phase calculation and density modification were carried out using the programs SOLVE and RESOLVE (Terwilliger & Berendzen, 1999 ▶). Molecular replacement was performed using CNS (Brünger et al., 1998 ▶). The crystals of the XccFimXEAL–c-di-GMP complex belonged to space group P3221, while those of the XccFimXEAL–c-di-GMP–XccPilZ1028 complex belonged to space group P6322. The diffraction statistics are summarized in Table 1 ▶.
Figure 3.
Diffraction patterns of the native XccFimXEAL–c-di-GMP and SeMet-XccFimXEAL–-c-di-GMP–XccPilZ1028 complexes collected on a MAR CCD system using synchrotron radiation on the 13B1 beamline at NSRRC in Taiwan and the BL12B1 beamline at SPring-8 in Japan. The exposure time was 1 s, the oscillation range was 1° per frame and the crystal-to-detector distance was 250 mm. The edge of the detector corresponds to a resolution of 2.5 and 3.2 Å, respectively.
Table 1. Summary of the native and Se-SAD crystallographic data for the XccFimXEAL–c-di-GMP and SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complexes.
Values in parentheses are for the outermost shell.
| XccFimXEAL–c-di-GMP | SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028† | |
|---|---|---|
| Native | Peak | |
| Beamline | NSRRC BL13B1 | Spring-8 |
| Wavelength (Å) | 1.00000 | 0.97934 |
| Space group | P3221 | P6322 |
| Unit-cell parameters (Å, °) | a = b = 65.67, c = 121.29, γ = 120 | a = b = 158.22, c = 64.81, γ = 120 |
| Resolution range (Å) | 30–2.5 (2.59–2.50) | 30–2.7 (2.80–2.70) |
| Total reflections | 68379 (5890) | 189837 (16170) |
| Unique reflections | 19685 (1899) | 49419 (4899) |
| Multiplicity | 3.5 (3.1) | 3.8 (3.3) |
| Completeness (%) | 97.1 (93.5) | 99.4 (98.7) |
| Rmerge‡ (%) | 3.6 (22.1) | 13.8 (55.2) |
| 〈I/σ(I)〉 | 30.5 (5.4) | 7.8 (1.8) |
| Matthews coefficient (Å3 Da−1) | 2.74 | 2.93 |
| Solvent content (%) | 55.2 | 58.0 |
The FOM (figure of merit) for the SAD data for this complex was 0.37.
R
merge = 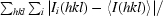
 , where I
i(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is its average.
, where I
i(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is its average.
3. Results and discussion
In this manuscript, we report the successful cloning, protein expression and purification of the XccFimXEAL and XccPilZ1028 proteins and the crystal screening and preliminary X-ray data analyses of the native XccFimXEAL–c-di-GMP and SeMet-substituted XccFimXEAL–c-di-GMP–XccPilZ1028 complexes. Since the FimX protein is a large bacterial protein containing a tandem of REC, PAS, GGDEF and EAL domains, we tried to construct clones from different combinations of these domains as shown in Fig. 1 ▶(a) in order to express the domains and determine their structures using X-ray crystallography. Unfortunately, most of these constructs gave proteins in inclusion bodies and only XccFimXEAL gave soluble protein. As shown in Fig. 1 ▶, the His6 tag and linker of the XccFimXEAL target could be successfully cleaved by TEV (tobacco etch virus) protease at 289 K for 10 h to obtain target protein that is more than 99% pure. It contains only an extra tripeptide (SNA) at the N-terminal end after further gel-filtration chromatography. However, in the absence of c-di-GMP no crystal formation was observed for the XccFimXEAL domain and only a poor diffraction pattern was detected for the XccFimXEAL–XccPilZ1028 complex even though it formed seemingly good crystals. These results indicated that c-di-GMP was crucial in forming compact crystals for both the XccFimXEAL domain and the XccFimXEAL–XccPilZ1028 complex.
We were surprised by the apparent existence of data to higher resolution once data collection for the native XccFimXEAL-c-di-GMP complex began (Table 1 ▶) and are considering an experimental effort to collect these data for refinement of the structure at higher resolution.
Interestingly, although the EAL-domain structure has been solved and found to be conserved (Minasov et al., 2009 ▶; Navarro et al., 2009 ▶; Tchigvintsev et al., 2010 ▶), we were unable to solve the crystal structure of XccFimXEAL–c-di-GMP using a molecular-replacement approach. Fortunately, we were able to crystallize the XccFimXEAL–XccPilZ1028 complex using an SeMet-substituted XccFimXEAL domain in the presence of c-di-GMP. Although the resolution of the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complex was a little poorer (2.7 Å) than that of native XccFimXEAL–c-di-GMP (2.5 Å), it should be possible to perform successful phasing of the protein using an Se-SAD approach based on the figure-of-merit statistic (Table 1 ▶). Indeed, the model of the XccFimXEAL domain in the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complex was almost complete and the initial structure of XccFimXEAL was used as a model for molecular replacement to determine the phases of the XccFimXEAL–c-di-GMP complex. The Matthews coefficient and solvent content were 2.74 Å3 Da−1 and 55.2%, respectively, for the XccFimXEAL–c-di-GMP complex and 2.93 Å3 Da−1 and 58.0%, respectively, for the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complex. The c-di-GMP was clearly identified in the electron-density maps of both the XccFimXEAL–c-di-GMP and the SeMet-XccFimXEAL–c-di-GMP–XccPilZ1028 complexes. Refinement of both complexes is now in progress.
Acknowledgments
This work was supported in part by the Ministry of Education, Taiwan, ROC under the ATU plan and by the National Science Council, Taiwan, ROC (grant 97-2113-M005-005-MY3 to S-HC). We appreciate the Structural Genomics Databases service provided by the GMBD Bioinformatics Core (http://www.tbi.org.tw), NRPGM, Taiwan, ROC. We would also like to thank the Core Facilities for Protein X-ray Crystallography in the Academia Sinica, Taiwan, ROC for help in crystal screening, the National Synchrotron Radiation Research Center (NSRRC) in Taiwan and the SPring-8 Synchrotron facility in Japan for assistance in X-ray data collection. The National Synchrotron Radiation Research Center is a user facility supported by the National Science Council, Taiwan, ROC and the Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine, Taiwan, ROC.
References
- Alm, R. A., Bodero, A. J., Free, P. D. & Mattick, J. S. (1996). J. Bacteriol. 178, 46–53. [DOI] [PMC free article] [PubMed]
- Amikam, D. & Galperin, M. Y. (2006). Bioinformatics, 22, 3–6. [DOI] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Benach, J., Swaminathan, S. S., Tamayo, R., Handelman, S. K., Folta-Stogniew, E., Ramos, J. E., Forouhar, F., Neely, H., Seetharaman, J., Camilli, A. & Hunt, J. F. (2007). EMBO J. 26, 5153–5166. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Guzzo, C. R., Salinas, R. K., Andrade, M. O. & Farah, C. S. (2009). J. Mol. Biol. 393, 846–866. [DOI] [PubMed]
- Huang, B., Whitchurch, C. B. & Mattick, J. S. (2003). J. Bacteriol. 185, 7068–7076. [DOI] [PMC free article] [PubMed]
- Jenal, U. & Malone, J. (2006). Annu. Rev. Genet. 40, 385–407. [DOI] [PubMed]
- Kazmierczak, B. I., Lebron, M. B. & Murray, T. S. (2006). Mol. Microbiol. 60, 1026–1043. [DOI] [PMC free article] [PubMed]
- Li, T.-N., Chin, K.-H., Fung, K.-M., Yang, M.-T., Wang, A. H.-J. & Chou, S.-H. (2011). PLoS One, 6, e22036. [DOI] [PMC free article] [PubMed]
- Li, T.-N., Chin, K.-H., Liu, J.-H., Wang, A. H.-J. & Chou, S.-H. (2009). Proteins, 75, 282–288. [DOI] [PubMed]
- Li, T.-N., Chin, K.-H., Shih, H.-L., Wang, A. H.-J. & Chou, S.-H. (2009). Acta Cryst. F65, 1056–1059. [DOI] [PMC free article] [PubMed]
- Mattick, J. S. (2002). Annu. Rev. Microbiol. 56, 289–314. [DOI] [PubMed]
- McCarthy, Y., Ryan, R. P., O’Donovan, K., He, Y.-Q., Jiang, B.-L., Feng, J.-X., Tang, J.-L. & Dow, J. M. (2008). Mol. Plant Pathol. 9, 819–824. [DOI] [PMC free article] [PubMed]
- Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y. & Lory, S. (2007). Mol. Microbiol. 65, 876–895. [DOI] [PubMed]
- Minasov, G., Padavattan, S., Shuvalova, L., Brunzelle, J. S., Miller, D. J., Baslé, A., Massa, C., Collart, F. R., Schirmer, T. & Anderson, W. F. (2009). J. Biol. Chem. 284, 13174–13184. [DOI] [PMC free article] [PubMed]
- Navarro, M. V., De, N., Bae, N., Wang, Q. & Sondermann, H. (2009). Structure, 17, 1104–1116. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pratt, J. T., Tamayo, R., Tischler, A. D. & Camilli, A. (2007). J. Biol. Chem. 282, 12860–12870. [DOI] [PMC free article] [PubMed]
- Qi, Y., Chuah, M. L.-C., Dong, X., Xie, K., Luo, Z., Tang, K. & Liang, Z.-X. (2011). J. Biol. Chem. 286, 2910–2917. [DOI] [PMC free article] [PubMed]
- Ramelot, T. A., Yee, A., Cort, J. R., Semesi, A., Arrowsmith, C. H. & Kennedy, M. A. (2007). Proteins, 66, 266–271. [DOI] [PubMed]
- Rao, F., Pasunooti, S., Ng, Y., Zhuo, W., Lim, L., Liu, A. W. & Liang, Z.-X. (2009). Anal. Biochem. 389, 138–142. [DOI] [PubMed]
- Römling, U. (2011). Environ. Microbiol., doi:10.1111/j.1462-2920.2011.02617.x.
- Römling, U. & Amikam, D. (2006). Curr. Opin. Microbiol. 9, 218–228. [DOI] [PubMed]
- Römling, U., Gomelsky, M. & Galperin, M. Y. (2005). Mol. Microbiol. 57, 629–639. [DOI] [PubMed]
- Ross, P., Mayer, R., Weinhouse, H., Amikam, D., Huggirat, Y., Benziman, M., de Vroom, E., Fidder, A., de Paus, P., Sliedregt, L. A. J. M., van der Marel, G. A. & van Boom, J. H. (1990). J. Biol. Chem. 265, 18933–18943. [PubMed]
- Ross, P., Weinhouse, H., Aloni, Y., Michaeli, D., Weinberger-Ohana, P., Mayer, R., Braun, S., de Vroom, E., van der Marel, G. A., van Boom, J. H. & Benziman, M. (1987). Nature (London), 325, 279–281. [DOI] [PubMed]
- Ryan, R. P., Fouhy, Y., Lucey, J. F., Crossman, L. C., Spiro, S., He, Y.-W., Zhang, L.-H., Heeb, S., Cámara, M., Williams, P. & Dow, J. M. (2006). Proc. Natl Acad. Sci. USA, 103, 6712–6717. [DOI] [PMC free article] [PubMed] [Retracted]
- Ryan, R. P., Fouhy, Y., Lucey, J. F., Jiang, B.-L., He, Y.-Q., Feng, J.-X., Tang, J.-L. & Dow, J. M. (2007). Mol. Microbiol. 63, 429–442. [DOI] [PubMed]
- Ryjenkov, D. A., Simm, R., Römling, U. & Gomelsky, M. (2006). J. Biol. Chem. 281, 30310–30314. [DOI] [PubMed]
- Simm, R., Morr, M., Kader, A., Nimtz, M. & Römling, U. (2004). Mol. Microbiol. 53, 1123–1134. [DOI] [PubMed]
- Slater, H., Alvarez-Morales, A., Barber, C. E., Daniels, M. J. & Dow, J. M. (2000). Mol. Microbiol. 38, 986–1003. [DOI] [PubMed]
- Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R. & Donnelly, M. I. (2001). Protein Expr. Purif. 25, 8–15. [DOI] [PubMed]
- Tal, R., Wong, H. C., Calhoon, R., Gelfand, D., Fear, A. L., Volman, G., Mayer, R., Ross, P., Amikam, D., Weinhouse, H., Cohen, A., Sapir, S., Ohana, P. & Benziman, M. (1988). J. Bacteriol. 180, 4416–4425. [DOI] [PMC free article] [PubMed]
- Tchigvintsev, A., Xu, X., Singer, A., Chang, C., Brown, G., Proudfoot, M., Cui, H., Flick, R., Anderson, W. F., Joachimiak, A., Galperin, M. Y., Savchenko, A. & Yakunin, A. F. (2010). J. Mol. Biol. 402, 524–538. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Tischler, A. D. & Camilli, A. (2004). Mol. Microbiol. 53, 857–869. [DOI] [PMC free article] [PubMed]
- Wu, Y.-Y., Chin, K.-H., Chou, C.-C., Lee, C.-C., Shr, H.-L., Gao, F. P., Lyu, P.-C., Wang, A. H.-J. & Chou, S.-H. (2005). Acta Cryst. F61, 902–905. [DOI] [PMC free article] [PubMed]