TPP3 is a class II plant defensin from tomato. Here, the expression, purification, crystallization and preliminary X-ray crystallographic analysis of recombinant TPP3 are reported in order to define its structure and function in relation to other class II plant defensins.
Keywords: TPP3, plant defensins, Pichia pastoris, recombinant expression
Abstract
Class II defensins have been shown to have potent antifungal activity and are being exploited to protect agricultural crops against fungal pathogens. TPP3 is a poorly characterized member of the class II plant defensin family from tomato. To gain structural insight into the function of TPP3, soluble recombinant TPP3 was expressed and purified using the Pichia pastoris expression system, and the crystallization and preliminary X-ray crystallographic analysis of the protein are reported. Crystals of rTPP3 were obtained using the sitting-drop vapour-diffusion method at 293 K. Diffraction data were collected to 1.7 Å resolution. The crystals belonged to the hexagonal space group P6122, with unit-cell parameters a = 64.97, b = 64.97, c = 82.40 Å, α = 90, β = 90, γ = 120°.
1. Introduction
Defensins (formerly referred to as γ-thionins) are small (∼5 kDa) cationic peptides that form an important defence component of the plant innate immune system (Lay & Anderson, 2005 ▶). They contain eight cysteine residues that form a disulfide array that results in a highly compact and stable structure known as the cysteine-stabilized αβ (CSαβ) motif (Lay, Schirra et al., 2003 ▶). This structural motif is also conserved in insect defensins and scorpion toxins (reviewed in Lay & Anderson, 2005 ▶). The first members of this family were isolated from the endosperm of barley (Mendez et al., 1990 ▶) and wheat (Colilla et al., 1990 ▶). The family has now grown to include members from various monocotyledonous and dicotyledonous plants (reviewed by Lay & Anderson, 2005 ▶). It is likely that defensins are present in all plant species.
All plant defensins characterized to date are produced as preproteins with a signal sequence (∼25 amino acids) for directing secretion into the endoplasmic reticulum (ER). Solanaceous plants, including tobacco, petunia, capsicum and tomato, express a small subset of defensins that contain an additional propeptide of ∼33 amino acids at their C-terminal end (CTPP; Milligan & Gasser, 1995 ▶; Brandstädter et al., 1996 ▶; Komori et al., 1997 ▶; Yamada et al., 1997 ▶; Aluru et al., 1999 ▶; Janssen et al., 2003 ▶; Lay, Brugliera et al., 2003 ▶; Lay, Schirra et al., 2003 ▶; Lay & Anderson, 2005 ▶). Plant defensins can thus be classified into one of two classes based on the absence (class I) or the presence (class II) of the CTPP. It is worth noting that plants that express class II defensins also produce class I defensins (Lay & Anderson, 2005 ▶).
While the majority of studies have been directed at the more abundant class I defensins, which are readily sourced from plant material such as seeds, far less is known about the class II defensins. The best characterized of these is NaD1, which was identified from the flowers of the ornamental tobacco (Lay, Brugliera et al., 2003 ▶). NaD1 is a potent antifungal protein against several agronomically important fungal pathogens such as Fusarium oxysporum (Lay, Brugliera et al., 2003 ▶; van der Weerden et al., 2008 ▶, 2010 ▶). Its mechanism of antifungal action is not fully understood, but relies at least in part on specific interactions with the fungal cell wall followed by plasma-membrane permeabilization and entry into the cytoplasm (van der Weerden et al., 2008 ▶, 2010 ▶).
Tomatoes express a class II defensin in their flowers known as tomato pistil-predominant 3 (TPP3; Milligan & Gasser, 1995 ▶). A cDNA clone encoding TPP3 was initially isolated from tomato pistils through differential hybridization screening. An additional clone encoding an identical protein to TPP3, referred to as AT2, was subsequently reported (Brandstädter et al., 1996 ▶). Milligan & Gasser (1995 ▶) demonstrated that TPP3 mRNA levels were highest in immature pistils and were moderate in pistils from mature flowers. TPP3 is closely related to other class II defensins, sharing 35.2% amino-acid identity with NaD1 from Nicotiana alata (Lay, Brugliera et al., 2003 ▶) over the entire precursor sequence and 64.5% identity over the mature defensin domain. While TPP3 gene expression has been studied, little is known about the biochemical characteristics, structure or function of the protein. In this study, we used the Pichia pastoris expression system to produce the mature domain of TPP3 in a secreted soluble form (hereafter referred to as rTPP3) and report the crystallization and preliminary X-ray crystallographic analysis of rTPP3.
2. Materials and methods
2.1. Expression of rTPP3 in P. pastoris
Mature TPP3 (GenBank accession No. SLU20591; amino acids 26–73) was recombinantly expressed in the methylotrophic yeast P. pastoris. To achieve this, DNA encoding the mature region of TPP3 was cloned into the pPIC9 expression vector (Invitrogen) directly in-frame with the yeast α-mating factor secretion signal using the restriction enzymes XhoI and NotI. An alanine was added to the N-terminus of the TPP3 sequence to ensure efficient cleavage of the signal at the Kex2 cleavage site. After transformation into Escherichia coli TOP10 cells, the pPIC9-TPP3 plasmid was isolated and linearized using SalI to allow integration at the his4 locus of the P. pastoris genome.
Linearized DNA was transformed into electrocompetent yeast as described by Chang et al. (2005 ▶) and His+ transformants were selected for by plating onto MD agar (1.34% YNB, 4 × 10−5% biotin, 1% dextrose, 1.5% agar). A single positive colony was used to inoculate 200 ml BMG (100 mM potassium phosphate pH 6.0, 1.34% YNB, 4 × 10−5% biotin, 1% glycerol) and incubated with constant shaking at 303 K until the OD600 reached ∼5.0. The cell mass was collected by centrifugation (1500g, 10 min) and resuspended in 1 l BMM (100 mM potassium phosphate pH 6.0, 1.34% YNB, 4 × 10−5% biotin, 0.5% methanol) to a final OD600 of 1.0 to induce expression. Expression was continued for 4 d with constant shaking at 303 K, after which the cell mass was removed by centrifugation (10 000g, 10 min) and the rTPP3-containing supernatant was collected.
A one-twentieth volume of 1 M potassium phosphate buffer pH 6.0 was added to the supernatant and the pH was adjusted to 6.0 by the addition of 10 M KOH. The supernatant was then applied onto an SP Sepharose column (GE Healthcare Biosciences) pre-equilibrated with 100 mM potassium phosphate buffer pH 6.0. Following extensive washing with 100 mM potassium phosphate buffer pH 6.0, the bound proteins were eluted with 100 mM potassium phosphate buffer pH 6.0 containing 0.5 M NaCl. The eluted proteins were subsequently concentrated using Amicon Ultra 3000 MWCO centrifugal filters (Millipore) and desalted in Milli-Q water using the same centrifugal filters. The protein concentration was determined using the BCA assay (Pierce) and the purity and identity of rTPP3 was assessed by reducing SDS–PAGE and mass spectrometry.
2.2. Crystallization
Crystallization trials were carried out with rTPP3 (at a concentration of 15 mg ml−1 in deionized water) at 293 K using the sitting-drop method by mixing 150 nl protein solution with an equal volume of mother liquor. The initial crystallization conditions were established using the Crystal Screen HT sparse-matrix protein crystallization screen (Hampton Research) at the CSIRO Collaborative Crystallization Centre (Melbourne, Australia). The initial crystals, which were obtained in 0.2 M ammonium acetate, 30%(w/v) PEG 4000, 0.1 M sodium citrate pH 5.6, were thin needle-shaped and appeared after 3 d. After further optimization using a randomized approach in which a range of ammonium acetate concentrations (0.1–0.3 M), PEG 4000 concentrations [24–32%(w/v)] and sodium citrate pH (4.8–6.4) were tested, large rod-like crystals of good diffraction quality were obtained in 0.16 M ammonium acetate, 24%(w/v) PEG 4000, 0.1 M sodium citrate pH 5.1 after 14 d.
2.3. Data collection and processing
Native diffraction data were collected at 100 K from crystals flash-cooled in mother liquor supplemented with 10%(w/v) PEG 400 at a wavelength of 0.9573 Å at the Australian Synchrotron (beamline MX2) and were processed with XDS (Kabsch, 2010 ▶).
3. Results and discussion
The TPP3 gene is known to be predominantly expressed in the pistils of tomato (Milligan & Gasser, 1995 ▶); however, prior to the current study the TPP3 protein had not been examined biochemically or structurally. Based on its homology to related class II defensins such as NaD1, it is likely to possess antifungal properties and to have potential application in agricultural crops to confer enhanced disease resistance against fungal pathogens. In this study, we expressed and purified recombinant TPP3 (rTPP3) and established crystallization conditions to determine its structure in order to gain insight into its mechanism of action.
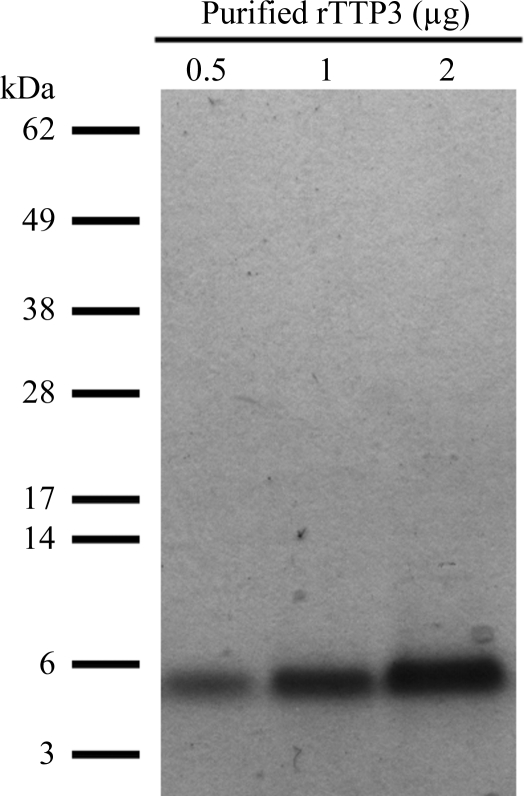
We used the pPIC9 vector for methanol-inducible expression of secreted soluble TPP3 in the yeast P. pastoris. 4 d post-induction, the expression culture medium was harvested and rTPP3 (with an overall net charge of +7) was purified by cation-exchange chromatography. The relative size and purity of rTPP3 was analyzed by reducing SDS–PAGE, where it appeared as a single protein band of ∼5.5 kDa (Fig. 1 ▶). Mass spectrometry was used to further verify the identity of rTPP3 and confirmed that the introduced N-terminal alanine was present (data not shown).
Figure 1.
Three different amounts (0.5, 1 and 2 µg) of purified rTPP3 were subjected to reducing SDS–PAGE prior to Coomassie Brilliant Blue staining.
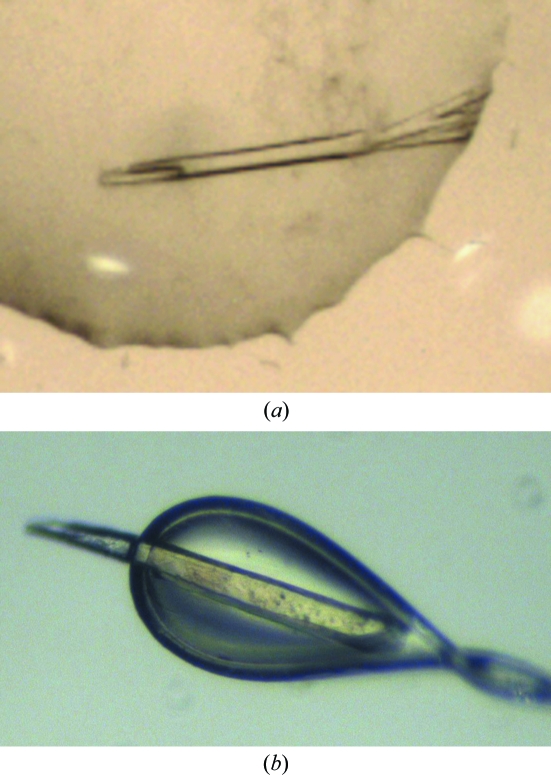
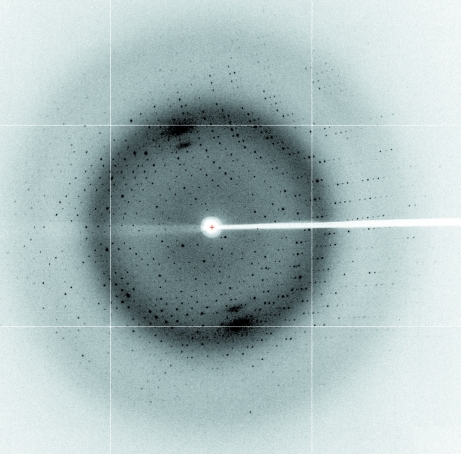
Crystallization of rTPP3 was performed using the sitting-drop vapour-diffusion method at 293 K. Diffraction-quality crystals (Fig. 2 ▶) were obtained after optimization of an initial crystallization condition obtained from a sparse-matrix screen: 24%(w/v) PEG 4000, 0.16 M ammonium acetate, 0.1 M sodium citrate pH 5.1. Native data were collected to 1.7 Å resolution (Fig. 3 ▶). The crystals belonged to the hexagonal space group P6122, with unit-cell parameters a = 64.97, b = 64.97, c = 82.40 Å, α = 90, β = 90, γ = 120°. The crystals contained two molecules in the asymmetric unit, with a calculated Matthews coefficient (V M; Matthews, 1968 ▶) of 2.28 Å3 Da−1 and a solvent content of 46%. The data-collection statistics are shown in Table 1 ▶. Phasing, model building and refinement are in progress.
Figure 2.
Crystals of rTPP3 grown in 24%(w/v) PEG 4000, 0.16 M ammonium acetate, 0.1 M sodium citrate pH 5.1. (a) rTPP3 crystals in the well and (b) rTPP3 crystals mounted on a 100 µm loop.
Figure 3.
X-ray diffraction image from the rTPP3 crystal.
Table 1. Data-collection statistics for rTPP3 crystals.
Values in parentheses are for the highest resolution shell.
| Space group | P6122 |
| Unit-cell parameters (Å, °) | a = b = 64.97, c = 82.40, α = β = 90.00, γ = 120.00 |
| Wavelength (Å) | 0.9537 |
| Resolution (Å) | 46.46–1.70 (1.79–1.70) |
| Rmerge | 0.072 (1.073) |
| Rp.i.m. | 0.012 (0.174) |
| 〈I/σ(I)〉 | 51.4 (5.3) |
| Completeness (%) | 100 (100) |
| Multiplicity | 34.7 (38.1) |
| No. of reflections | 412499 (64170) |
| No. of observed reflections | 11889 (1684) |
| Matthews volume (Å3 Da−1) | 2.28 |
| Molecules in asymmetric unit | 2 |
Acknowledgments
We would like to thank Professor Marilyn Anderson (La Trobe University) for the rTPP3 Pichia expression clone. We also thank the staff at the CSIRO Collaborative Crystallization Centre for the set-up of crystal screens and the staff of the MX team at the Australian Synchrotron for assistance with X-ray diffraction data collection. We gratefully acknowledge support from Balmoral Australia Pty Ltd, Hexima Ltd and the National Health and Medical Research Council of Australia (NHMRC fellowship 637372 to MK).
References
- Aluru, M., Curry, J. & O’Connell, M. A. (1999). Plant Physiol. 120, 633.
- Brandstädter, J., Rossbach, C. & Theres, K. (1996). Mol. Gen. Genet. 252, 146–154. [DOI] [PubMed]
- Chang, T., Schroder, L. A., Thomson, J. M., Klocman, A. S., Tomasini, A. J., Strømhaug, P. E. & Dunn, W. A. (2005). Mol. Biol. Cell, 16, 4941–4953. [DOI] [PMC free article] [PubMed]
- Colilla, F. J., Rocher, A. & Mendez, E. (1990). FEBS Lett. 270, 191–194. [DOI] [PubMed]
- Janssen, B. J., Schirra, H. J., Lay, F. T., Anderson, M. A. & Craik, D. J. (2003). Biochemistry, 42, 8214–8222. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Komori, T., Yamada, S. & Imaseki, H. (1997). Plant Physiol. 115, 314.
- Lay, F. T. & Anderson, M. A. (2005). Curr. Protein Pept. Sci. 6, 85–101. [DOI] [PubMed]
- Lay, F. T., Brugliera, F. & Anderson, M. A. (2003). Plant Physiol. 131, 1283–1293. [DOI] [PMC free article] [PubMed]
- Lay, F. T., Schirra, H. J., Scanlon, M. J., Anderson, M. A. & Craik, D. J. (2003). J. Mol. Biol. 325, 175–188. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mendez, E., Moreno, A., Colilla, F., Pelaez, F., Limas, G. G., Mendez, R., Soriano, F., Salinas, M. & de Haro, C. (1990). Eur. J. Biochem. 194, 533–539. [DOI] [PubMed]
- Milligan, S. B. & Gasser, C. S. (1995). Plant Mol. Biol. 28, 691–711. [DOI] [PubMed]
- van der Weerden, N. L., Hancock, R. E. & Anderson, M. A. (2010). J. Biol. Chem. 285, 37513–37520. [DOI] [PMC free article] [PubMed]
- van der Weerden, N. L., Lay, F. T. & Anderson, M. A. (2008). J. Biol. Chem. 283, 14445–14452. [DOI] [PubMed]
- Yamada, S., Komori, T. & Imaseki, H. (1997). Plant Physiol. 115, 314.