The subclass B3 metallo-β-lactamase SMB-1 was crystallized by the hanging-drop vapour-diffusion method. Two types of crystals belonging to space group P31 were obtained.
Keywords: metallo-β-lactamases, SMB-1, carbapenem resistance
Abstract
The carbapenem-hydrolyzing subclass B3 metallo-β-lactamase SMB-1 was expressed in Escherichia coli and purified. Diffraction data were collected from two types of SMB-1 crystals that were obtained under different conditions. One crystal (SMB-1a) belonged to the trigonal space group P31 with unit-cell parameters a = b = 67.83, c = 48.67 Å, while the other crystal (SMB-1b) also belonged to space group P31 but with unit-cell parameters a = b = 67.25, c = 46.83 Å. Both crystals contained one molecule per asymmetric unit. Initial phases were determined by molecular replacement; further refinement and model building are in progress.
1. Introduction
Metallo-β-lactamases (MBLs) catalyze the hydrolysis of amide bonds in a variety of β-lactam antibiotics, penicillins, cephalosporins and carbapenems, resulting in inactivation of the antimicrobial activity of β-lactam antibiotics. The MBLs hydrolyzing β-lactams are classified into three major groups based on their primary amino-acid sequence: subclasses B1, B2 and B3 (Garau et al., 2004 ▶). MBLs commonly contain one or two zinc ions that are bound by histidine, aspartic acid and cysteine residues in the active site (PDB entries 1sml and 1x8g; Garau et al., 2004 ▶). Recently, the emergence of carbapenem-resistant Enterobacteriaceae such as Escherichia coli and Klebsiella pneumoniae via the production of MBLs is becoming a great concern in clinical settings because carbapenem antibiotics remain important agents for the treatment of infectious diseases such as bacteraemia as well as infections of the respiratory tract and urinary tract caused by Enterobacteriaceae.
The most prevalent MBLs among the Enterobacteriaceae are the subclass B1 MBLs, which include IMP-type, VIM-type, NDM-type and SPM-type MBLs. The responsible genes are often procured with a mobile genetic element such as a transposon and are further embedded with transferable plasmids (Cornaglia et al., 2011 ▶). Therefore, these MBLs can easily be spread among different bacterial species. The genes of several subclass B1 MBLs such as the IND-type and BlaB-type MBLs are intrinsically presented on the chromosome of human opportunistic pathogens (Cricco & Vila, 1999 ▶). The genes of subclass B3 MBLs identified to date are from human opportunistic pathogens and environmental microbial genomes (Cricco & Vila, 1999 ▶; Stoczko et al., 2006 ▶). We recently identified a novel subclass B3 MBL named SMB-1 from the human opportunistic pathogen Serratia marcescens, which was isolated from a urine sample of an inpatient (Wachino et al., 2011 ▶). The gene for SMB-1 was flanked by two copies of an ISCR1 element, which is known to mediate the translocation of a variety of antibiotic-resistance genes in Gram-negative bacteria. As such, we hypothesized that the SMB-1 gene was integrated into the chromosome of S. marcescens from another bacterial species through a genetic recombination event caused by ISCR1 (Toleman & Walsh, 2011 ▶; Wachino et al., 2011 ▶).
Structure-based design of a potent specific inhibitor that can repress MBL activity could be one solution to overcome β-lactam resistance conferred by MBLs. To date, the X-ray crystal structures of subclass B1 MBLs in complex with various inhibitory agents have been reported, including IMP-1–mercaptocarboxylate derivatives (Concha et al., 2000 ▶), IMP-1–dansylC4SH (Kurosaki et al., 2006 ▶), VIM-2–phenylC3SH (Yamaguchi et al., 2007 ▶), BlaB–d-captopril (García-Saez et al., 2003 ▶) and CcrA–biphenyltetrazoles (Toney et al., 1998 ▶). In addition, the crystal structures of subclass B3 MBLs in complex with inhibitory agents have been reported, including L1–d-captopril (Nauton et al., 2008 ▶) and BJP-1–4-nitrobenzenesulfonamide (Docquier et al., 2010 ▶). To develop potent MBL inhibitors, further information on the three-dimensional structure of MBLs, especially subclass B3 MBLs, will be required. As such, we report here the crystallization and preliminary X-ray analysis of the subclass B3 MBL SMB-1.
2. Methods and results
2.1. Protein expression, purification and crystallization
The expression of SMB-1 (GenBank accession No. AB636283) in E. coli and purification were performed according to a previously described method (Wachino et al., 2011 ▶). In brief, full-length SMB-1 protein (residues 1–280) including the predicted signal peptide (residues 1–18) was overexpressed using a pET30a vector and E. coli strain BL21(DE3)pLysS. The expressed protein was purified by three chromatographic steps using a HiTrap SP HP column (GE Healthcare), a HiLoad 16/60 Superdex 200 pg column (GE Healthcare) and a HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) (Fig. 1 ▶). The purified protein was then exchanged into 20 mM HEPES–NaOH buffer pH 7.5 by diafiltration using an Amicon Ultra-15 Centricon (Millipore) and finally concentrated to 15 mg ml−1. The N-terminal sequence of mature SMB-1 was determined to be QDRDW (residues 19–23), which corresponds to the sequence after cleavage of the predicted signal peptide described in our previous study (Wachino et al., 2011 ▶). Mature SMB-1 was calculated to have a molecular mass of 27.7 kDa.
Figure 1.
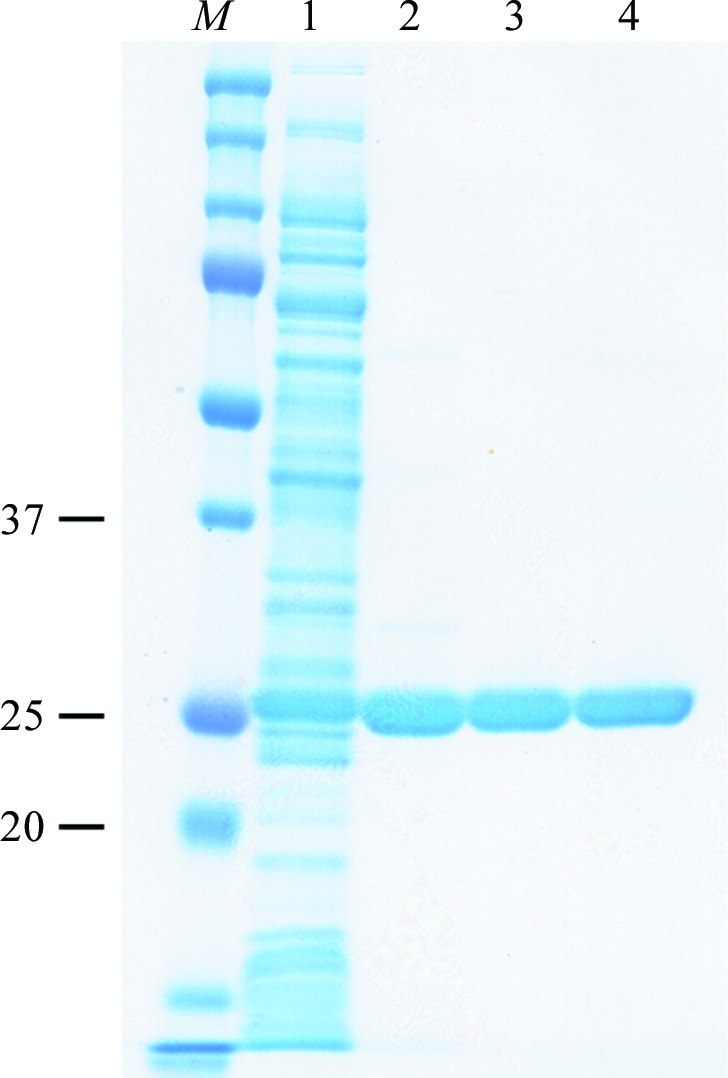
Purification of SMB-1. Lane M, protein size markers (labelled in kDa); lane 1, soluble cell extract; lane 2, fraction after cation-exchange chromatography; lane 3, fraction after size-exclusion chromatography with a HiLoad 16/60 Superdex 200 pg column; lane 4, fraction after size-exclusion chromatography with a HiPrep 16/60 Sephacryl S-200 HR column. The protein was subjected to SDS–PAGE on 12.5% polyacrylamide gel and stained with Coomassie Brilliant Blue.
Initial screening for crystallization was performed manually using 290 different reservoir conditions from commercially available screening kits, including Crystal Screen, Crystal Screen 2, PEG/Ion Screen and Grid Screens (PEG/LiCl, Sodium Chloride, Ammonium Sulfate and PEG 6000) from Hampton Research and the Wizard III random sparse-matrix crystallization screen from Emerald BioSystems. All initial crystallization experiments were performed at 293 K using the sitting-drop vapour-diffusion method. 1 µl purified SMB-1 protein solution and 1 µl reservoir solution were mixed in 96-well Intelli-Plates (Art Robbins Instruments) and equilibrated against 100 µl reservoir solution. Crystals were obtained within one week using several reservoir solutions, including Crystal Screen 2 condition No. 13 [0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.6, 30%(w/v) PEG monomethyl ether (MME) 2000] and Grid Screen PEG/LiCl condition D3 [1.0 M lithium chloride, 0.1 M MES pH 6.0, 30%(w/v) PEG 6000]. The pH and the salt and precipitant concentrations in the reservoir solution were further optimized manually using the hanging-drop vapour-diffusion method. Purified protein (3 µl) and reservoir solution (3 µl) were mixed and equilibrated against 500 µl reservoir solution.
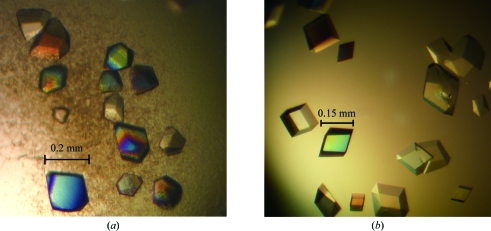
As a result, two types of crystals that were suitable for the collection of diffraction data were obtained under different conditions. One crystal (SMB-1a) was obtained using a reservoir solution consisting of 0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 5.3, 28%(w/v) PEG MME 2000. The crystals grew to dimensions of approximately 0.2 × 0.2 × 0.1 mm after one week (Fig. 2 ▶ a). A second crystal (SMB-1b) was obtained using a reservoir solution consisting of 1.0 M lithium chloride, 0.1 M MES pH 5.8, 26%(w/v) PEG 6000. The crystals grew to dimensions of approximately 0.15 × 0.15 × 0.1 mm after one week (Fig. 2 ▶ b).
Figure 2.
Optimized crystals: (a) SMB-1a and (b) SMB-1b.
2.2. X-ray diffraction, data collection and analysis
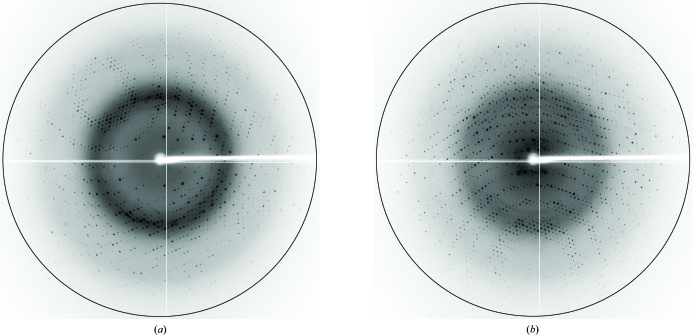
Prior to data collection, the SMB-1a crystals were soaked in a cryoprotectant solution consisting of 0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 5.3, 28%(w/v) PEG MME 2000, 20%(v/v) glycerol for a few seconds. X-ray diffraction data were collected at 100 K using a Quantum 315 CCD detector (Area Detector Systems Corp.) on beamline BL-5A at the Photon Factory (PF; Tsukuba, Japan; Fig. 3 ▶ a). The collected diffraction data were processed and scaled at 1.60 Å resolution using DENZO and SCALEPACK from the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). The space group was determined to be P31 or P32, with unit-cell parameters a = b = 67.83, c = 48.67 Å. The number of molecules in each asymmetric unit was estimated to be one from the Matthews coefficient (2.33 Å3 Da−1), corresponding to a solvent content of 47.3%. Scaling and merging of the crystallographic data showed an overall R merge of 5.1%. The SMB-1b crystals were soaked for cryoprotection in a solution consisting of 1.0 M lithium chloride, 0.1 M MES pH 5.8, 26%(w/v) PEG 6000, 20%(v/v) glycerol for a few seconds. The diffraction data were collected (Fig. 3 ▶ b), processed and scaled at 1.60 Å resolution. The SMB-1b crystals belonged to space group P31 or P32, with unit-cell parameters a = b = 67.25, c = 46.83 Å. The asymmetric unit of the SMB-1b crystal was estimated to contain one molecule from the Matthews coefficient (2.21 Å3 Da−1), corresponding to a solvent content of 44.2%. Scaling and merging of the crystallographic data showed an overall R merge of 5.8%. A summary of the data statistics is shown in Table 1 ▶.
Figure 3.
X-ray diffraction images of (a) SMB-1a and (b) SMB-1b crystals. The black circles indicate a resolution of 1.65 Å.
Table 1. Data-collection statistics.
Values in parentheses are for the last shell.
| Crystal | SMB-1a | SMB-1b |
|---|---|---|
| Beamline | BL-5A, PF | BL-5A, PF |
| Wavelength (Å) | 1.00000 | 1.00000 |
| Oscillation angle (°) | 1 | 1 |
| Crystal-to-detector distance (mm) | 142.3 | 142.3 |
| Exposure time (s) | 1.0 | 1.0 |
| Temperature (K) | 100 | 100 |
| Resolution range (Å) | 50.0–1.60 (1.63–1.60) | 50.0–1.60 (1.63–1.60) |
| VM (Å3 Da−1) | 2.33 | 2.21 |
| Solvent content (%) | 47.3 | 44.2 |
| Space group | P31 | P31 |
| Unit-cell parameters | ||
| a (Å) | 67.83 | 67.25 |
| b (Å) | 67.83 | 67.25 |
| c (Å) | 48.67 | 46.83 |
| No. of observed reflections | 367548 | 338253 |
| No. of unique reflections | 32909 (1654) | 31322 (1567) |
| Completeness (%) | 99.4 (100.0) | 99.7 (100.0) |
| Multiplicity | 11.2 (11.0) | 10.8 (10.2) |
| Rmerge† (%) | 5.1 (21.6) | 5.8 (23.3) |
| Mean I/σ(I) | 73.4 (17.3) | 71.1 (15.4) |
R
merge = 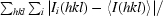
 , where I
i(hkl) is the observed intensity for reflection hkl and 〈I(hkl)〉 is the average intensity calculated for reflection hkl from replicate data.
, where I
i(hkl) is the observed intensity for reflection hkl and 〈I(hkl)〉 is the average intensity calculated for reflection hkl from replicate data.
The structure of SMB-1 was solved by molecular replacement using the MOLREP program (Vagin & Teplyakov, 2010 ▶) from the CCP4 suite (Winn et al., 2011 ▶). The L1 MBL from Stenotrophomonas maltophilia, which has 31% amino-acid identity to SMB-1, was used as the search model (Ullah et al., 1998 ▶). The structure of L1 solved using 1.7 Å resolution diffraction data (Nauton et al., 2008 ▶; PDB entry 1sml) was modified by the deletion of water molecules, other ions and several loop structures; mismatched amino acids were replaced by alanine. The molecular replacement was performed using the modified L1 structure as the search model and it was confirmed that both the SMB-1a and the SMB-1b crystals belonged to space group P31. Further refinement and model building are in progress.
Acknowledgments
This study was supported by the Ministry of Health, Labour and Welfare of Japan (grant H21-Shinkou-Ippan-008). We are grateful to the beamline BL-5A staff at Photon Factory, Tsukuba, Japan.
References
- Concha, N. O., Janson, C. A., Rowling, P., Pearson, S., Cheever, C. A., Clarke, B. P., Lewis, C., Galleni, M., Frère, J.-M., Payne, D. J., Bateson, J. H. & Abdel-Meguid, S. S. (2000). Biochemistry, 39, 4288–4298. [DOI] [PubMed]
- Cornaglia, G., Giamarellou, H. & Rossolini, G. M. (2011). Lancet Infect. Dis. 11, 381–393. [DOI] [PubMed]
- Cricco, J. A. & Vila, A. J. (1999). Curr. Pharm. Des. 5, 915–927. [PubMed]
- Docquier, J.-D., Benvenuti, M., Calderone, V., Stoczko, M., Menciassi, N., Rossolini, G. M. & Mangani, S. (2010). Antimicrob. Agents Chemother. 54, 4343–4351. [DOI] [PMC free article] [PubMed]
- Garau, G., García-Sáez, I., Bebrone, C., Anne, C., Mercuri, P., Galleni, M., Frère, J.-M. & Dideberg, O. (2004). Antimicrob. Agents Chemother. 48, 2347–2349. [DOI] [PMC free article] [PubMed]
- García-Saez, I., Hopkins, J., Papamicael, C., Franceschini, N., Amicosante, G., Rossolini, G. M., Galleni, M., Frère, J. M. & Dideberg, O. (2003). J. Biol. Chem. 278, 23868–23873. [DOI] [PubMed]
- Kurosaki, H., Yamaguchi, Y., Yasuzawa, H., Jin, W., Yamagata, Y. & Arakawa, Y. (2006). ChemMedChem, 1, 969–972. [DOI] [PubMed]
- Nauton, L., Kahn, R., Garau, G., Hernandez, J. F. & Dideberg, O. (2008). J. Mol. Biol. 375, 257–269. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Stoczko, M., Frère, J.-M., Rossolini, G. M. & Docquier, J.-D. (2006). Antimicrob. Agents Chemother. 50, 1973–1981. [DOI] [PMC free article] [PubMed]
- Toleman, M. A. & Walsh, T. R. (2011). FEMS Microbiol. Rev. 35, 912–935. [DOI] [PubMed]
- Toney, J. H., Fitzgerald, P. M. D., Grover-Sharma, N., Olson, S. H., May, W. J., Sundelof, J. G., Vanderwall, D. E., Cleary, K. A., Grant, S. K., Wu, J. K., Kozarich, J. W., Pompliano, D. L. & Hammond, G. G. (1998). Chem. Biol. 5, 185–196. [DOI] [PubMed]
- Ullah, J. H., Walsh, T. R., Taylor, I. A., Emery, D. C., Verma, C. S., Gamblin, S. J. & Spencer, J. (1998). J. Mol. Biol. 284, 125–136. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wachino, J., Yoshida, H., Yamane, K., Suzuki, S., Matsui, M., Yamagishi, T., Tsutsui, A., Konda, T., Shibayama, K. & Arakawa, Y. (2011). Antimicrob. Agents Chemother. 55, 5143–5149. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yamaguchi, Y., Jin, W., Matsunaga, K., Ikemizu, S., Yamagata, Y., Wachino, J., Shibata, N., Arakawa, Y. & Kurosaki, H. (2007). J. Med. Chem. 50, 6647–6653. [DOI] [PubMed]