The crystallization of Tic22 from P. falciparum is reported.
Keywords: Tic22, Plasmodium falciparum, apicoplasts, import apparatus
Abstract
Tic22 is a component of the protein-import apparatus of the chloroplasts of plants and algae and the apicoplasts of the Apicomplexa, a large group of organisms that includes the parasites that cause malaria. Tic22 is important for protein import into these organelles and for organelle biogenesis. It lies between the two membranes of chloroplasts, making interactions with components of both the TIC and TOC complexes. In the apicoplast, it is predicted to be located between the inner two membranes and to play a similar role in import. Although Tic22 is ubiquitous, its function is as yet uncertain. Tic22 from Plasmodium falciparum was therefore overproduced, purified and crystallized. A data set extending to 2.15 Å resolution has been collected from a crystal containing selenomethionine-labelled protein and structure determination is under way.
1. Introduction
The presence of a chloroplast is the defining feature of the cells of photosynthetic organisms, including plants, mosses and algae. This essential organelle is the site of light capture and the location for the conversion of light into chemical energy and reducing power. Although chloroplasts contain a circular genome, thought to be the remnant of the genome of an engulfed endosymbiont, hundreds of proteins required for chloroplast function are encoded in the nucleus (van Wijk, 2004 ▶). Chloroplast biogenesis therefore requires the targeting of these proteins to the chloroplast and their movement through two chloroplast membranes to their correct location. This requires a complex signal peptide at the N-terminus of the imported protein (Zhang & Glaser, 2002 ▶) and is mediated by the TIC and TOC complexes (Soll & Schleiff, 2004 ▶; Inaba & Schnell, 2008 ▶).
The TIC and TOC complexes both contain integral membrane components that form translocons across the inner and outer membranes, respectively. The core of the TOC complex is formed from two GTPases, Toc34 and Toc159, and the Toc75 channel. This complex recognizes transit peptides and translocates the associated protein across the outer membrane using a GTP-driven mechanism. The TIC complex is centred around Tic20, Tic21 and Tic110, which are proposed to come together to form channels across the inner membrane (Soll & Schleiff, 2004 ▶; Inaba & Schnell, 2008 ▶; Kovács-Bogdán et al., 2011 ▶). Tic22 is a soluble component located in the space between the two membranes (Kouranov et al., 1998 ▶) and has been shown to interact loosely with components of both the TIC (Kouranov et al., 1998 ▶) and TOC complexes (Becker et al., 2004 ▶).
The Apicomplexa are a group of protists, many of which contain a plastid, the apicoplast (McFadden et al., 1996 ▶; Wilson et al., 1996 ▶; Köhler et al., 1997 ▶; Waller & McFadden, 2005 ▶). These include Plasmodium species, the parasites that cause malaria, together with other parasites of medical and veterinary importance, including Toxoplasma, Theileria, Eimeria, Babesia and Crytosporidium. The apicoplast is thought to be the result of a secondary endosymbiotic event (Gould et al., 2008 ▶) involving the engulfment of photosynthetic red algae. It is essential for the survival of Plasmodium species in both the liver and erythrocyte stages, with a delayed-death phenotype resulting from apicoplast disruption (Goodman et al., 2007 ▶; Vaughan et al., 2008 ▶; Yu et al., 2008 ▶; McConkey et al., 1997 ▶; Ralph et al., 2001 ▶). This may result from the role of the apicoplast in fatty-acid synthesis and a need for fatty acids for both the formation of the parasitophorous vacuole in which the parasite resides within host cells and the formation of the invasive merozoite form of the parasite (He et al., 2001 ▶; Dahl et al., 2006 ▶; Goodman et al., 2007 ▶; Ramya et al., 2007 ▶; Dahl & Rosenthal, 2008 ▶, Waller et al., 1998 ▶). The apicoplast is therefore an important drug target for the development of treatments for malaria.
The apicoplast has four membranes; the outer of these is derived from the endoplasmic reticulum, while the inner two are equivalent to the two membranes of the engulfed algal chloroplast (Köhler et al., 1997 ▶; McFadden, 1999 ▶). Some 466 nuclear-encoded proteins are predicted to be targeted to the apicoplast owing to the presence of a bipartite peptide at their amino-terminus (Foth et al., 2003 ▶). This peptide contains a signal sequence required for initial entry into the outer layer of the apicoplast and a transit peptide required for passage through the inner two ‘chloroplast-like’ membranes (Waller et al., 1998 ▶; Foth et al., 2003 ▶). While many of the components of the TIC and TOC complexes cannot be identified in the genomes of the Apicomplexa (Tonkin et al., 2008 ▶), Tic20 and Tic22 have been found (Lim et al., 2009 ▶; Kalanon et al., 2009 ▶) and Tic20 has been shown to be essential for growth of Toxoplasma gondii (van Dooren et al., 2008 ▶).
Despite the ubiquitous presence of Tic22 in plants and the Apicomplexa, a role for this protein has not been identified, although it may be involved in linking the TIC and TOC complexes or in guiding proteins between the two (Soll & Schleiff, 2004 ▶). We have therefore expressed, purified and crystallized the Tic22 protein and aim to provide structural insights into the function of this molecule in protein import.
2. Experimental procedures
2.1. Cloning, expression and purification
A construct containing the gene for Tic22 (accession code NP_195061) from the 3D7 strain of P. falciparum (PfTic22) was provided by Dr Ming Kalanon (Kalanon et al., 2009 ▶). Residues 68–279 were selected, omitting those residues thought to form the transit and signal peptides, and were inserted into the pEt30b vector in frame with an N-terminal six histidine tag and an enterokinase cleavage site.
Rosetta Eschericha coli cells (Novagen) were transformed with the plasmid encoding PfTic22.
To produce selenomethione-labelled protein, a metabolic poisoning strategy was used (van Duyne et al., 1993 ▶). Cells were initially grown in 2×YT medium to an optical density at 600 nm of 0.5 and were pelleted before resuspension in M9 medium (20% glucose, 40 mM Na2HPO4, 20 mM KH2PO4, 20 mM NH4Cl, 8.5 mM NaCl, 1 mM MgSO4, 100 mg l−1 each of l-lysine, l-phenylalanine and l-threonine and 50 mg l−1 each of l-isoleucine, l-leucine, l-valine and l-selenomethionine). They were incubated at 310 K for a further 1 h, induced with 1 mM IPTG and incubated overnight at 298 K to allow expression.
The cells were pelleted, resuspended in solubilization buffer (20 mM Tris pH 8.0, 100 mM NaCl, 15 mM imidazole, 0.5% Triton X-100) and lysed by sonication. The cell lysate was centrifuged for 30 min at 45 000g and purified by affinity chromatography using Ni–NTA Sepharose (Qiagen). The protein was loaded onto the Ni–NTA resin, washed with solubilization buffer and eluted with 20 mM Tris pH 8.0, 150 mM NaCl, 200 mM imidazole.
The protein was buffer-exchanged into 20 mM Tris pH 8.0, 150 mM NaCl, 2 mM CaCl2 and cleaved by incubation overnight at 277 K in the presence of 0.001%(w/w) enterokinase. The cleaved protein was passed through an Ni–NTA affinity column. The eluant was concentrated using an Amicon Ultra centifugal filter device (10 000 molecular-weight cutoff) and further purified using a Superdex 75 16/60 (GE Healthcare) column run using 20 mM Tris pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol. The protein was concentrated to 17.5 mg ml−1.
2.2. Crystallization and crystal optimization
Crystals were grown using the hanging-drop vapour-diffusion technique by mixing 1 µl protein solution with 1 µl reservoir solution and equilibrating against 1 ml reservoir solution. A single initial crystal appeared in 0.1 M MES pH 6, 20% PEG 4000 after 7 d at 291 K. Nucleation could be increased by initially incubating the crystallization plate at 303 K for 15 h before transferring it to 291 K, but this resulted in stepped and pitted crystals. These crystals were harvested in well solution and were vortexed in the presence of a Seed Bead (Hampton Research) to generate microseeds. Crystallization droplets consisting of 1 µl protein solution at 8–10 mg ml−1 and 1 µl 0.1 M MES pH 6.0, 20% PEG 4000 were incubated overnight at 291 K before introduction of microseeds using a horse hair. Single crystals appeared after 1 d and grew to final dimensions of 0.3 × 0.3 × 0.2 mm.
2.3. Data collection and processing
Crystals were cryoprotected by adding 0.1 M MES pH 6.0, 20% PEG 4000, 25% glycerol to the crystallization droplet over the course of 5 min and were flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on beamline I04 at the Diamond Light Source. A fluorescence scan was employed to locate the absorbance edge for selenium, with the peak and inflection wavelengths obtained using CHOOCH (Evans & Pettifer, 2001 ▶). A three-wavelength anomalous dispersion data set was collected at wavelengths of 0.9800 Å (peak), 0.9802 Å (inflection) and 0.9537 Å (remote). A total of 360° of data were collected at each wavelength with a 0.5° oscillation angle and 0.3 s exposure with 38% transmission.
Data were processed using iMOSFLM (Battye et al., 2011 ▶; Leslie, 1992 ▶) and SCALA (Evans, 1993 ▶) from the CCP4 suite (Winn et al., 2011 ▶) and were consistent with a primitive tetragonal lattice. Systematic absences indicated that the crystals belonged to space group P41212. A complete data set was collected to a resolution limit of 2.15 Å.
3. Results and discussion
We have expressed and purified the import-apparatus component Tic22 from P. falciparum. The protein was subjected to hanging-drop crystallization trials and a crystal was observed after 7 d. Structure-based sequence alignment using FUGUE (Shi et al., 2001 ▶) identified no homologues of Tic22 with known structure suitable for molecular replacement. With this knowledge, and also the experience that the optimization to produce diffraction-quality crystals of PfTic22 was challenging, we decided to optimize crystal formation using selenomethionine-labelled protein rather than unlabelled protein.
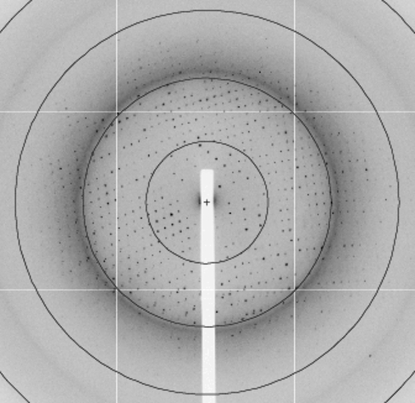
Crystal nucleation was aided by incubation of the plate for 15 h at 303 K before moving to 291 K. The resulting crystals were unsuitable for structure determination, but could be used to generate microseeds to produce diffraction-quality crystals from selenomethionine-labelled protein. These grew to maximum dimensions of 0.3 × 0.3 × 0.2 mm (Fig. 1 ▶) and diffracted to 2.15 Å resolution (Fig. 2 ▶). A complete multiwavelength data set has been collected (Table 1 ▶). Preliminary analysis of the peak data set using SHELXC (Sheldrick, 2008 ▶) showed a high value of d′′/sig, indicating significant differences between the intensities of the Bijvoet pairs and the presence of usable anomalous signal to 2.8 Å resolution (Table 2 ▶). We are currently working to complete this structure and to use the insight that it provides to investigate the role of Tic22 in protein import during apicoplast biogenesis.
Figure 1.
Crystals of Tic22 from P. falciparum.
Figure 2.
Diffraction pattern from a crystal of Tic22. Resolution rings are at 7.7, 3.9, 2.6 and 1.9 Å resolution.
Table 1. Data-collection statistics.
| Peak | Inflection | Remote | Remote wedge (290–430°) | |
|---|---|---|---|---|
| Space group | P41212 | |||
| Unit-cell parameters (Å, °) | a = b = 66.25, c = 129.37, α = β = γ = 90.00 | |||
| Molecules per asymmetric unit | 1 | |||
| Matthews coefficient (Å3 Da−1) | 2.75 | |||
| Solvent content (%) | 55.4 | |||
| Resolution (Å) | 59–2.34 (2.47–2.34) | 59–2.34 (2.47–2.34) | 59–2.34 (2.47–2.34) | 46.9–2.15 (2.27–2.15) |
| Wavelength (Å) | 0.9800 | 0.9802 | 0.9537 | 0.9537 |
| Mosaicity (°) | 0.67 | 0.69 | 0.70 | 0.67 |
| Rmerge | 0.158 (0.707) | 0.149 (0.509) | 0.163 (0.611) | 0.100 (0.627) |
| Rmeas (within I+/I−) | 0.164 (0.736) | 0.155 (0.529) | 0.169 (0.632) | 0.111 (0.695) |
| Observed reflections | 238848 (39070) | 332088 (41035) | 339770 (50988) | 84566 (12492) |
| Unique reflections | 12618 (1618) | 12610 (1620) | 12830 (1842) | 16317 (2316) |
| Mean I/σ(I) | 14.6 (4.5) | 15.3 (5.6) | 14.1 (5.2) | 7.9 (2.1) |
| Completeness (%) | 99.7 (98.4) | 99.8 (99.8) | 99.8 (100) | 99.6 (100) |
| Multiplicity | 26.1 (24.1) | 26.3 (25.3) | 26.5 (27.7) | 5. 2 (5.4) |
| Anomalous completeness (%) | 99.7 (98.3) | 99.9 (99.8) | 99.9 (100) | 99.2 (99.3) |
| Anomalous multiplicity | 14.4 (12.9) | 14.6 (13.5) | 14.6 (14.9) | 2.8 (2.8) |
Table 2. Indication of the quality of the anomalous signal from the peak data set determined using SHELXC .
| Resolution (Å) | ∞–8.0 | 8.0–6.0 | 6.0–5.0 | 5.0–4.0 | 4.0–3.6 | 3.6–3.4 | 3.4–3.2 | 3.2–3.0 | 3.0–2.8 | 2.8–2.6 | 2.6–2.35 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of data | 361 | 469 | 581 | 1280 | 949 | 659 | 817 | 1051 | 1370 | 1812 | 3194 |
| 〈I/σ(I)〉 | 23.5 | 22.6 | 23.7 | 24.3 | 23.0 | 21.2 | 18.0 | 16.0 | 12.1 | 8.4 | 5.0 |
| Completeness (%) | 89.6 | 98.7 | 99.3 | 99.5 | 99.5 | 99.4 | 99.8 | 99.6 | 99.8 | 99.7 | 99.7 |
| 〈d′′/sig〉 | 3.15 | 3.34 | 2.88 | 2.31 | 2.06 | 2.02 | 1.84 | 1.59 | 1.35 | 1.12 | 0.88 |
Acknowledgments
We would like to thank the Medical Research Council and the Royal Society for funding and Dr Ming Kalanon for providing the PfTic22 gene.
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Becker, T., Hritz, J., Vogel, M., Caliebe, A., Bukau, B., Soll, J. & Schleiff, E. (2004). Mol. Biol. Cell, 15, 5130–5144. [DOI] [PMC free article] [PubMed]
- Dahl, E. L. & Rosenthal, P. J. (2008). Trends Parasitol. 24, 279–284. [DOI] [PubMed]
- Dahl, E. L., Shock, J. L., Shenai, B. R., Gut, J., DeRisi, J. L. & Rosenthal, P. J. (2006). Antimicrob. Agents Chemother. 50, 3124–3131. [DOI] [PMC free article] [PubMed]
- Dooren, G. G. van, Tomova, C., Agrawal, S., Humbel, B. M. & Striepen, B. (2008). Proc. Natl Acad. Sci. USA, 105, 13574–13579. [DOI] [PMC free article] [PubMed]
- Evans, P. R. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 114–133. Warrington: Daresbury Laboratory.
- Evans, G. & Pettifer, R. F. (2001). J. Appl. Cryst. 34, 82–86.
- Foth, B. J., Ralph, S. A., Tonkin, C. J., Struck, N. S., Fraunholz, M., Roos, D. S., Cowman, A. F. & McFadden, G. I. (2003). Science, 299, 705–708. [DOI] [PubMed]
- Goodman, C. D., Su, V. & McFadden, G. I. (2007). Mol. Biochem. Parasitol. 152, 181–191. [DOI] [PubMed]
- Gould, S. B., Waller, R. F. & McFadden, G. I. (2008). Annu. Rev. Plant Biol. 59, 491–517. [DOI] [PubMed]
- He, C. Y., Shaw, M. K., Pletcher, C. H., Striepen, B., Tilney, L. G. & Roos, D. S. (2001). EMBO J. 20, 330–339. [DOI] [PMC free article] [PubMed]
- Inaba, T. & Schnell, D. J. (2008). Biochem. J. 413, 15–28. [DOI] [PubMed]
- Kalanon, M., Tonkin, C. J. & McFadden, G. I. (2009). Eukaryot. Cell, 8, 1146–1154. [DOI] [PMC free article] [PubMed]
- Köhler, S., Delwiche, C. F., Denny, P. W., Tilney, L. G., Webster, P., Wilson, R. J., Palmer, J. D. & Roos, D. S. (1997). Science, 275, 1485–1489. [DOI] [PubMed]
- Kouranov, A., Chen, X., Fuks, B. & Schnell, D. J. (1998). J. Cell Biol. 143, 991–1002. [DOI] [PMC free article] [PubMed]
- Kovács-Bogdán, E., Benz, J. P., Soll, J. & Bölter, B. (2011). BMC Plant Biol. 11, 133. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Lim, L., Kalanon, M. & McFadden, G. I. (2009). Trends Parasitol. 25, 197–200. [DOI] [PubMed]
- McConkey, G. A., Rogers, M. J. & McCutchan, T. F. (1997). J. Biol. Chem. 272, 2046–2049. [DOI] [PubMed]
- McFadden, G. I. (1999). J. Eukaryot. Microbiol. 46, 339–346. [DOI] [PubMed]
- McFadden, G. I., Reith, M. E., Munholland, J. & Lang-Unnasch, N. (1996). Nature (London), 381, 482. [DOI] [PubMed]
- Ralph, S. A., D’Ombrain, M. C. & McFadden, G. I. (2001). Drug Resist. Updat. 4, 145–151. [DOI] [PubMed]
- Ramya, T. N., Mishra, S., Karmodiya, K., Surolia, N. & Surolia, A. (2007). Antimicrob. Agents Chemother. 51, 307–316. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, J., Blundell, T. L. & Mizuguchi, K. (2001). J. Mol. Biol. 310, 243–257. [DOI] [PubMed]
- Soll, J. & Schleiff, E. (2004). Nature Rev. Mol. Cell Biol. 5, 198–208. [DOI] [PubMed]
- Tonkin, C. J., Kalanon, M. & McFadden, G. I. (2008). Traffic, 9, 166–175. [DOI] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol. 229, 105–124. [DOI] [PubMed]
- Vaughan, A. M., Aly, A. S. & Kappe, S. H. (2008). Cell Host Microbe, 4, 209–218. [DOI] [PMC free article] [PubMed]
- Waller, R. F., Keeling, P. J., Donald, R. G., Striepen, B., Handman, E., Lang-Unnasch, N., Cowman, A. F., Besra, G. S., Roos, D. S. & McFadden, G. I. (1998). Proc. Natl Acad. Sci. USA, 95, 12352–12357. [DOI] [PMC free article] [PubMed]
- Waller, R. F. & McFadden, G. I. (2005). Curr. Issues Mol. Biol. 7, 57–79. [PubMed]
- Wijk, K. J. van (2004). Plant Physiol. Biochem. 42, 963–977. [DOI] [PubMed]
- Wilson, R. J., Denny, P. W., Preiser, P. R., Rangachari, K., Roberts, K., Roy, A., Whyte, A., Strath, M., Moore, D. J., Moore, P. W. & Williamson, D. H. (1996). J. Mol. Biol. 261, 155–172. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yu, M. et al. (2008). Cell Host Microbe, 4, 567–578. [DOI] [PMC free article] [PubMed]
- Zhang, X.-P. & Glaser, E. (2002). Trends Plant Sci. 7, 14–21. [DOI] [PubMed]