The SH2 domain of the human protein tyrosine kinase Fyn has been expressed, purified and crystallized in the unbound state and in complex with a high-affinity phosphotyrosine peptide.
Keywords: SH2 domains, Fyn, Src kinases
Abstract
SH2 domains are widespread protein-binding modules that recognize phosphotyrosines and play central roles in intracellular signalling pathways. The SH2 domain of the human protein tyrosine kinase Fyn has been expressed, purified and crystallized in the unbound state and in complex with a high-affinity phosphotyrosine peptide. X-ray data were collected to a resolution of 2.00 Å for the unbound form and 1.40 Å for the protein in complex with the phosphotyrosine peptide.
1. Introduction
Selective protein–protein interactions play a critical role in cell-cycle regulation and are generally mediated by modular protein domains. Src homology 2 (SH2) domains are one of the paradigm modules involved in regulatory processes. They consist of about 100 amino acids and are uniquely dedicated to recognition of phosphotyrosine-modified polypeptides. Depending on the nature of the neighbouring units, SH2-containing proteins play many different roles in numerous signal transduction pathways (Pawson, 2004 ▶; Ishizawar & Parsons, 2004 ▶; Machida & Mayer, 2005 ▶; Dierck et al., 2009 ▶). Despite being found in a wide range of proteins, SH2 domains are highly structurally conserved (Schlessinger & Lemmon, 2003 ▶; Liu et al., 2006 ▶, 2011 ▶).
In this work, we present the crystallization and preliminary X-ray data analysis of the SH2 domain from the human Fyn protein (Fyn SH2) in the apo form and in complex with a phosphotyrosine peptide. Fyn is part of the Src family of nonreceptor tyrosine kinases; it is found in most cell types and mediates cytoplasmic signalling pathways for a variety of membrane receptors (Arold et al., 2001 ▶; Schlessinger & Lemmon, 2003 ▶; Kinoshita et al., 2006 ▶; Engen et al., 2008 ▶).
The Src-family kinases have a modular architecture consisting of a unique N-terminal sequence, three protein modules (the SH3, SH2 and kinase domains) and a C-terminal tail. In the inactive form the SH2 domain binds to the phosphorylated tyrosine in the C-terminal tail, whereas the SH3 domain docks onto a particular region of the kinase domain via a proline-containing motif. One of the activation mechanisms consists of dephosphorylation of this discrete tyrosine in the C-terminal tail, which releases both the SH2 and SH3 domains (Young et al., 2001 ▶). Solution structures of the Fyn SH2 and SH3 domains showed its binding specificity for certain peptide inhibitors (Morton et al., 1996 ▶; Pintar et al., 1996 ▶; Mulhern et al., 1997 ▶). However, the significance of the fine-tuning of intramolecular and intermolecular interactions is still of extreme importance for understanding the full functionality of the Src kinases (Aleshin & Finn, 2010 ▶; Wadhawan et al., 2011 ▶).
In order to reveal the structural determinants for the underlying functional mechanism of human Fyn kinase, we report here the purification, crystallization and preliminary crystallographic analysis of the Fyn SH2 domain in the apo form and in complex with a phosphotyrosine peptide corresponding to residues 321–331 of hamster middle-T antigen, which has been shown to bind Fyn with high affinity (Cantley et al., 1991 ▶; Bradshaw et al., 1998 ▶).
2. Materials and methods
2.1. Cloning and expression
PCR primers were designed based on the DNA sequence of the SH2 domain from human p59fyn (residues 149–248) using the DNAWorks online software (Hoover & Lubkowski, 2002 ▶). The complete list of oligonucleotides used for Fyn SH2 gene synthesis is given in Table 1 ▶. The target gene was amplified by PCR using 0.2 ng µl−1 of each oligonucleotide, 5 µl Ex Taq buffer (10×), 0.2 mM of each dNTP and 1 U TaKaRa Ex Taq DNA polymerase. The PCR protocol for gene assembly began with 1 min denaturation at 368 K, followed by 25 cycles of 50 s at 368 K for denaturation, 30 s at 339 K for annealing and 90 s at 345 K for extension. The last step of the protocol was an incubation cycle at 345 K for 10 min. The PCR product (approximately 400 nucleotides) was digested with NdeI and XbaI and the fragment containing the Fyn SH2 gene was inserted into two different vectors: pET15b (Novagen) and pColdII (TaKaRa). In the case of pET15b a thrombin cleavage site situated between the tag and the construct allowed the removal of the His tag, leaving the sequence GSHM at the N-terminus of the protein after cleavage. Escherichia coli BL21 (DE3) cells were subsequently transformed with the pET15b-Fyn SH2 construct and with the pColdII-Fyn SH2 construct.
Table 1. The 12 oligonucleotides used for Fyn SH2 gene synthesis.
| 1 | GAGGTAATACACCATGAATCACAAAGTGCATCATCATCATCATCATATGGAAT |
| 2 | CTTTACGACCCAGTTTACCAAAGTACCATTCCATATGATGATGATGATGATGCAC |
| 3 | GTACTTTGGTAAACTGGGTCGTAAAGACGCGGAACGTCAGCTCCTGAGCTTCGGT |
| 4 | TCAGATTCACGGATCAGGAAGGTACCACGCGGGTTACCGAAGCTCAGGAGCTGAC |
| 5 | ACCTTCCTGATCCGTGAATCTGAAACCACCAAAGGTGCGTACTCCCTGAGCATCC |
| 6 | CTTAACGTGGTCACCTTTCATATCGTCCCAGTCGCGGATGCTCAGGGAGTACGCA |
| 7 | CGATATGAAAGGTGACCACGTTAAGCATTATAAGATTCGCAAACTGGATAATGGT |
| 8 | TGCGCACGGGTGGTGATATAGTAGCCACCATTATCCAGTTTGCGAATCTTATAAT |
| 9 | ATCACCACCCGTGCGCAGTTCGAAACCCTGCAGCAGCTCGTCCAGCACTACTCTG |
| 10 | ACTACCAGACGGCAGCAGAGGCCCGCAGCACGTTCAGAGTAGTGCTGGACGAGCT |
| 11 | CTCTGCTGCCGTCTGGTAGTGCCGTGCCATAAATAGTCTAGATAGGTAATCTCTG |
| 12 | CAGGGATCTTAGATTCTGTGCTTTTAAGCAGAGATTACCTATCTAGACTATTTATGGCA |
The pET15b-Fyn SH2 construct was used to obtain crystals of the free Fyn SH2 domain without the His tag. 1 l minimal growth medium containing 4 g glucose, 1 g ammonium chloride and 100 µg ml−1 ampicillin was inoculated with an overnight preculture. Cells were grown to an OD600 of 0.7 at 310 K and induced with 1 mM IPTG. The overexpression temperature was modified to 303 K. 18 h after induction, the culture was harvested by centrifugation.
Overexpression of the pColdII-Fyn SH2 domain was performed using a modified protocol based on the single protein production system, which provides protein expression following a 10–40-fold condensation of cells (Suzuki et al., 2005 ▶, 2006 ▶). In this case the Fyn SH2 gene was synthesized with an N-terminal His tag (consisting of NHKVHHHHHHM). This construct was used to obtain crystals of the complex with the phosphotyrosine peptide. Cell cultures were grown at 310 K in 5 l minimal growth medium supplemented with 4 g glucose, 1 g ammonium chloride and 100 µg ml−1 ampicillin. When the OD at 600 nm reached 0.5, the culture was chilled in an ice–water bath to quickly reach 288 K. 45 min of incubation at 288 K allowed the cells to adapt to the new conditions and the culture was then harvested by centrifugation and resuspended in 250 ml minimal growth medium containing 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside; Sigma). The overexpression temperature was maintained at 288 K. 24 h after IPTG and cold-shock induction, cells were harvested by centrifugation.
2.2. Purification of free Fyn SH2
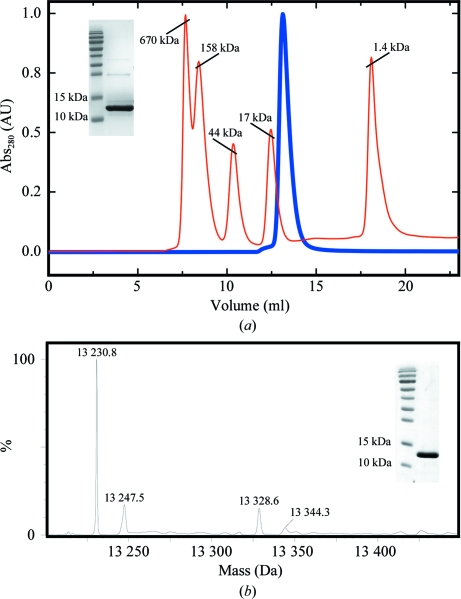
Harvested cells were resuspended in a lysis buffer consisting of 100 mM HEPES pH 8.0, 1 M NaCl, 20 mM imidazole and the protease inhibitors leupeptin (N-acetyl-l-leucyl-l-leucyl-l-argininal; MP Biomedicals) and AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride; Carl Roth GmbH]. Cells were broken at 277 K by passage through a French press three times and cell debris was removed by centrifugation. The supernatant was applied onto a 5 ml pre-packed Ni column (GE Healthcare) and eluted with 100 mM HEPES pH 8.0, 1 M NaCl, 1 M imidazole. In the case of the Fyn SH2 domain obtained from pET15b expression vector, an additional purification step was required after His-tag cleavage. Cleavage was performed at 299 K for 16 h using 2 U bovine thrombin (Calbiochem) per milligram of protein. The noncleaved Fyn SH2 domain was separated from the cleaved protein using a pre-packed Ni column. In the last purification step, the protein was loaded onto a Superdex 75 16/90 gel-filtration column and eluted in 50 mM sodium phosphate buffer pH 6.50, 5 mM DTT (dithiothreitol; Duchefa Biochemie). As previously shown by Pintar and coworkers, the Fyn SH2 domain is slightly prone to aggregation (Pintar et al., 1996 ▶). However, this small dimeric fraction can be efficiently removed by gel filtration, as shown in our case. The purity of the sample was analyzed by running a 20% SDS–PAGE gel in the presence of β-mercaptoethanol and size-exclusion analysis of the monomeric form. Bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B12 (1.35 kDa) were used as molecular-weight standards (Fig. 1 ▶ a).
Figure 1.
Purification of the human Fyn SH2 domain. (a) Analytical gel-filtration profile of non-His-tagged Fyn SH2 domain on a Superdex 75 HR column (loaded onto the column at 14 mg ml−1). The molecular-weight standards and protein chromatograms are represented as red and blue lines, respectively. The insert shows a 20% SDS–PAGE under reducing conditions of the molecular-weight marker and the purified Fyn SH2 domain. (b) Mass-spectrometric analysis of N-terminally His-tagged Fyn SH2 domain (from pColdII expression vector; expected mass based on the sequence, 13 231.0 Da). The insert shows 15% SDS–PAGE under reducing conditions of the molecular-weight marker and the purified Fyn SH2 domain.
In the case of the Fyn SH2 domain obtained from pColdII expression vector, no additional purification step was needed. After the final gel-filtration step, the sample quality was verified by 15% SDS–PAGE in the presence of β-mercaptoethanol and the protein identity was confirmed by mass spectrometry (Fig. 1 ▶ b).
2.3. Preparation of the complex
A phosphotyrosine peptide corresponding to residues 321–331 of hamster middle-T antigen has been shown to bind Fyn with high affinity (Cantley et al., 1991 ▶; Bradshaw et al., 1998 ▶). The peptide, with sequence acetyl-EPQ-pY-EEIPIYL-NH2, was purchased from JPT Peptide Technologies GmbH with more than 95% purity and was dissolved in 50 mM sodium phosphate buffer pH 6.50, 5 mM DTT.
The Fyn SH2–phosphotyrosine peptide complex was prepared by adding an excess of 18.5 mg ml−1 phosphotyrosine peptide to 10.6 mg ml−1 His-tagged Fyn SH2 domain (from pColdII expression vector) to give a molar ratio of 1.75:1. No further purification was necessary.
2.4. Crystallization
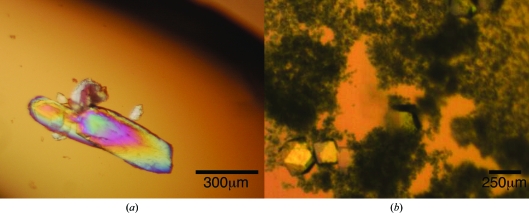
Crystals of the free Fyn SH2 domain were observed after one week of storage at 277 K of a concentrated stock of non-His-tagged protein at 23.3 mg ml−1 in 50 mM sodium phosphate pH 6.5 (Fig. 2 ▶ a).
Figure 2.
Crystals of Fyn SH2: (a) unbound and (b) in complex with the phosphotyrosine peptide.
For the Fyn SH2–phosphotyrosine peptide complex, crystallization conditions were screened using the hanging-drop vapour-diffusion method in 48-well plates using the Morpheus screen (Molecular Dimensions; Gorrec, 2009 ▶) at 293 K. The drops were prepared by mixing 1.5 µl protein solution and 1.5 µl reservoir solution and were equilibrated against 150 µl reservoir solution. Crystals of the complex were observed in a condition consisting of 0.1 M MOPS/Na HEPES pH 7.5, 0.12 M monosaccharide mixture (0.02 M d-glucose, 0.02 M d-mannose, 0.02 M d-galactose, 0.02 M l-fucose, 0.02 M d-xylose, 0.02 M N-acetyl-d-glucosamine), 12.5%(w/v) PEG 1000, 12.5%(w/v) PEG 3350 and 12.5%(v/v) MPD (Fig. 2 ▶ b).
2.5. Data collection and analysis
A crystal of free Fyn SH2 was cryoprotected by transferring it to drops consisting of 30 mM sodium phosphate, 5 mM DTT and increasing amounts of ethylene glycol. The concentration of ethylene glycol was increased in 5% steps to a final value of 35%. The crystal was subsequently vitrified in liquid nitrogen. X-ray data were collected on the PROXIMA-1 beamline of the SOLEIL synchrotron (Gif-Sur-Yvette, France) using an ADSC Quantum Q315 CCD detector and a wavelength of 0.98 Å.
A crystal of the Fyn SH2–phosphotyrosine peptide complex was directly vitrified in liquid nitrogen from the crystallization solution [0.1 M MOPS/Na HEPES pH 7.5, 0.12 M monosaccharide mixture (0.02 M d-glucose, 0.02 M d-mannose, 0.02 M d-galactose, 0.02 M l-fucose, 0.02 M d-xylose, 0.02 M N-acetyl-d-glucosamine), 12.5%(w/v) PEG 1000, 12.5%(w/v) PEG 3350 and 12.5%(v/v) MPD] without the need for any additional cryoprotectant. X-ray data were collected on beamline ID14-1 of the ESRF synchrotron (Grenoble, France) using an ADSC Quantum 4 CCD detector and a wavelength of 0.93 Å.
Data for free Fyn-SH2 were indexed and integrated using DENZO and subsequently scaled and merged using SCALEPACK (Otwinowski & Minor, 1997 ▶). The data for the Fyn SH2–phosphotyrosine peptide complex were indexed with MOSFLM (Leslie, 1992 ▶) and scaled with SCALA (Evans, 2006 ▶). Intensities were converted to structure-factor amplitudes using the CCP4 program TRUNCATE (Winn et al., 2011 ▶). Matthews coefficients were calculated with the program MATTHEWS_COEF for cell-content analysis (Winn et al., 2011 ▶). Calculations of self-rotation functions were performed with MOLREP (Vagin & Teplyakov, 2010 ▶).
3. Results and discussion
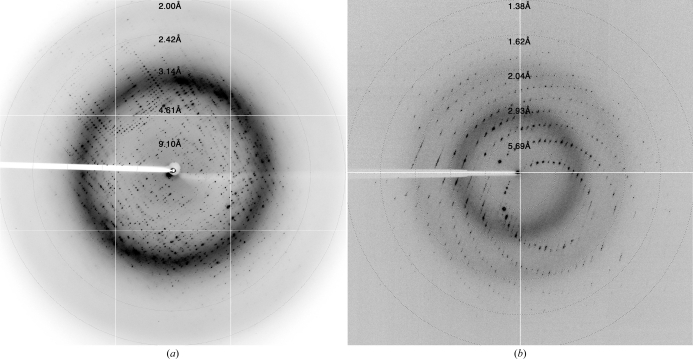
Two versions of the human Fyn SH2 domain (non-His-tagged and N-terminally His-tagged) were overexpressed and purified to high homogeneity for crystallization. Size-exclusion analysis of Fyn SH2 without a His tag suggests an apparent molecular weight of 12 kDa, which is very similar to that expected from its amino-acid sequence (12.21 kDa). The His-tagged version of Fyn SH2 was confirmed by mass-spectrometric analysis (Fig. 1 ▶). The purified His-tagged protein was stored at 277 K as a concentrated stock at 23.3 mg ml−1 in 50 mM sodium phosphate pH 6.5. Very large crystals of free Fyn SH2 grew in the storage container after a week (Fig. 2 ▶ a). These crystals deteriorated rapidly after being removed from the storage liquor. Therefore, we harvested several and directly cryocooled them in liquid N2. Several cryoprotectant solutions were tested, with the best results being obtained using 35% ethylene glycol and stepwise transfer of the crystals from the mother liquor to drops with increasing concentration of the cryoprotectant. The best crystals diffracted to approximately 2 Å resolution (Fig. 3 ▶ a) and belonged to space group C2, with unit-cell parameters a = 89.0, b = 57.9, c = 101.2 Å, β = 90.6°. Statistics of data collection are given in Table 2 ▶.
Figure 3.
Diffraction patterns of Fyn SH2: (a) unbound and (b) in complex with the phosphotyrosine peptide. The resolution at the edge of the detector is 1.62 Å for unbound Fyn SH2 and 1.11 Å for the phosphotyrosine peptide complex.
Table 2. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Fyn SH2 | Fyn SH2–pTyr peptide | |
|---|---|---|
| Beamline | PROXIMA-1 | ID14-1 |
| High-resolution pass | ||
| Distance (mm) | 292.07 | 127.78 |
| Δϕ (°) | 0.5 | 0.3 |
| Low-resolution pass | ||
| Distance (mm) | 464.73 | 320.76 |
| Δϕ (°) | 0.7 | 1.0 |
| No. of images per pass | 360 | 600 |
| Mosaicity (°) | 1.07 | 0.30 |
| Wavelength (Å) | 0.98011 | 0.9334 |
| Resolution (Å) | 48.5–1.99 (2.09–1.99) | 50.0–1.40 (1.48–1.40) |
| Completeness (%) | 98.2 (92.3) | 99.9 (100.0) |
| No. of measured reflections | 136861 (16997) | 293052 (40685) |
| No. of unique reflections | 35049 (4785) | 23431 (3304) |
| 〈I/σ(I)〉 | 7.8 (3.1) | 29.1 (8.1) |
| Multiplicity | 3.9 | 12.5 |
| Rmerge† | 0.087 (0.308) | 0.051 (0.438) |
| Rmeas‡ | 0.098 (0.361) | 0.052 (0.455) |
| Rp.i.m.§ | 0.045 (0.186) | 0.012 (0.121) |
| Wilson B factor (Å2) | 25.82 | 12.49 |
| Space group | C2 | P43212 |
| Unit-cell parameters | ||
| a (Å) | 89.0 | 39.2 |
| b (Å) | 57.9 | 39.2 |
| c (Å) | 101.2 | 145.2 |
| α (°) | 90.0 | 90.0 |
| β (°) | 90.6 | 90.0 |
| γ (°) | 90.0 | 90.0 |
R
merge = 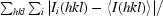
 .
.
R
meas = 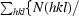
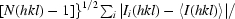
 .
.
R
p.i.m. = 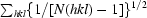
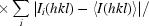
 .
.
Crystals of the Fyn SH2–phosphotyrosine peptide complex were obtained in 0.1 M MOPS/Na HEPES pH 7.5, 0.12 M monosaccharide mixture, 12.5%(w/v) PEG 1000, 12.5%(w/v) PEG 3350 and 12.5%(v/v) MPD. The crystals grew to dimensions of 0.3 × 0.2 × 0.3 mm after several weeks and could be cryocooled directly in the N2 cryostream without any additional cryoprotectant for data collection. They belonged to space group P43212, with unit-cell parameters a = b = 39.2, c = 145.2 Å. The best crystals diffracted to 1.4 Å resolution (Fig. 3 ▶ b). Statistics of data collection are given in Table 2 ▶.
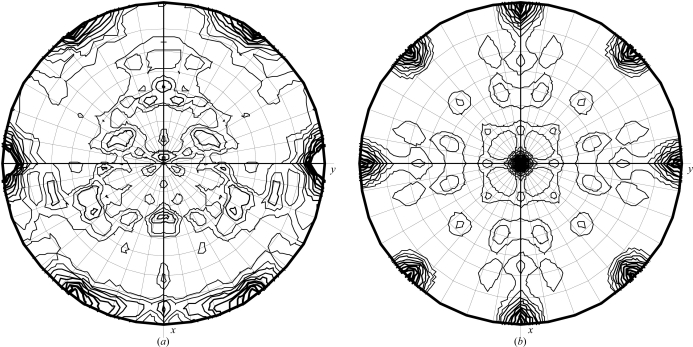
Analysis of the unit-cell contents of the crystals of free Fyn SH2 using the CCP4 program MATTHEWS_COEF suggested that the asymmetric unit contained between three (Matthews coefficient of 3.81 Å3 Da−1; 68% solvent) and five (Matthews coefficient of 2.3 Å3 Da−1; 46% solvent) Fyn SH2 units. An analysis of the self-rotation function (Fig. 4 ▶ a) showed two prominent peaks (besides the crystallographic twofold symmetry from the C2 space group) in the κ = 180° section. This agrees with the presence of four molecules in the asymmetric unit. The κ = 120° and κ = 90° sections did not show any significant peaks.
Figure 4.
κ = 180° section of the self-rotation function for (a) unbound Fyn SH2 and (b) its complex with the phosphotyrosine peptide.
Analysis of the unit-cell contents of the crystals of the Fyn SH2–peptide complex strongly suggested that the asymmetric unit contained only one Fyn SH2–peptide complex unit (Matthews coefficient of 2.15 Å3 Da−1; 43% solvent). The self-rotation function (Fig. 4 ▶ b) shows only peaks from crystallographic twofold axes, which confirms that there is indeed a single molecule in the asymmetric unit.
Determination of three-dimensional structures of SH2-containing proteins is crucial for understanding and modifying many pathways in the human interactome where these small domains play important roles as information carriers and molecular sensors. SH2 domains play central roles in mediating intracellular signalling by receptor tyrosine kinases and cytoplasmic protein tyrosine kinases. Signalling proteins such as phospholipase C-γ and Ras-GTPase (guanosine trisphosphatase) activating protein become tyrosine-phosphorylated in response to cell stimulation by epidermal growth factor or platelet-derived growth factor (Ellis et al., 1990 ▶; Schlessinger & Lemmon, 2003 ▶).
Given the importance of signalling mediated by tyrosine phosphorylation, there is great interest in strategies to define or profile the global state of tyrosine phosphorylation in the cell. Many tyrosine-phosphorylation sites exert their biological activity through binding to other proteins containing SH2 domains. Therefore, by characterizing SH2 domains we can very efficiently capture information relevant to the activation state of signalling pathways (Pawson et al., 2001 ▶; Liu et al., 2006 ▶; Machida et al., 2007 ▶; Filippakopoulos et al., 2009 ▶; Lu et al., 2010 ▶). High-resolution structural information of Fyn SH2 domain in the apo form and in complex with a phosphotyrosine peptide will extend our understanding of the mechanisms of action and functioning of SH2 domains from the Src kinase family.
Acknowledgments
The authors acknowledge the use of synchrotron beamtime at the SOLEIL synchrotron (Gif-Sur-Yvette, France) and the ESRF (Grenoble, France). We thank Jef Rozenski from Rega Institute–K. U. Leuven for measuring the mass-spectrometric data. This work was funded by the Vlaams Interuniversitair Instituut voor Biotechnologie (VIB), the Onderzoeksfonds of the Vrije Universiteit Brussel (OZR) and the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO). RH is funded through FWO grant G.0116.09N. AG-P and LB are postdoctoral fellows of the FWO. Some of the materials were paid for by FNRS Crédit au Chercheurs No. 1.5.081.10.
References
- Aleshin, A. & Finn, R. S. (2010). Neoplasia, 12, 599–607. [DOI] [PMC free article] [PubMed]
- Arold, S. T., Ulmer, T. S., Mulhern, T. D., Werner, J. M., Ladbury, J. E., Campbell, I. D. & Noble, M. E. M. (2001). J. Biol. Chem. 276, 17199–17205. [DOI] [PubMed]
- Bradshaw, J. M., Grucza, R. A., Ladbury, J. E. & Waksman, G. (1998). Biochemistry, 37, 9083–9090. [DOI] [PubMed]
- Cantley, L. C., Auger, K. R., Carpenter, C., Duckworth, B., Graziani, A., Kapeller, R. & Soltoff, S. (1991). Cell, 64, 281–302. [DOI] [PubMed]
- Dierck, K., Machida, K., Mayer, B. J. & Nollau, P. (2009). Methods Mol. Biol. 527, 131–155. [DOI] [PubMed]
- Ellis, C., Moran, M., McCormick, F. & Pawson, T. (1990). Nature (London), 343, 377–381. [DOI] [PubMed]
- Engen, J. R., Wales, T. E., Hochrein, J. M., Meyn, M. A. III, Banu Ozkan, S., Bahar, I. & Smithgall, T. E. (2008). Cell. Mol. Life Sci. 65, 3058–3073. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Filippakopoulos, P., Müller, S. & Knapp, S. (2009). Curr. Opin. Struct. Biol. 19, 643–649. [DOI] [PMC free article] [PubMed]
- Gorrec, F. (2009). J. Appl. Cryst. 42, 1035–1042. [DOI] [PMC free article] [PubMed]
- Hoover, D. M. & Lubkowski, J. (2002). Nucleic Acids Res. 30, e43. [DOI] [PMC free article] [PubMed]
- Ishizawar, R. & Parsons, S. J. (2004). Cancer Cell, 6, 209–214. [DOI] [PubMed]
- Kinoshita, T., Matsubara, M., Ishiguro, H., Okita, K. & Tada, T. (2006). Biochem. Biophys. Res. Commun. 346, 840–844. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Liu, B. A., Jablonowski, K., Raina, M., Arcé, M., Pawson, T. & Nash, P. D. (2006). Mol. Cell, 22, 851–868. [DOI] [PubMed]
- Liu, B. A., Shah, E., Jablonowski, K., Stergachis, A., Engelmann, B. & Nash, P. D. (2011). Sci. Signal. 4, ra83. [DOI] [PMC free article] [PubMed]
- Lu, X.-L., Cao, X., Liu, X. Y. & Jiao, B.-H. (2010). Curr. Med. Chem. 17, 1117–1124. [DOI] [PubMed]
- Machida, K. & Mayer, B. J. (2005). Biochim. Biophys. Acta, 1747, 1–25. [DOI] [PubMed]
- Machida, K. et al. (2007). Mol. Cell, 26, 899–915. [DOI] [PubMed]
- Morton, C. J., Pugh, D. J., Brown, E. L., Kahmann, J. D., Renzoni, D. A. & Campbell, I. D. (1996). Structure, 4, 705–714. [DOI] [PubMed]
- Mulhern, T. D., Shaw, G. L., Morton, C. J., Day, A. J. & Campbell, I. D. (1997). Structure, 5, 1313–1323. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pawson, T. (2004). Cell, 116, 191–203. [DOI] [PubMed]
- Pawson, T., Gish, G. D. & Nash, P. (2001). Trends Cell Biol. 11, 504–511. [DOI] [PubMed]
- Pintar, A., Hensmann, M., Jumel, K., Pitkeathly, M., Harding, S. E. & Campbell, I. D. (1996). Eur. Biophys. J. 24, 371–380. [DOI] [PubMed]
- Schlessinger, J. & Lemmon, M. A. (2003). Sci. STKE, 2003, re12. [DOI] [PubMed]
- Suzuki, M., Roy, R., Zheng, H., Woychik, N. & Inouye, M. (2006). J. Biol. Chem. 281, 37559–37565. [DOI] [PubMed]
- Suzuki, M., Zhang, J., Liu, M., Woychik, N. A. & Inouye, M. (2005). Mol. Cell, 18, 253–261. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wadhawan, A., Smith, C., Nicholson, R. I., Barrett-Lee, P. & Hiscox, S. (2011). Cancer Treat. Rev. 37, 234–241. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Young, M. A., Gonfloni, S., Superti-Furga, G., Roux, B. & Kuriyan, J. (2001). Cell, 105, 115–126. [DOI] [PubMed]