This parasite may be transmitted from macaques to humans.
Keywords: malaria, epidemiology, Plasmodium knowlesi, parasites, mixed-species malaria infection, Thailand, merozoite surface protein 1, small subunit ribosomal RNA, cross-transmission, humans, macaques, research
Abstract
Naturally acquired human infections with Plasmodium knowlesi are endemic to Southeast Asia. To determine the prevalence of P. knowlesi malaria in malaria-endemic areas of Thailand, we analyzed genetic characteristics of P. knowlesi circulating among naturally infected macaques and humans. This study in 2008–2009 and retrospective analysis of malaria species in human blood samples obtained in 1996 from 1 of these areas showed that P. knowlesi accounted for 0.67% and 0.48% of human malaria cases, respectively, indicating that this simian parasite is not a newly emergent human pathogen in Thailand. Sequence analysis of the complete merozoite surface protein 1 gene of P. knowlesi from 10 human and 5 macaque blood samples showed considerable genetic diversity among isolates. The sequence from 1 patient was identical with that from a pig-tailed macaque living in the same locality, suggesting cross-transmission of P. knowlesi from naturally infected macaques to humans.
Plasmodium knowlesi circulates mainly among long-tailed macaques (Macaca fascicularis) and pig-tailed macaques (M. nemestrina) that inhabit a wide area of Southeast Asia (1). Microscopy-based detection of P. knowlesi has failed because morphologic features of young trophozoites of P. knowlesi resemble those of P. falciparum and characteristic band-shaped growing trophozoites resemble those of P. malariae (2–4). To date, the effective tool for diagnosing P. knowlesi infection is PCR specific for multicopy genes, such as small subunit rRNA and mitochondrial cytochrome b (3–5).
Human infections with P. knowlesi vary by geographic location (highest prevalence in Malaysian Borneo), but individual cases have been increasingly identified in countries in Southeast Asia (6). Our large-scale molecular-based survey of malaria in Thailand during 2006–2007 showed that P. knowlesi was widely distributed at a low prevalence (in 0.57% of all malaria cases identified) in several malaria-endemic areas bordering Myanmar, Cambodia, and Malaysia (7). Correct diagnosis of malaria has a major effect on malaria control in terms of treatment outcomes, disease transmission, and interpretation of efficiency of a given control measure.
Although malaria caused by P. knowlesi is generally benign and responsive to chloroquine treatment, severe and fatal cases similar to complicated P. falciparum malaria cases have been documented (6,8). To date, it has been unknown whether human infections with P. knowlesi in Thailand were caused by a new emergence of this parasite species or whether the parasite had been circulating cryptically with other human malaria parasites. Furthermore, it would be useful to explore spatiotemporal distribution of malaria species in humans and analyze genetic characteristics of P. knowlesi circulating among naturally infected macaques and humans. These data could lead to a better understanding of malaria transmission and provide information for a more effective malaria control policy at a nationwide level. Therefore, we sought to determine the prevalence of this simian malaria in malaria-endemic regions of Thailand.
Materials and Methods
Prospective Study and Sample Collection
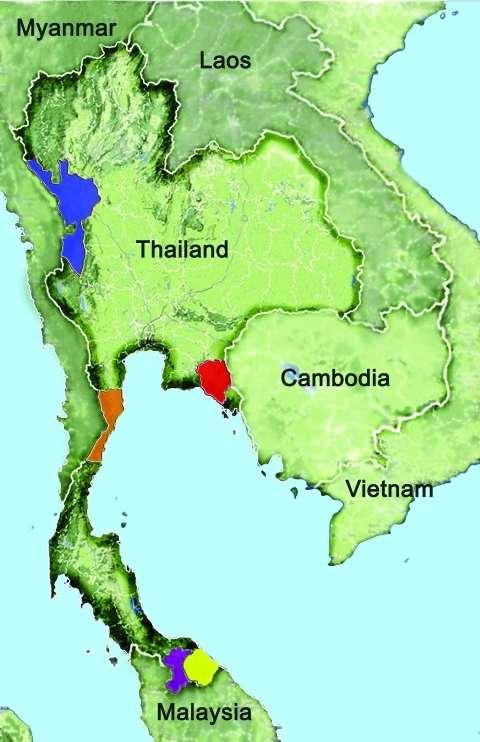
Most malaria infections in Thailand occur in forests or forest fringes along its borders with other countries, and malaria transmission exhibits a bimodal pattern that peaks in May–July and October–November (9,10). During October 2008–September 2009, venous or finger prick blood samples were obtained from 3,770 febrile persons (2,577 male and 1,193 female; mean age 27.4 years, range 1–87 years) who came to malaria clinics in northwestern (Tak Province, n = 1,354), eastern (Chantaburi Province, n = 401), and southern (Yala Province, n = 1,552, and Narathiwat Province, n = 463) Thailand (Figure 1). These 3,770 persons represented 12.4% of the 30,425 malaria cases in these areas during the study period (10). A total of 470 blood samples from these persons were negative for malaria parasites by microscopy (153 in Tak, 179 in Yala, and 138 in Narathiwat). The study was reviewed and approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University.
Figure 1.
Provinces of Thailand where blood samples were obtained and tested for malaria, 1996–2009. Tak: blue, n = 210 in 1996, n = 681 in 2006–2007, and n = 1,216 in 2008–2009; Prachuab Khirikhan: orange, n = 215 in 2006–2007; Yala: purple, n = 286 in 2006–2007 and n = 1,408 in 2008–2009; Narathiwat: yellow, n = 370 in 2006–2007 and n = 421 in 2008–2009; and Chantaburi: red, n = 261 in 2006–2007 and n = 401 in 2008–2009.
Retrospective Study
A total of 210 blood samples obtained in 1996 from microscopy-positive symptomatic malaria patients (139 male and 71 female; mean age 25.1 years, range 12–72 years) living in Tak Province were included for comparative analysis. Of these samples, 143 were obtained during May–July; the remaining 67 samples were obtained during October–December. Blood samples were preserved in EDTA and stored at −40°C until use. Results of malaria species distribution during 2006–2007 from our analysis (7) were included for comparison.
Microscopic Diagnosis
Thin and thick smears were prepared from each blood sample, stained with Giemsa solution, and examined by experienced microscopists for >200 leukocytes for a thick film and >200 microscopic fields with a 100× objective. Microscopists were blinded to clinical information and results of PCR detection.
PCR-based Diagnosis
DNA was extracted from 200 μL of blood by using a DNA Minikit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Malaria species was identified by using a nested PCR with Plasmodium genus–specific outer primers (M18SF0: 5′-CCATTAATCAAGAACGAAAGTTAAGG-3′ and M18SR0c: 5′-TGTTGTTGCCTTAAACTTCCTTG-3′) derived from small subunit rRNA for a primary PCR (7). Nested PCR was conducted in separate tubes for each pair of species-specific primers: P. falciparum, PF18SF: 5′-CATCTTTCGAGGTGACTTTTAG-3′ and PF18SRc: 5′-GAAGAGAGAAATAGAGTAAAAAAC-3′; P. vivax, PV18SF: 5′-GAATTTTCTCTTCGGAGTTTATTC-3′ and PV18SRc: 5′-GTAGAAAAGGGAAAGGGAAACTGTTA-3′; P. malariae, PM18SF: 5′-GAGACATTCATATATATGAGTG-3′ and PM18SRc: 5′-GGGAAAAGAACGTTTTTATTAAAAAAAAC-3′; P. ovale, PO18SF: 5′-AATTCCTTTCGGGGAAATTTCTTA-3′ and PO18SRc: 5′-GGGAAAAGGACACATTAATTGTAT-3′; and P. knowlesi, PK18SF: 5′-GAGTTTTTCTTTTCTCTCCGGAG-3′ and PK18SRc: 5′-GGGAAAGGAATCACATTTAACGT-3′. Thermal cycling profiles for primary and nested PCRs contained 35 and 25 cycles (94°C for 40 s, 60°C for 30 s, and 72°C for 1 min), respectively. PCR products were analyzed by 2% agarose gel electrophoresis.
Sequencing the Complete Merozoite Surface Protein 1 Gene of P. knowlesi
During December 2008–June 2009, a prospective survey of malaria in monkeys inhabiting Yala Province (n = 70) and Narathiwat Province (n = 566) showed that 5 M. nemestrina monkeys, 1 M. fascicularis monkey, and 1 Semnopithecus obscurus monkey in Narathiwat Province had P. knowlesi infections (11). The complete nucleotide sequences of the merozoite surface protein–1 gene of P. knowlesi (Pkmsp-1) from these pig-tailed macaques and humans in the current survey were obtained by direct sequencing of PCR-amplified products as described (12). Sequences have been deposited in GenBank under accession nos. JF837339–JF837353.
Data Analysis
Sequences were aligned by using ClustalX with minor manual adjustments made by visual inspection (13). Phylogenetic construction was inferred from maximum-likelihood methods by using the Hasegawa, Kishino, and Yano model with 1,000 bootstrap iterations as implemented in MEGA version 5.01 (14). Differences between numbers of malaria cases were computed by using the Mann-Whitney U test, χ2 test, or Fisher exact test. A 2-tailed p value <0.05 indicated statistical significance.
Results
Malaria Species Distribution
During October 2008–September 2009, microscopic examinations of blood samples from 3,770 febrile patients showed that 3,300 had malaria parasites and 470 did not. Most malaria cases diagnosed by microscopy were either P. falciparum (51.55%) or P. vivax (48.21%); the prevalence of co-infections with both species was 0.18%. Of 432 samples containing co-infections with P. falciparum and P. vivax detected by PCR, only 3 (0.69%) samples were correctly diagnosed by microscopy. Microscopy showed negative results for 37 (8.56%) samples and failed to detect cryptic P. falciparum for 181 (41.90%) samples and cryptic P. vivax for 211 (48.84%) samples (Table 1). Conversely, 186 of 470 microscopy-negative samples were positive by nested PCR (79 P. falciparum, 68 P. vivax, 37 P. falciparum and P. vivax, 1 co-infection with P. falciparum and P. malariae, and 1 P. knowlesi); PCR failed to detect malaria in 46 microscopy-positive samples.
Table 1. Distribution of Plasmodium species in 3,770 febrile patients, Thailand, October 2008–September 2009.
| Species detected by PCR |
Microscopy-based detection | PCR detection |
|||||
|---|---|---|---|---|---|---|---|
|
P. falciparum
|
P. vivax
|
P. malariae
|
P. ovale
|
P. falciparum and P. vivax |
Negative |
||
| P. falciparum | 1,397 | 78 | 0 | 0 | 2 | 79 | 1,556 |
| P. vivax | 52 | 1,301 | 0 | 0 | 1 | 68 | 1,422 |
| P .malariae | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| P .ovale | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P. knowlesi | 3 | 3 | 1 | 0 | 0 | 1 | 8 |
| P. falciparum and P. vivax | 211 | 181 | 0 | 0 | 3 | 37 | 432 |
| P. falciparum and P. malariae | 1 | 0 | 0 | 0 | 0 | 1 | 2 |
| P. falciparum and P. ovale | 3 | 0 | 0 | 0 | 0 | 0 | 3 |
| P. falciparum and P. knowlesi | 6 | 0 | 0 | 0 | 0 | 0 | 6 |
| P. vivax and P. malariae | 0 | 3 | 0 | 0 | 0 | 0 | 3 |
| P. vivax and P. ovale | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| P. vivax and P. knowlesi | 3 | 1 | 0 | 0 | 0 | 0 | 4 |
| P. falciparum, P. vivax, and P. knowlesi | 2 | 3 | 0 | 0 | 0 | 0 | 5 |
| Negative |
21 |
19 |
0 |
0 |
0 |
284 |
324 |
| Total | 1,701 | 1,591 | 2 | 0 | 6 | 470 | 3,770 |
A total of 3,446 blood samples contained malaria parasite DNA and showed greater differences in species distribution and mixed species infections than did microscopy (Table 1). Mixed species infections accounted for 13.26% of all PCR-positive cases and were most common in Tak Province (Table 2). Microscopy failed to detect P. knowlesi in these samples. In contrast, PCR identified 23 (0.67%) patients with P. knowlesi infections (8 monoinfections and 15 co-infections with other Plasmodium species) (Table 1).
Table 2. Distribution of Plasmodium species by malaria-endemic areas of Thailand, October 2008–September 2009*.
| Infection | Region, no. (%) |
Total no. (%) | |||
|---|---|---|---|---|---|
| Northwestern (Tak) | Eastern (Chantaburi) | Southern (Yala) | Southern (Narathiwat) | ||
| Monoinfection | 989 (81.33) | 351 (87.53) | 1,297 (92.12) | 352 (83.61) | 2,989 (86.74) |
| P. falciparum | 507 (41.69) | 27 (6.73) | 810 (57.53) | 212 (50.36) | 1,556 (45.15) |
| P. vivax | 481 (39.56) | 320 (79.80) | 485 (34.45) | 136 (32.30) | 1,422 (41.27) |
| P. malariae | 1 (0.08) | 1 (0.25) | 0 | 1 (0.24) | 3 (0.09) |
| P. ovale | 0 | 0 | 0 | 0 | 0 |
|
P. knowlesi
|
0 |
3 (0.75) |
2 (0.14) |
3 (0.71) |
8 (0.23) |
| Co-infection | 227 (18.67) | 50 (12.47) | 111 (7.88) | 69 (16.39) | 457 (13.26) |
| P. falciparum and P. vivax | 217 (17.85) | 46 (11.47) | 105 (7.46) | 64 (15.20) | 432 (12.54) |
| P. falciparum and P. malariae | 1 (0.08) | 0 | 0 | 1 (0.24) | 2 (0.06) |
| P. falciparum and P. ovale | 2 (0.16) | 0 | 1 (0.07) | 0 | 3 (0.09) |
| P. falciparum and P. knowlesi | 2 (0.16) | 0 | 3 (0.21) | 1 (0.24) | 6 (0.17) |
| P. vivax and P. malariae | 1 (0.08) | 0 | 2 (0.14) | 0 | 3 (0.09) |
| P. vivax and P. ovale | 1 (0.08) | 0 | 0 | 1 (0.24) | 2 (0.06) |
| P. vivax and P. knowlesi | 2 (0.16) | 1 (0.25) | 0 | 1 (0.24) | 4 (0.12) |
|
P. falciparum, P. vivax, and P. knowlesi |
1 (0.08) |
3 (0.75) |
0 |
1 (0.24) |
5 (0.15) |
| Total | 1,216 | 401 | 1,408 | 421 | 3,446 |
*Data from nested PCR. Percentages are of the total number in that region or infection category.
P. knowlesi in Blood Samples Obtained in 1996
PCR analysis of 210 blood samples from microscopy-positive patients in Tak Province obtained during 1996 identified 55 and 96 patients as having monoinfections with P. falciparum (26.19%) and P. vivax (45.71%), respectively. The remaining 59 patients (28.10%) had mixed species infections with P. falciparum and P. vivax (n = 50), P. vivax and P. malariae (n = 7), P. vivax and P. knowlesi (n = 1), and P. falciparum, P. vivax, and P. malariae (n = 1). These findings indicate that P. knowlesi has circulated among humans in Thailand for at least 12–13 years.
Spatial Variation
The distribution of P. falciparum and P. vivax displayed spatial variation in Thailand. PCR assays showed that P. falciparum and P. vivax contributed almost equally to malaria cases in Tak Province during 2008–2009 (a total of 50.55% and 48.68%, respectively), P. vivax was more prevalent than P. falciparum in Chantaburi Province, and P. falciparum was more prevalent than P. vivax in Yala and Narathiwat Provinces. Despite a low overall prevalence of the remaining malaria species (<2%), P. knowlesi could be detected more often than P. malariae and P. ovale in these malaria-endemic areas (Table 3).
Table 3. Temporal variation in distribution of Plasmodium species by malaria-endemic region of Thailand, 2006–2009*.
| Species | % Distribution |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Northwestern (Tak) |
Eastern (Chantaburi) |
Southern (Yala) |
Southern (Narathiwat) |
||||||||
| 2006–2007, n = 681 | 2008–2009, n = 1,216 | 2006–2007, n = 261 | 2008–2009, n = 401 | 2006–2007, n = 286 | 2008–2009, n = 1,408 | 2006–2007, n = 370 | 2008–2009, n = 421 | ||||
| P. falciparum | 44.54 | 50.55 | 8.42 | 16.74 | 57.14 | 60.50 | 23.31 | 56.82 | |||
| P. vivax | 52.63 | 48.68 | 91.03 | 81.50 | 41.86 | 38.97 | 76.17 | 41.34 | |||
| P. malariae | 1.16 | 0.21 | 0 | 0.22 | 0 | 0.13 | 0 | 0.41 | |||
| P. ovale | 1.41 | 0.21 | 0.27 | 0 | 0 | 0.07 | 0 | 0.20 | |||
| P. knowlesi | 0.26 | 0.35 | 0.27 | 1.54 | 1.00 | 0.33 | 0.52 | 1.22 | |||
*Data from nested PCR. Temporal variation in distribution in 1996 (n = 210) was 39.26 for Plasmodium falciparum, 57.41 for P. vivax, 2.96 for P. malariae, 0 for P. ovale, and 0.37 for P. knowlesi.
Temporal Variation
Distribution of P. falciparum, P. vivax, P. malariae, and P. ovale in Thailand among samples obtained during 2006–2007 and 2008–2009 exhibited significant temporal variation (χ2 99.9, 92.2, 24.6, and 20.5, respectively; all p values <0.0001). Conversely, distribution of P. knowlesi remained stable over these periods (χ2 0.17, p = 0.68). If one considers that samples from 3 periods (1999, 2006–2007, and 2008–2009) were available for comparison only for Tak Province, the contribution of P. knowlesi to malaria cases was not different in all pairwise comparisons, which suggested stable prevalence of this simian malaria for more than a decade in this malaria-endemic area. In contrast, a significant difference was found in the distribution of P. falciparum, P. vivax, and P. malariae obtained during the 3 periods in each malaria-endemic area (p<0.05 for all pairwise comparisons), and P. ovale was not detected in samples obtained during 1996 but comprised 1.41% and 0.21% of samples obtained during 2006–2007 and 2008–2009, respectively (Table 3).
Prevalence of mixed species infections showed regional and seasonal variations characterized by a significantly higher prevalence during the dry season than in the rainy season in isolates from Chantaburi Province (χ2 5.94, p = 0.015; odds ratio [OR] 2.59, 95% confidence interval [CI] 1.20–5.60). The opposite result was observed for isolates from Yala Province (χ2 101.7, p<0.001; OR 7.2, 95% CI 4.65–10.90). No significant seasonal difference in prevalence of mixed species infections occurred in isolates from Tak Province (χ2 1.09, p = 0.60; OR 1.09, 95% CI 0.81–1.46) and Narathiwat Province (χ2 0.10, p = 0.76; OR 1.09, 95% CI 0.64–1.87).
Characteristics of P. knowlesi–infected Patients
Of 5,254 PCR-positive samples obtained during 1996, 2006–2007, and 2008–2009, P. knowlesi was detected in 34 patients (0.65%) (Table 2, Table 3) (7). All patients infected with P. knowlesi, including the first case-patient identified in 2000 (n = 35), had uncomplicated malaria symptoms (3,7). Age range of P. knowlesi–infected patients was 4–59 years (mean 30 years, mode 19 years, median 33.5 years), and most (73.50%) cases occurred in persons 16–45 years of age. P. knowlesi malaria was diagnosed in male patients ≈2× more often than in female patients (M:F ratio 2.18:1), which was similar to the sex distribution of total malaria cases (M:F ratio 2.16:1). Thus, no sex preference was observed for P. knowlesi (p = 0.97).
Almost half of the P. knowlesi–infected malaria patients acquired infections in southern Thailand near Malaysia. Monoinfections with P. knowlesi were observed in 10 patients whose parasite densities ranged from 0 to 145,000 parasites/μL. The remaining 24 patients had co-infections with P. falciparum (n = 16) or P. vivax (n = 9) or triple infections with P. falciparum and P. vivax (n = 5). Although the geometric mean parasite density for P. knowlesi moninfections was higher than that for mixed species infections, no significant difference was observed (Table 4).
Table 4. Parasite densities of patients with Plasmodium knowlesi monoinfection and co-infection with other malaria species, Thailand, October 2008–September 2009*.
| Category | Parasite density, parasites/μL† |
||
|---|---|---|---|
| Geometric mean | Ratio of geometric means‡ | Range | |
|
P. knowlesi monoinfection, n = 10 |
4,165 |
NA |
0–145,000 |
| P. knowlesi co-infection with | |||
| P. falciparum, n = 11 | 1,632 | 2.55 | 440–6,560 |
| P. vivax, n = 9 | 1,686 | 2.47 | 320–13,120 |
|
P. falciparum and P. vivax, n = 5 |
487 |
8.55 |
320–1,520 |
| All co-infections, n = 25 | 1,271 | 3.28 | 320–13,120 |
*NA, not applicable. †Parasite densities between categories were not significantly different (p>0.05, by Mann-Whitney U test). ‡Ratio of parasite densities of monoinfection to co-infection.
Initial microscopic diagnosis identified P. malariae for patients with moninfections and P. falciparum or P. vivax for patients with mixed species infections. Approximately two thirds (23/35) of P. knowlesi malaria occurred in the rainy season. However, the ratio of P. knowlesi–infected patients to total malaria patients for the rainy and dry seasons was not significantly different (χ2 0.25, p = 0.62; OR 1.20, 95% CI 0.60–2.38). Most (73.50%) human infections with P. knowlesi occurred in areas with macaques living nearby.
Msp-1 Gene Sequence of P. knowlesi
Fifteen complete Pkmsp-1 gene sequences were analyzed for 10 isolates from humans and 5 isolates from pig-tailed macaques. These human isolates were obtained from 8 patients in the current survey (3 from Narathiwat Province, 3 from Chantaburi Province, and 2 from Yala Province) and from 2 patients from Prachuab Khirikhan Province obtained during 2006–2007 (7). The monkey isolates were obtained from 5 naturally infected pig-tailed macaques from Narathiwat Province during the same period as the current malaria survey in humans (11). Each isolate had single Pkmsp-1 gene sequences because no superimposed results were observed in electropherograms. Size variation (range 5,430–5,613 bp) was observed among Pkmsp-1 genes of these isolates.
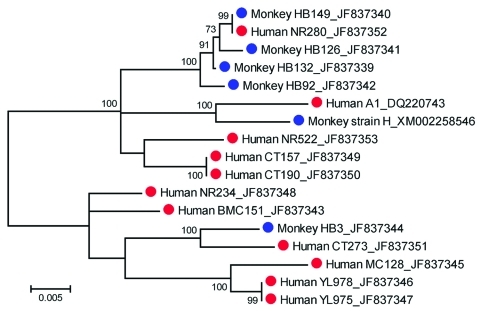
Sequence analysis identified conserved and variable domains in Pkmsp-1 genes, similar to those found in the msp-1 gene of P. vivax (15). Nucleotide substitutions in conserved regions showed a dimorphic pattern. Phylogenetic analysis showed that human and macaque isolates had genetic diversity in the Pkmsp-1 gene that could be located in 2 clusters with 100% bootstrap support, which confirmed dimorphism of this gene. One cluster contained 6 human isolates (BMC151, CT273, MC128, NR234, YL975, and YL978) and a monkey isolate from Narathiwat Province (isolate HB3), and the remaining isolates belonged to the other cluster (Figure 2). The Pkmsp-1 gene sequence from 1 patient (NR280) in Narathiwat Province was identical with that isolated from a pig-tailed macaque (HB149) living in the same locality. Furthermore, isolates from 2 patients (YL975 and YL978) who lived in Yala Province had identical Pkmsp-1 gene sequences. The patients were concurrently infected with P. knowlesi in the same malaria-endemic areas. The other identical sequences were found in isolates from 2 patients (CT157 and CT190) in Chantaburi Province who had onset of febrile illness a few days apart (Figure 2).
Figure 2.
Maximum-likelihood tree inferred from the complete merozoite surface protein 1 gene sequences of Plasmodium knowlesi from humans (red circles) and macaques (blue circles). The tree is drawn to scale, and branch lengths are measured in number of substitutions per site by using MEGA version 5.01 (14). Bootstrap values >50% from 1,000 iterations are shown. Human isolates are from the following provinces: Narathiwat (NR280, NR234, and NR522); Yala (YL975 and YL978); Chantaburi (CT157, CT190, and CT273); and Prachuab Khirikhan (BMC151, MC128, and DQ220743). Isolates HB3, HB92, HB126, HB132, and HB149 are from macaques in Narathiwat Province. GenBank accession nos. are shown after isolate names.
Discussion
Our PCR analysis identified spatiotemporal heterogeneity in the prevalence of P. falciparum and P. vivax, which is consistent with that of previous analyses by microscopy-based detection (16,17). Prevalence of P. falciparum and P. vivax in adjacent Yala and Narathiwat Provinces also showed a spatial difference in species distribution during 2006–2007 but no difference during 2008–2009. These results supported a spatiotemporal pattern on a microgeographic level as reported for other malaria-endemic areas in Thailand (16). Furthermore, the prevalence of P. malariae and P. ovale varied according to location and time.
Cross-border migration of infected persons, predominantly along the Thailand–Myanmar and Thailand–Cambodia borders, could contribute spatial and temporal variation in the prevalence of these malaria parasite species (16,17). However, migration per se could not explain similar findings in Yala and Narathiwat Provinces because almost all malaria cases in these areas were autochthonous (10). A dramatic increase in P. vivax prevalence in malaria-endemic areas bordering Cambodia during the past decade has reportedly been caused by relative changes in species distribution of local vectors and differences in vector capacity (18–20). Furthermore, intrinsic differences in parasite biology such as hypnozoites in P. vivax and P. ovale could contribute to additional episodes of infections if these infections are not radically treated. Therefore, spatiotemporal variation in these human cases of malaria in Thailand likely results from multiple variable factors.
Our previous survey in 2006–2007 showed that co-infections with different malaria parasite species displayed spatial variation, characterized by a high prevalence (≈23%–24%) along the Thailand–Myanmar border that contrasts with a low prevalence of 3% and 5% in malaria-endemic areas bordering Cambodia and Malaysia, respectively (7). Our current survey has detected different prevalences of mixed species infections than during 2006–2007 in these same malaria-endemic areas that could have been caused by different prevalences of each malaria parasite species.
Microscopy usually did not detect cryptic P. falciparum or cryptic P. vivax in samples containing both species identified by PCR (41.90% and 48.84%, respectively). When 1 of these 2 species coexisted with other malaria species (including P. knowlesi), only P. falciparum or P. vivax infection was diagnosed by microscopy. Failure to diagnose mixed species infections could result in repeated diagnosis and treatment, economic losses, misinterpretation of drug or vaccine efficacies, and inadequate control policies (21).
Co-infections with P. falciparum and P. vivax could be advantageous to hosts in terms of reduced disease severity (22), less chance for gametocyte carriage (23), and decreased parasite density (24). However, detrimental outcomes have been observed in other studies (25–27). Although parasite densities of P. knowlesi monoinfections and co-infections with other species were not different, the geometric mean of monoinfections was 2× that of co-infections (Table 4). Therefore, interference by a high prevalence of mixed species infections (≈14% for the current survey) and cross-species immunity may contribute to the low prevalence of P. knowlesi in Thailand. In contrast, mixed species infections were less prevalent in Sarawak, where most (≈91%) malaria patients had P. knowlesi monoinfections (4,6).
No difference in temporal and spatial distribution of P. knowlesi was found in the study areas, which suggested that transmission patterns could be different from those for human malarias caused by other parasites. The natural vectors for P. knowlesi in the Malay Peninsula are Anopheles cracens and An. latens mosquitoes, which are members of the Leucosphyrus group (28,29). The main vectors for human malaria in Thailand are An. minimus, An. maculates, and An. dirus mosquitoes (30). Although An. dirus mosquitoes also belong to the Leucosphyrus group and have been identified as potential vectors for P. knowlesi in Vietnam (31), this vector species has drastically decreased in abundance in all major malaria-endemic areas of Thailand during the past decade (C. Putaporntip et al., unpub. data), although feeding patterns of these mosquitoes are zoonotic rather than anthropophilic in certain areas (32). Therefore, transmission of P. knowlesi to humans could be limited and distinct from transmission by other species that cause malaria in humans in Thailand. Conversely, identification of P. knowlesi cryptically circulating among malaria patients in Tak Province obtained during 1996 has implied its occurrence at least 12–13 years ago in Thailand and a relatively stable prevalence. Likewise, recent analysis of archive blood samples for patients in Sarawak obtained more than a decade ago has supported the suggestion that P. knowlesi is not a newly emergent zoonotic malaria species in humans (33).
Our recent survey of simian malaria in long-tailed and pig-tailed macaques in Thailand (n = 754) showed that P. knowlesi had a prevalence of 5.6% and 2.3%, respectively (11). Sequence analysis of Pkmsp-1 genes from 5 P. knowlesi–infected macaques living near infected humans has shown that all parasites had unique sequences, which reaffirmed genetic heterogeneity of P. knowlesi in its natural hosts in Thailand (7). P. knowlesi isolated from a patient in Narathiwat Province shared an identical Pkmsp-1 gene sequence with that from a pig-tailed macaque living nearby, which suggested that P. knowlesi could be transmitted from macaques to humans and vice versa through anopheline vectors. However, the higher prevalence of P. knowlesi in macaques than in humans and the chronic course of parasitemia in asymptomatic macaques could increase the likelihood of transmission from macaques to humans. Furthermore, the acute clinical course of malaria infection in humans would be rapidly eliminated by antimalarial treatment and result in a lower likelihood of additional transmission of P. knowesi gametocytes to vectors (1,7).
Identical Pkmsp-1 gene sequences found in 2 patients in Chantaburi Province who lived in the same area and had symptomatic malaria during the same week suggests that both patients acquired the infections from a common source. Alternatively, parasites harboring this identical Pkmsp-1 allele could predominate among infected macaques in the region. Unfortunately, no extensive survey of malaria in wild macaques in Chantaburi Province was performed during the study. Despite limited number of samples in our study, Pkmsp-1 gene sequences from humans in eastern and southern Thailand were found in both clusters of the phylogenetic tree, which suggested that both dimorphic types are widely distributed in Thailand. Whether both dimorphic types of Pkmsp-1 genes from infected persons co-exist in other malaria-endemic regions of Southeast Asia remains to be investigated.
A spectrum of clinical manifestations has been observed in persons infected with P. knowlesi (6,34). However, most patients with P. knowlesi malaria had febrile illness and associated symptoms that were indistinguishable from those caused by other malaria species (2,3). P. knowlesi requires 24 hours to complete its asexual erythrocytic cycle, which results in a unique quotidian type of fever pattern different from that for the other 4 human malaria species (1). However, such paroxysms have no practical value for presumptive diagnosis because characteristic fever patterns would not be observed during early phase of infections, and mixed species infections could further complicate febrile symptoms (35). Furthermore, P. knowlesi malaria is responsive to chloroquine treatment and would likely be responsive to other antimalarial drugs (4). Therefore, reappearance of P. knowlesi among patients with mixed infections after initial treatment for infection with another malaria species in Thailand seems less likely. P. knowlesi infections in humans caused severe symptoms and deaths in 6.5% and 1.8%, respectively, in the Kapit population in Sarawak where it comprised 70% of all malaria cases (4). However, if P. knowlesi malaria had a high prevalence of complicated symptoms, the probability of observing such severe and fatal consequences from P. knowlesi infections among patients in Thailand would be low.
In conclusion, human malaria caused by P. knowlesi has occurred in Thailand for more than a decade. Despite variations in the prevalence of all 4 human malaria species, P. knowlesi has shown stable prevalence rates, which suggests different transmission cycles. Human infections with P. knowlesi in Thailand could result from macaques, as shown by identical Pkmsp-1 gene sequences of human and macaque origins.
Acknowledgments
We thank all patients who providing blood samples for this survey and the staff of the Bureau of Vector Borne Disease, Department of Disease Control, Ministry of Public Health, Thailand, for assistance in field work.
This study was supported by the National Research Council of Thailand (grant for fiscal years 2008–2009) and CU-Cluster-Emerging Fund from Chulalongkorn University (grant no. H-6-66-53).
Biography
Dr Jongwutiwes is a molecular parasitologist and physician at Chulalongkorn University, Bangkok, Thailand. His research interest is molecular characterization of medically relevant protozoa and helminths.
Footnotes
Suggested citation for this article: Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, Sirichaisinthop J, et al. Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg Infect Dis [serial on the Internet]. 2011 Oct [date cited]. http://dx.doi.org/10.3201/eid1710.110349
References
- 1.Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias [CD-ROM]. Version 1.0 (originally published in 1971). Atlanta: Centers for Disease Control and Prevention; 2003.
- 2.Chin W, Contacos PG, Coatney GR, Kimball HR. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science. 1965;149:865. 10.1126/science.149.3686.865 [DOI] [PubMed] [Google Scholar]
- 3.Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. 2004;10:2211–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24. 10.1016/S0140-6736(04)15836-4 [DOI] [PubMed] [Google Scholar]
- 5.Putaporntip C, Buppan P, Jongwutiwes S. Improved performance with saliva and urine as alternative DNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays. Clin Microbiol Infect. 2011; [Epub ahead of print]. [DOI] [PubMed]
- 6.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–71. 10.1086/524888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, Cui L, et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis. 2009;199:1143–50. 10.1086/597414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daneshvar C, Davis TM, Cox-Singh J, Rafa’ee MZ, Zakaria SK, Divis PC, et al. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis. 2009;49:852–60. 10.1086/605439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thimasarn K, Jatapadma S, Vijaykadga S, Sirichaisinthop J, Wongsrichanalai C. Epidemiology of malaria in Thailand. J Travel Med. 1995;2:59–65. 10.1111/j.1708-8305.1995.tb00627.x [DOI] [PubMed] [Google Scholar]
- 10.Annual statistics, Division of Vector-Borne Diseases, Ministry of Public Health, Thailand [cited 2011 Feb 14]. www.thaivbd.org/cms/index.php?option=com_frontpage&Itemid=1
- 11.Putaporntip C, Jongwutiwes S, Thongaree S, Seethamchai S, Grynberg P, Hughes AL. Ecology of malaria parasites infecting Southeast Asian macaques: evidence from cytochrome b sequences. Mol Ecol. 2010;19:3466–76. 10.1111/j.1365-294X.2010.04756.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putaporntip C, Jongwutiwes S, Iwasaki T, Kanbara H, Hughes AL. Ancient common ancestry of the merozoite surface protein 1 of Plasmodium vivax as inferred from its homologue in Plasmodium knowlesi. Mol Biochem Parasitol. 2006;146:105–8. 10.1016/j.molbiopara.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 15.Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho WG, Kaneko A, et al. Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc Natl Acad Sci U S A. 2002;99:16348–53. 10.1073/pnas.252348999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou G, Sirichaisinthop J, Sattabongkot J, Jones J, Bjørnstad ON, Yan G, et al. Spatio-temporal distribution of Plasmodium falciparum and P. vivax malaria in Thailand. Am J Trop Med Hyg. 2005;72:256–62. [PubMed] [Google Scholar]
- 17.Childs DZ, Cattadori IM, Suwonkerd W, Prajakwong S, Boots M. Spatiotemporal patterns of malaria incidence in northern Thailand. Trans R Soc Trop Med Hyg. 2006;100:623–31. 10.1016/j.trstmh.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 18.Sattabongkot J, Tsuboi T, Zollner GE, Sirichaisinthop J, Cui L. Plasmodium vivax transmission: chances for control? Trends Parasitol. 2004;20:192–8. 10.1016/j.pt.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 19.Somboon P, Lines J, Aramrattana A, Chitprarop U, Prajakwong S, Khamboonruang C. Entomological evaluation of community-wide use of lambdacyhalothrin-impregnated bed nets against malaria in a border area of north-west Thailand. Trans R Soc Trop Med Hyg. 1995;89:248–54. 10.1016/0035-9203(95)90525-1 [DOI] [PubMed] [Google Scholar]
- 20.Apiwathnasor C, Prommongkol S, Samung Y, Limrat D, Rojruthai B. Potential for Anopheles campestris (Diptera: Culicidae) to transmit malaria parasites in Pa Rai subdistrict (Aranyaprathet, Sa Kaeo Province), Thailand. J Med Entomol. 2002;39:583–6. 10.1603/0022-2585-39.4.583 [DOI] [PubMed] [Google Scholar]
- 21.Mayxay M, Pukrittayakamee S, Newton PN, White NJ. Mixed-species malaria infections in humans. Trends Parasitol. 2004;20:233–40. 10.1016/j.pt.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 22.McKenzie FE, Bossert WH. Mixed-species Plasmodium infections of humans. J Parasitol. 1997;83:593–600. 10.2307/3284229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price RN, Simpson JA, Nosten F, Luxemburger C, Hkirjaroen L, ter Kuile F, et al. Factors contributing to anemia in uncomplicated falciparum malaria. Am J Trop Med Hyg. 2001;65:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason DP, McKenzie FE. Blood-stage dynamics and clinical implications of mixed Plasmodium vivax–Plasmodium falciparum infections. Am J Trop Med Hyg. 1999;61:367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinathan VP, Subramanian AR. Pernicious syndromes in Plasmodium infections. Med J Aust. 1982;2:568–72. [PubMed] [Google Scholar]
- 26.Mayxay M, Pukritrayakamee S, Chotivanich K, Imwong M, Looareesuwan S, White NJ. Identification of cryptic coinfection with Plasmodium falciparum in patients presenting with vivax malaria. Am J Trop Med Hyg. 2001;65:588–92. [DOI] [PubMed] [Google Scholar]
- 27.May J, Falusi AG, Mockenhaupt FP, Ademowo OG, Olumese PE, Bienzle U, et al. Impact of subpatent multi-species and multi-clonal plasmodial infections on anaemia in children from Nigeria. Trans R Soc Trop Med Hyg. 2000;94:399–403. 10.1016/S0035-9203(00)90119-6 [DOI] [PubMed] [Google Scholar]
- 28.Vythilingam I, Noorazian YM, Huat TC, Jiram AI, Yusri YM, Azahari AH, et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit Vectors. 2008;1:26. 10.1186/1756-3305-1-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vythilingam I, Tan CH, Asmad M, Chan ST, Lee KS, Singh B. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans R Soc Trop Med Hyg. 2006;100:1087–8. 10.1016/j.trstmh.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 30.Chareonviriyaphap T, Bangs MJ, Ratanatham S. Status of malaria in Thailand. Southeast Asian J Trop Med Public Health. 2000;31:225–37. [PubMed] [Google Scholar]
- 31.Nakazawa S, Marchand RP, Quang NT, Culleton R, Manh ND, Maeno Y. Anopheles dirus co-infection with human and monkey malaria parasites in Vietnam. Int J Parasitol. 2009;39:1533–7. 10.1016/j.ijpara.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 32.Sungvornyothin S, Kongmee M, Muenvorn V, Polsomboon S, Bangs MJ, Prabaripai A, et al. Seasonal abundance and blood-feeding activity of Anopheles dirus sensu lato in western Thailand. J Am Mosq Control Assoc. 2009;25:425–30. 10.2987/09-5907.1 [DOI] [PubMed] [Google Scholar]
- 33.Lee KS, Cox-Singh J, Brooke G, Matusop A, Singh B. Plasmodium knowlesi from archival blood films: Further evidence that human infections are widely distributed and not newly emergent in Malaysian Borneo. Int J Parasitol. 2009;39:1125–8. 10.1016/j.ijpara.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van den Eede P, Van HN, Van Overmeir C, Vythilingam I, Duc TN, Hung le X, et al. Human Plasmodium knowlesi infections in young children in central Vietnam. Malar J. 2009;8:249. 10.1186/1475-2875-8-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenzie FE, Smith DL, O’Meara WP, Forney JR, Magill AJ, Permpanich B, et al. Fever in patients with mixed-species malaria. Clin Infect Dis. 2006;42:1713–8. 10.1086/504330 [DOI] [PMC free article] [PubMed] [Google Scholar]