Abstract
Self-incompatibility has been considered by geneticists a model system for reproductive biology and balancing selection, but our understanding of the genetic basis and evolution of this molecular lock-and-key system has remained limited by the extreme level of sequence divergence among haplotypes, resulting in a lack of appropriate genomic sequences. In this study, we report and analyze the full sequence of eleven distinct haplotypes of the self-incompatibility locus (S-locus) in two closely related Arabidopsis species, obtained from individual BAC libraries. We use this extensive dataset to highlight sharply contrasted patterns of molecular evolution of each of the two genes controlling self-incompatibility themselves, as well as of the genomic region surrounding them. We find strong collinearity of the flanking regions among haplotypes on each side of the S-locus together with high levels of sequence similarity. In contrast, the S-locus region itself shows spectacularly deep gene genealogies, high variability in size and gene organization, as well as complete absence of sequence similarity in intergenic sequences and striking accumulation of transposable elements. Of particular interest, we demonstrate that dominant and recessive S-haplotypes experience sharply contrasted patterns of molecular evolution. Indeed, dominant haplotypes exhibit larger size and a much higher density of transposable elements, being matched only by that in the centromere. Overall, these properties highlight that the S-locus presents many striking similarities with other regions involved in the determination of mating-types, such as sex chromosomes in animals or in plants, or the mating-type locus in fungi and green algae.
Author Summary
Self-incompatibility is a common genetic system preventing selfing through recognition and rejection of self-pollen in hermaphroditic flowering plants. In the Brassicaceae family, this system is controlled by a single genomic region, called the S-locus, where many distinct specificities segregate in natural populations. In this study, we obtained genomic sequences comprising the S-locus in two closely related Brassicaceae species, Arabidopsis lyrata and A. halleri, and analyzed their diversity and patterns of molecular evolution. We report compelling evidence that the S-locus presents many similar properties with other genomic regions involved in the determination of mating-types in mammals, insects, plants, or fungi. In particular, in spite of their diversity, these genomic regions all show absence of similarity in intergenic sequences, large depth of genealogies, highly divergent organization, and accumulation of transposable elements. Moreover, some of these features were found to vary according to dominance of the S-locus specificities, suggesting that dominance/recessivity interactions are key drivers of the evolution of this genomic region.
Introduction
Sexual reproduction entails the combination of genetic material from different individuals to produce offspring. Yet in many species mating is not entirely random, being only possible between individuals with either distinct sexes or distinct mating-types [1]. Sexes or mating-types are typically determined by very distinctive genomic tracts known as sex chromosomes in animals [2], [3] and plants [4], [5], sex-determining loci in honeybees [6], mating-type loci in green algae [7], [8] and fungi [9]–[12] or self-incompatibility (SI) loci in plants [1]. In spite of the wide diversity of organisms and types of molecular and genetic systems involved, these genomic regions typically share several common features. In particular, the genes that directly determine the sexes or the mating-types are often tightly linked, sometimes with a large genomic region containing many genes, in which recombination is suppressed. Such regions can include most of a chromosome (e.g. the male-determining region of mammalian Y chromosomes). Recombination suppression in these genomic regions is typically accompanied by a variety of degeneration signatures [13], [10], [2], [14] such as low efficacy of natural selection, low gene density and accumulation of repeated DNA such as transposable elements (TEs).
At present, a comprehensive understanding of the forces driving evolution of these genomic regions is still missing [15]. In particular, two sets of issues remain unanswered. First, the process by which recombination is suppressed and the shape of the transition between recombining and non-recombining regions is not known. In sex chromosomes of mammals and those of the plant Silene latifolia, the level of X-Y divergence increases with increasing distance from the boundary with the recombining (pseudo-autosomal) region. Recombination suppression is therefore thought to have occurred in successive and discrete steps [3], [14], [16]–[20], possibly involving large chromosomal inversions. Second, the factors determining the size of the non-recombining region remain poorly understood. In mammals, the size of the Y chromosome is 37% that of the X [3], [14], while in Silene latifolia it is 150% that of the X [5].
Homomorphic self-incompatibility (SI) is a highly relevant genetic system to address these issues. SI functions to prevent self-fertilization in hermaphroditic plants [21]. While relatively widespread (being present in at least 94 flowering plant families [22]), homomorphic SI has been described at the molecular level in only a handful of taxa (reviewed in [23], [24]). The genetics of SI involves a single genomic region or a small number of regions. All of the few incompatibility loci that have been characterized at the molecular level contain at least two genes, one expressed in pistils and the other in anthers for sporophytic SI; in gametophytic SI systems, the pollen-S gene is expressed in pollen, and there are sometimes multiple genes [25]. These genes encode proteins that physically interact in a haplotype-specific manner, ultimately allowing normal cross-pollen germination and/or growth when proteins are produced by haplotypes carrying different specificities, but preventing it when pollen and pistils express cognate specificities, in particular avoiding self-fertilization.
Evolutionary properties of the genes controlling SI have been studied in several taxa, including the Brassicaceae, Solanaceae and Papaveraceae species [26], [27]. In accordance with negative frequency-dependent selection theory [28], these genes show remarkable evolutionary features. First, the S-locus typically has very high haplotype diversity, with up to >100 distinct specificities in natural populations within species (see [29] for a review). Second, because they are maintained within species for extended periods of time, these haplotypes show high nucleotide divergence among specificities within species [30] and trans-specific polymorphism between closely related species [31]. Third, to maintain specific recognition, the pollen and pistil genes are expected to be in strong linkage disequilibrium and hence to constitute co-adapted haplotypic combinations [32]. Indeed, recombination between the two component genes would disrupt specific recognition, leading to self-compatible haplotypes [33], [34]. Several studies in different SI systems confirmed that recombination among haplotypes in the S-locus is highly infrequent [35], [33], [36], [34], [37], [30], and consequently that pollen and pistil genes are expected to follow the same evolutionary history. Fourth, in species whose SI system is sporophytic [21], complex dominance relationships have been described among S-haplotypes controlling both pollen and pistil phenotypes [38]. Sporophytic SI has been described at the molecular level in a single family, the Brassicaceae. In both Brassica and Arabidopsis, the dominance relationships among haplotypes are partly related to their phylogenetic distance, with roughly four different classes in A. lyrata, corresponding to four phylogenetic groups [39] and two dominance classes in Brassica corresponding to two phylogenetic groups [40], [41]. In line with theoretical expectations [42], [43], dominant and recessive S-haplotypes appear to experience contrasted evolutionary dynamics [30]. In particular, recessive haplotypes generally occur at higher frequency and may form homozygotes. Since molecular polymorphism has been reported among gene copies within a given S-allele [30], homozygote combinations may allow recombination between these highly similar genes copies.
Because of linkage to the targets of negative frequency-dependent selection, the surrounding genomic region is also expected to show deeper coalescence than the genomic background, and hence high sequence divergence among haplotypes [44]. The physical extent of this genomic region is potentially large, in inverse proportion to the extent of local recombination restriction within the S-locus. Analysis of the S-locus in different species belonging to different SI systems confirmed that this genomic region is indeed highly heteromorphic in terms of sequence similarity among haplotypes [45]–[49]. However detailed analyses of the patterns of molecular evolution in the S-locus region are lacking because full sequences of the region are available for just a handful of haplotypes and for a few taxa belonging to different SI systems. In the best documented SI system, that of the Brassicaceae, twelve S-haplotypes have been sequenced in the cultivated species of the Brassica genus [50]–[53], [46], [54], [55]. However, many of these sequences lack the flanking regions, hence preventing comparative analysis. In addition, three haplotypes of the S-locus were sequenced in A. thaliana, one of which is a recombinant haplotype between two of the three main haplogroups currently segregating in the species [56], [57], [49]. However, although the breakdown of SI is arguably recent in A. thaliana [58], the three available sequences encode non-functional haplotypes and may have decayed substantially, especially in light of the rapid genomic changes that occurred since the split with A. lyrata [59]. Only five haplotypes from natural populations have been sequenced in Brassicaceae with functional SI, all from A. lyrata [60]–[62]. Additionally, two haplotypes with truncated SCR sequence, consequently carrying non-functional specificities, were also reported and sequenced in this species [62].
Here, we obtained full sequences for a sample of 11 S-haplotypes from natural populations of A. halleri and A. lyrata, distributed across the four phylogenetic classes described in these species. We first used these data to determine accurately the boundaries of the non-recombining S-locus region and evaluated its extent, by studying the breakdown of sequence similarity and changes in inter-haplotype phylogenetic patterns at the interface between the flanking regions and the S-locus. We then investigated patterns of variation among haplotypes in the genomic distance between SCR and SRK, in their relative orientation and in the occurrence of additional ORFs or pseudogenes. We also compared the complement of transposable elements across haplotypes and asked whether the different evolutionary processes acting on dominant and recessive haplotypes had left different molecular signatures. Finally, we took advantage of the complete haplotypic combinations of the two component genes SCR and SRK in A. lyrata and A. halleri to investigate their pattern of co-divergence in natural populations.
Results
The genomic sequences of seven A. halleri and four A. lyrata S-locus haplotypes were obtained through sequencing of bacterial artificial chromosome (BAC) clones extracted from 9 individual genomic libraries. Libraries were screened with probes from the two genes immediately flanking the S-locus region (U-box and ARK3). Positive clones were checked using BAC-end sequencing and further validated by PCR targeted on SRK sequences using haplotype-specific primers [63]. Full BAC sequences were then obtained using 454 pyrosequencing technology. Because of the large sequence divergence among haplotypes, individual sequencing reads were assembled de novo, resulting in two to nine large contigs for each clone, with an average clone size of 98 kb and mean coverage of 57×. Attempts to increase coverage did not eliminate the gaps, suggesting that they may contain repetitive sequences. To reject the hypothesis of non-functional SCR or SRK genes, we used long-range PCR to validate the proposed assemblies when assembly gaps occurred within SCR or SRK introns (AhSRK15, AlSRK01, AlSCR39 and AhSCR03). All these PCR resulted in successful amplifications and the different exons of SCR or SRK were thus confirmed to be consecutive. Detailed characteristics of the BAC clone sequences are reported in Table S1.
Recombination suppression and the boundaries of the S-locus
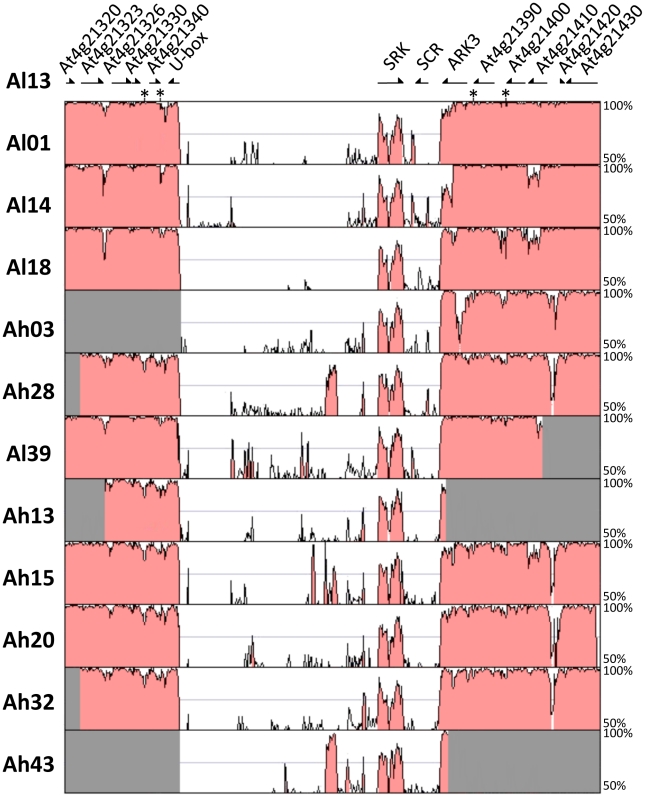
To determine the precise location of the boundaries of the non-recombining S-locus region, we compared sequences from twelve S-locus haplotypes (additionally including the reference haplotype Al13 from the A. lyrata full genome sequence [59]) using the VISTA software [64], looking for a transition in the levels of sequence similarity among haplotypes. As shown in Figure 1 and Figure S1, the sequence conservation among different haplotypes is fairly high in flanking regions on both sides of the S-locus, but plummets sharply between about 300 bp upstream of the start codon of the U-box gene on one side and near the stop codon of ARK3 on the other side. Hence, we define the S-locus as this region of very low similarity lying between these two breakpoints. Synteny is remarkably well conserved outside the S-locus region, except for the presence or absence of some transposable elements in intergenic regions (which were removed from the reference sequence in Figure 1 for clarity). High sequence similarity among haplotypes and high collinearity of flanking genes in the region outside of the S-locus suggest that recombination among haplotypes does occur outside the region delimited by these breakpoints. Additional evidence comes from the observation that elevated diversity, as expected for neutral sites linked to sites under balancing selection [44], is mostly apparent for the two immediately flanking genes (the U-box and ARK3), while levels of synonymous nucleotide diversity are comparable with that of the genomic background (≈2%, [65], [66]) for genes located further away on the chromosome (Figure S2), as previously reported [37], [65]. In contrast, within the S-locus, sequence similarity is almost completely lacking, the only notable exceptions being the seven exons of SRK and some transposable elements of the same family. Interestingly, a pseudogenized partial duplicate of the ARK3 gene (from the end of the first exon to the end of the gene) is found within the S-locus in three different haplotypes: Al01, Ah15 and Ah43. These partial duplicates of ARK3 within the S-locus region could be responsible for the observation by Hagenblad et al. [67] of the occurrence of a pseudogenized paralog of ARK3 in some haplotypes, including one carrying allele Al01 at SRK. A similar partial duplicate sequence of ARK3 was found in the S-locus region of the recombinant C24 haplotype of A. thaliana, and it was hypothesized that this motif acted as the recombination breakpoint between the two common haplotypes A and C [57]. Interestingly, the duplicated ARK3 sequences in Al01, Ah15 and Ah43 are more similar to ARK3 gene copies present in haplotypes other than their own (Figure S3). Assuming that this second copy initially originated through gene duplication from the same chromosome, this observation implies that inter-haplotype recombination does occur at the genomic position of this gene, and hence supports our conclusion that ARK3 indeed lies outside the non-recombining region. Moreover, while the partial duplicates of ARK3 in Ah15 and Ah43 are closely related, that of Al01 is not phylogenetically close, suggesting at least two independent duplication events.
Figure 1. Sequence conservation in the S-locus region between Al13 (the reference A. lyrata genome) and each of the other haplotypes.
Note that the figure is not to scale except for the reference sequence. Portions of sequences not available for some haplotypes were colored in gray. For clarity, transposable elements outside of the S-locus in Al13 were extracted from the sequence, and their locations are indicated by an asterisk.
The S-locus has low gene density and shows important structural rearrangements
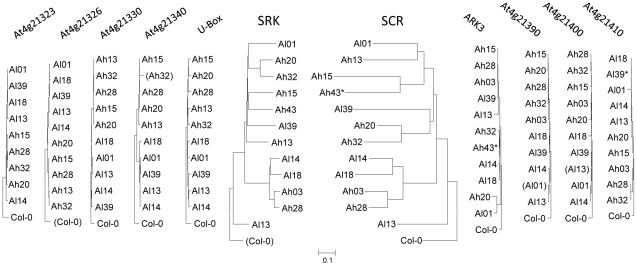
Annotation of the S-locus region revealed only the two incompatibility genes, SCR and SRK, plus TEs (see below). A single copy of SCR and of SRK was found in each haplotype, whereas a previous study [60] described two copies of SCR in one haplotype from A. lyrata (Al20). Multiple gene copies are therefore the exception rather than the rule in the S-locus of Arabidopsis. Sequencing of the 206.7 Mb A. lyrata genome predicted 32,670 genes [59], i.e. approximately 0.16 genes per kb. With only two genes in about 60 kb, the S-locus appears to have very low gene density (ca. 4.8 times lower than the genomic background). Striking differences in the timescales of gene genealogies for the S-locus genes SCR and SRK as compared to the flanking genes were observed (Figure 2), with much deeper genealogies for SCR and SRK, as expected for genes under strong frequency-dependent selection [68]. Moreover, the gene genealogies of SCR and SRK (Figure 2) were found to be more congruent than expected by chance (Icong = 1.53; P-value = 0.0014 [69]). Specifically, the phylogenetic classes defined based on SRK sequences [39] (class I: Al01; class II: Ah03, Ah28, Al18 and Al14; class III: Al13; class IV: all other haplotypes) are conserved in the SCR tree.
Figure 2. Gene phylogenies in and around the S-locus region.
Phylogenies were obtained by the Minimum Evolution method, and are based on coding sequences, with the A. thaliana reference sequence (Col-0) as an outgroup. Asterisks indicate partial sequences, and brackets non functional sequences. The inversion in the SCR coding sequence of Col-0 was de-inverted (i.e. restored to its original functional configuration in A. halleri) according to Tsuchimatsu et al. [70]. Separate phylogenies for each gene are available in Figure S4.
In contrast, the phylogenetic relationships among haplotypes were strikingly different for the flanking genes (Figure S4), as reported for haplotypes of the U-box and the ARK3 genes in A. thaliana [70]. Indeed, in our dataset gene genealogies of the flanking genes tend to cluster according to species overall, rather than to S-locus phylogenetic classes. This observation further supports the conclusion that the non-recombining region is confined to the S-locus and is determined by the two breakpoints identified based on sequence similarity.
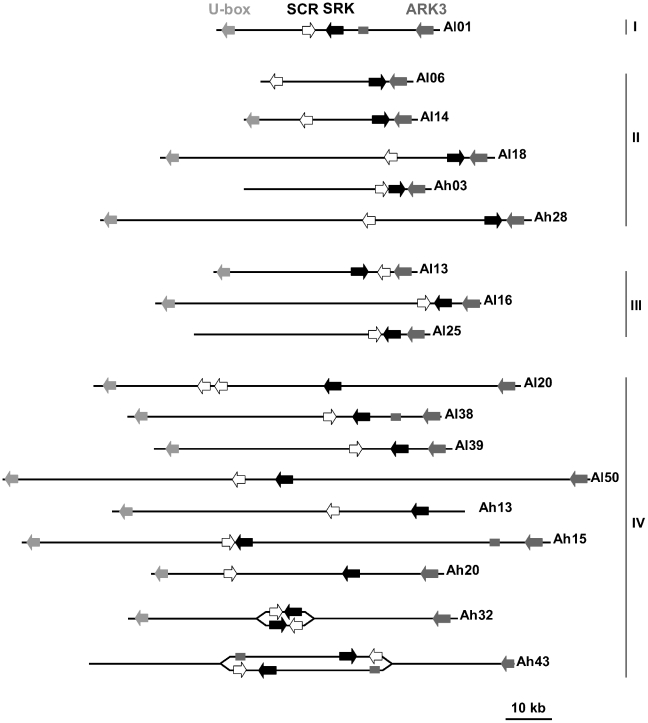
The S-locus region is variable in size across haplotypes, spanning from 31 kb (haplotype Al14) to 110 kb (haplotype Ah15) with an average size of 62 kb. Given that BAC sequences do not cover the totality of the S-locus from haplotypes Ah03, Ah13 and Ah43, these estimates are lower bounds. Also, several libraries that we constructed could not be exploited because no single clone showed both flanking genes used for screening, suggesting that the S-locus haplotypes they contain may have been larger than the average 100 kb typical of the BAC clones in our libraries. With an average size of 74 kb, haplotypes from SRK phylogenetic class IV are generally larger than haplotypes from classes I to III, showing an average size of 50 kb (Table 1). Figure 3 summarizes the gene organization within the S-locus and includes data from Kusaba et al. [60], Boggs et al. [61] and Guo et al. [62]. Globally, we found that gene organization within the S-locus is highly variable with regard to gene order (SRK located either on the ARK3 or the U-box side as compared to SCR, although the latter order was only found in a single haplotype, Al13), relative orientation of SCR and SRK (tail-to-tail, head-to-head or in the same direction), and distance separating them (from less than 1 kb to about 26 kb; Table 1). These patterns also vary among haplotypes within each of the SRK phylogenetic classes, with the exception of class II haplotypes showing mostly SCR and SRK oriented tail-to-tail and a location of SRK consistently very close to the flanking gene ARK3 in head-to-head orientation. Strikingly, these class II haplotypes were already reported to show common features that distinguish them from other phylogenetic classes [71], [39]. We found here that the strong sequence similarity previously noted in the kinase domain of these haplotypes [71] is extended to the whole intergenic region (about 900 bp in length) between SRK and ARK3 (Figure S5), in contrast to comparisons with other classes of haplotypes or between classes (Figure S1). As suggested by [39], these class II haplotypes could have originated by a gene conversion event implying unlinked members of the SRK gene family. Interestingly, this same intergenic region is also conserved between class II haplotypes and haplotypes Ah15 and Ah43, two of the three haplotypes carrying a pseudogeneized duplicated copy of ARK3. This observation strongly suggests that the duplication involved a recombination event between these haplotypes and a class II haplotype. Interestingly, while [62] suggested that haplotypes Al38 and Al50 lack the second exon of the SCR gene, we were able to detect the second exon upon closer examination applying the same approach than in our own data, suggesting that these haplotypes are indeed functional. In addition, while previous studies failed to detect a kinase domain for AlSRK01 [30], our genomic approach confirmed that all SRK sequences we observed contained a full-length kinase domain.
Table 1. Description of the different haplotypes.
| Haplotype | Phylogenetic class | Size of the S-locus | SCR - SRK distance |
| Al01 | I | 42 614 | 2 906 |
| Al14 | II | 30 909 | 8 671a |
| Al18 | II | 65 495 | 12 227 |
| Ah03 | II | 34 512 | 742 |
| Ah28 | II | 87 805 | 25 748 |
| Al13 | III | 37 013 | 1 752 |
| Al39 | IV | 55 787 | 6 601 |
| Ah13 | IV | 73 401 | 17 028 |
| Ah15 | IV | 109 864 | 618 |
| Ah20 | IV | 56 764 | 3 636a |
| Ah32 | IV | 52 987 | 1 974 |
| Ah43 | IV | 93 791 | 4 147 |
Because of the uncertainty on the orientation of some contigs, the indicated distance is the minimum distance between SCR and SRK.
Figure 3. Structural variation within the S-locus.
The direction of SCR, SRK and the two flanking genes is shown taking into account their approximate distances. Both possibilities are depicted when the orientation of genes remains unknown due to unoriented contigs. The presence of a pseudo-ARK3 sequence is represented by a dark gray rectangle. Organization of haplotypes Al20, Al06, Al25, Al16, Al38 and Al50 are based on Kusaba et al. [60], Boggs et al. [61] and Guo et al. [62].
Invasion by transposable elements and the effect of dominance
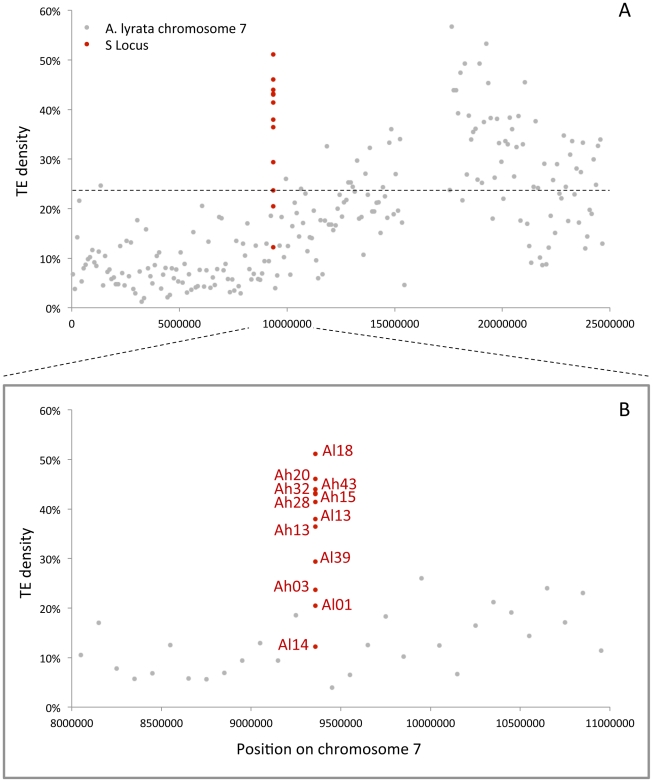
Transposable elements annotation with the CENSOR [72] and PLOTREP [73] programs revealed a strong density and diversified complements of TEs in the S-locus, with a representation of most families known in the A. thaliana genome (detailed annotation and a complete list of TEs for each haplotype are shown in Figure S6 and Table S2). In order to determine whether these observations are uncommon in the genomic background, we also used CENSOR [72] to estimate TE density along the A. lyrata genome divided in non-overlapping windows of 100 kb. Variation of TE density along chromosome 7 confirmed that the TE density of the S-locus sharply departs from its chromosomal background, being matched only by the centromeric region (Figure 4, and Figure S7 for the other chromosomes). This difference is not due to an invasion by a single class of TEs, since the quantitative difference in density was observed for most TE families (Figure S8).
Figure 4. TE density along A. lyrata chromosome 7, comparison with the S-locus data, zoom on the 3 Mbp region around the S-locus.
A. TE density along A. lyrata chromosome 7, and comparison with the S-locus data. Transposable elements contents were calculated using CENSOR [72] for non overlapping windows of 100 kb. B. Zoom on the 3 Mbp region around the S-locus. The dashed line represents a 95% confidence interval on the TE densities of this 3 Mbp genomic region.
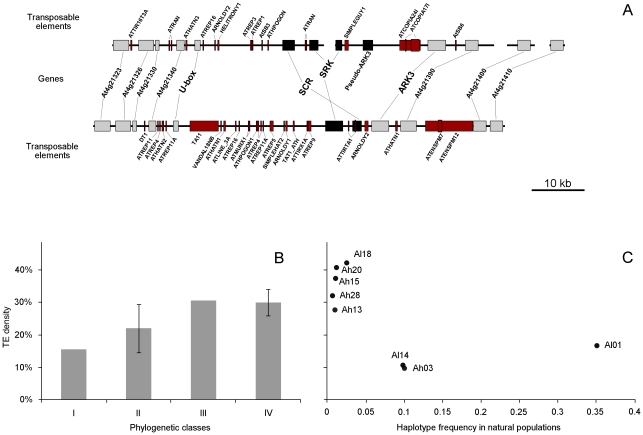
While most haplotypes have higher TE density than the genomic background, there is striking variability in TE density among haplotypes. Indeed, TE density depends on SRK phylogenetic classes, which are themselves associated with dominance with higher density in the more dominant haplotypes (Figure 5A and 5B). Since levels of dominance are in turn expected to correlate with S-haplotype frequency in natural populations [74], [42], [75], we plotted TE density against haplotype frequency, as estimated from S-locus genotype surveys in A. lyrata [76] and A. halleri (P. Goubet et al. unpublished data). We find that variation in TE density is even better captured by haplotype frequencies, with rare haplotypes being more enriched in TEs than more frequent haplotypes (Figure 5C).
Figure 5. Comparative annotation of genes and transposable elements, mean TE density, and TE density according to frequency.
A. Comparative annotation of genes and transposable elements for a recessive haplotype, Al01, and a dominant one, Al13. The S-locus genes are represented by black rectangles, and other genes by light gray ones. Transposable elements, annotated using CENSOR [72] and PLOTREP [73], are indicated in red. B. Mean TE density in the different phylogenetic classes. TE density corresponds to the percentage of a sequence which is covered by transposable elements. Standard deviation is indicated for classes II and IV, in which multiple haplotypes could be analysed. C. TE density of the different haplotypes according to their frequency in natural populations. Haplotype frequencies are based on SRK fragments. A. lyrata data concern 12 icelandic populations [76] and A. halleri data concern 39 european populations (unpublished data).
Discussion
Our results confirm that the S-locus in A. halleri and A. lyrata differs significantly from its genomic background in several respects: gene density is particularly low, gene genealogies are much deeper as compared to the flanking genes, gene order and orientation vary extensively, sequence similarity among haplotypes in intergenic sequences is completely lacking and the density of transposable elements is particularly elevated, being matched only by that in the centromere. Most of these properties are shared with many genetic systems controlling patterns of mating such as sex chromosomes, sex-determining loci or mating-type loci.
Size of genomic regions involved in mating-type determination
Contrasted patterns of conservation between the S-locus and its flanking regions are in line with two previous investigations comparing three and five haplotypes in A. thaliana [49] and A. lyrata [62], respectively. Based on a more extensive collection of S-haplotypes, we could precisely map the breakdown of synteny to two narrow regions very close to the 5′ or 3′ ends of the coding regions of the flanking genes U-box or ARK3, respectively. Note that [37] and [65] reported some level of co-segregation of flanking genes with variation at SRK in local A. lyrata populations, while our global sample from two related species might have left sufficient time for recombination to uncouple the S-locus region from its genomic background. Using this objective criterion to define the S-locus itself, we find that the S-locus has an average size of 62 kb, ranging from 31 to 110 kb among haplotypes, much larger than the average distance of 7 kb between the two S-locus genes, SCR and SRK, ranging from 1 kb to 26 kb. An orthologous sporophytic SI system occurs in the genus Brassica, although the S-locus is located in a different genomic region than in Arabidopsis. Based on the available sequences of four B. rapa haplotypes [53], [55] and using a similar criterion to define the S-locus we determined that the S-locus was somewhat smaller than in Arabidopsis, ranging from 28 to more than 60 kb. In contrast, the distance separating SCR and SRK was less variable, ranging from 2 to 11 kb. In Brassica, however, the S-locus generally comprises a third gene, SLG which is a paralog of SRK lacking the kinase domain, and the overall region comprising these three genes ranged from 23 to 43 kb. In Ipomoea trifida (Convolvulaceae), which also exhibits sporophytic SI but of a different molecular nature, the S-haplotype-specific divergent region between the only two sequenced haplotypes (S1 and S10) extends over 50 and 34 kb, respectively [77]. In the gametophytic SI system of Prunus dulcis and P. mume (Rosaceae), the S-locus was estimated as being a divergent genomic region of about 70 kb [78] and 15 to 27 kb [45], respectively. In Antirrhinum hispanicum (Plantaginaceae), the distance between the two component genes of haplotype S2 is 9 kb [79]. However, a major difference between the S-locus of the Brassicaceae and that of the Plantaginaceae and Solanaceae is that in the latter the pollen phenotype can be encoded by different members of the gene family to which the male determinant belongs, so that the S-locus comprises more than two genes [25], making this comparison tricky. Overall, in spite of the large diversity of species and molecular mechanisms involved in the different SI systems, the size of S-loci seems to be fairly constant across taxa, ranging from 27 to about 110 kb, with Arabidopsis species apparently in the upper part of the range.
Beyond the comparison with S-loci of other plants, the size of the S-locus can also be compared with that of the mating-type loci in fungi or green algae. In the basidiomycete Cryptococcus neoformans, sex determination is controlled by a locus including genes encoding a pheromone and its receptor. Haplotypes of this mating-type locus, α and a, represent a genomic region of approximately 105 to 130 kb [80], hence slightly larger than the S-locus in A. halleri and A. lyrata. In another basidiomycete, Ustilago hordei, the mating-type locus consists of a single region comprising two complexes, a and b, between which recombination is suppressed. The distance between these two complexes was estimated to be 500 kb and 413 kb in the MAT-1 and MAT-2 strains, respectively [9]. In the ascomycete Neurospora tetrasperma, the non-recombining region comprising the mating-type locus covers 78.4% of the chromosome length, i.e. 6.9 Mbp [11]. In green algae, the mating-type locus of the unicellular Chlamydomonas reinhardtii consists of a highly rearranged 200-kb region [81] while that of the multicellular Volvox carteri is about 500% larger and contains many ORFs. Interestingly, C. reinhardtii is an isogamous species with two morphologically indistinguishable mating-types [7] while V. carteri shows morphological differentiation between the mating-types, suggesting the general conclusion that genomic regions involved in mating-type systems that are not associated with morphological differences between mates may span smaller genomic regions. In other words, the accumulation of genes with a role in expression of the morphological differences between mating-types [8] may contribute to some extent to the variation in size of the mating-type locus, in addition to transposable elements and non coding DNA accumulating in these regions. Because in homomorphic SI the mating-types are not associated with morphological differences, the S-loci may retain a smaller size.
Structural rearrangements, yet shared evolutionary history, between SCR and SRK
Only six sequences of SCR were previously described in Arabidopsis because of the difficulty of finding conserved regions to perform PCR amplifications [60], [61], [82], [70], [62]. Our important sequencing effort of the S-locus region resulted in the successful identification of full SCR sequences in ten new S-haplotypes in A. halleri and A. lyrata and only the second exon of SCR in one haplotype (haplotype Ah43, for which we could not obtain the full S-locus sequence), along with their cognate SRK partner. These results do not support the hypothesis of existence of non-functional haplotypes carrying only partial SCR sequences, as proposed by Guo et al. [62], as we were able to localize the missing coding sequence for their two putative non-functional haplotypes when applying the ALN [83] software fed with all known SCR sequences. Congruence of SCR and SRK phylogenies reflects the coevolution necessary to maintain the specific SCR-SRK protein-protein recognition, and clearly indicates that recombination between the two SI genes has been precluded. Comparison of phylogenies between SCR and the S domain of SRK was already investigated by Sato et al. [32] for twelve haplotypes in Brassica oleraceae. They found that the hypothesis of an identical topology for the two trees was not rejected. Edh et al. [84] also compared SCR and SRK phylogenies in Brassica rapa, Brassica oleraceae and Brassica cretica class II haplotypes, but congruence between topologies could not be clearly demonstrated, perhaps as a consequence of the concerted evolution of the SLG and SRK genes within haplotypes, or of the more recent history of diversification within the class II lineage. In contrast, in the ascomycete Neurospora [85], the non-self recognition system is controlled by two tightly linked genes, het-c and pin-c. In agreement with our results in the S-locus, congruence was found between topologies of the phylogenies of these two genes, but not with those of the flanking genes. When more SCR/SRK sequences become available, it will be interesting to study in more details the co-evolutionary process.
Based on the study of nine haplotypes in A. thaliana, A. lyrata and Capsella rubella, Guo et al. [62] proposed that head-to-head orientation of SCR and SRK was the ancestral orientation in the Arabidopsis/Capsella lineage. However, the lack of conserved orientation pattern in our results based on a much larger number of haplotypes suggests that, in spite of the shared evolutionary history of SCR and SRK, the S-locus has experienced a history of frequent inversions and genomic rearrangements. At this stage, we argue that the ancestral orientation cannot be deduced. However, our results confirm that with a single exception SCR always occurs at the U-box side and SRK at the ARK3 side. Interestingly, the exception to this rule concerns haplotype Al13, which was obtained from an A. lyrata strain (MN47) with non-functional SI. This suggests the intriguing possibility that the observed inversion may have been associated with the breakdown of SI in this strain used for sequencing the A. lyrata genome. Strong structural variation among haplotypes seems to be a common feature of S-loci [86] and genomic rearrangements, particularly inversions, are known to be frequent in low recombination regions such as in sex chromosomes of mammals [3], [14], [87] and plants [19] or in the mating-type locus of green algae [81]. Evidence suggesting gradual suppression of recombination was found in sex chromosomes, and led to the concept of evolutionary strata [16]–[18], [3], [14], [19], [20]. These strata, composed of genes which stop recombining and therefore start diverging presumably at the same time, could have been caused by large inversions in the non-recombining sex chromosome [16]. As in sex chromosomes, inversions in the S-locus could have contributed to the reduction in recombination among haplotypes. However, no discrete strata of divergence among haplotypes can be identified. Instead, the proportion of sequence similarity changes abruptly to mostly zero within the S-locus region.
Transposable elements accumulation in sex-determining regions
Our results show that transposable elements are a major component of the S-locus region, as previously noted in other taxa [47], [48], [54]. On a wide scale, their density is higher in most S-haplotypes than in the genomic background. Such accumulation has already been observed in other genomic regions involved in mating-type and gender determination, and is not exclusive to the S-locus. Bachtrog [2] investigated four regions of the neo-sex chromosomes, containing homologous gene pairs, in Drosophila miranda. In each case, the neo-Y showed several transposable elements insertions that were absent from the neo-X. Similarly, Marais et al. [5] analyzed genetic degeneration of the Y chromosome in Silene latifolia, by examining seven sex-linked genes. Comparison of Y-linked genes and their X-linked homologs provided evidence that some of the Y-linked genes showed higher intron sizes, due to the accumulation of transposable elements. In the mating-type locus of the basidiomycete Ustilago hordei, sequencing of one of the two haplotypes, MAT-1, revealed that this genomic region was particularly rich in both retroelements and repetitive DNA compared to U. maydis, in which the a and b complexes are unlinked [88]. Similarly, the chromosome carrying the mating-type locus in the fungus Microbotryum violaceum was found to be enriched in transposable elements as compared to autosomal chromosomes [10]. In A. thaliana, Wright et al. [89] compared the transposable elements accumulation in chromosome arms and in low-recombining regions surrounding the centromeres, i.e. centromeres, pericentromeric regions and heterochromatic knobs. These regions of reduced recombination were shown to exhibit greater TE copy numbers than chromosome arms, particularly for Gypsy retrotransposons and EnSpm transposons. Interestingly, our results showed that precisely these two TE families present densities twice higher in the S-locus than in the overall genome of A. lyrata, suggesting that the increased TE density noticed in the S-locus is effectively linked to the restricted recombination.
TE accumulation: Driven by recombination suppression and mutational hazard?
Strikingly, we found that not all haplotypes present the same TE coverage, with dominant S-haplotypes (SRK phylogenetic classes III and IV) having higher TE density than those belonging to recessive classes (I and II). Signatures of intragenic recombination have been found in SRK only in S-haplotypes belonging to recessive classes I and II [30]. It was suggested that recombination can occur only in individuals carrying two copies of the same functional S-haplotype, which is most probable for recessive haplotypes, because they are predicted to have high frequencies in natural populations [42]. Indeed, in A. lyrata, the most recessive haplotype was 12.75 times commoner than the most dominant haplotypes in Icelandic natural populations [90]. Our observation that TE density is inversely related to haplotype population frequency also suggests that recombination plays a role in preventing TE accumulation in the S-locus. In addition, haplotype frequency also influences the effective population size of gene copies within S-haplotypes [68], so that genetic drift will be stronger in low-frequency dominant haplotypes (in agreement with the mutational-hazard model of Lynch and Conery [91]), and this may also affect TE accumulation. Sex chromosomes in mammals also differ in opportunities for recombination and in effective population sizes [91]. Recessive S-haplotypes tend to behave like the X chromosome, and dominant ones are more like the Y chromosome. These differences may be an important source of variation of the size of the S-locus among haplotypes.
Methods
Construction of BAC libraries
High Molecular Weight (HMW) DNA was prepared from young leaves of seven A. halleri and four A. lyrata haplotypes. For each extraction, approximately 20 grams of frozen leaf tissue was ground to powder in liquid nitrogen with a mortar and pestle used to prepare megabase-size DNA embedded in agarose plugs. HMW DNA of the various genotypes was prepared as described by Peterson et al. [92] and modified as described in [93]. Embedded HMW DNA was partially digested with HindIII (New England Biolabs, Ipswich, Massachusetts), subjected to two size selection steps by pulsed- field electrophoresis, using a BioRad CHEF Mapper system (Bio-Rad Laboratories, Hercules, California), and ligated to pIndigoBAC-5 HindIII-Cloning Ready vector (Epicentre Biotecnologies, Madison, Wisconsin). Pulsed-field migration programs, electrophoresis buffer, and ligation desalting conditions were performed according to [94].
To evaluate the average insert size of each library, BAC DNA was isolated from about 384 randomly selected clones in each library, restriction enzyme digested with the rare cutter NotI, and analyzed by Pulsed-Field Gel Electrophoresis (PFGE). All fragments generated by NotI digestion contained the 7.5 kb vector band and various insert fragments. Analysis of the insert sizes from the various BAC libraries showed a mean insert size comprised between 80 kb and 175 kb. Since the haploid genome of A. lyrata and A. halleri is estimated around 230 Mb and 250 Mb respectively, we picked the number of BAC clones required to obtain a library coverage of 5 genome equivalents.
Screening the BAC libraries
High-density colony filters were prepared from all the nine BAC libraries constructed using a robotic workstation QPix2 XT (Genetix). BAC clones were spotted in double using a 5×5 or 6×6 pattern onto 22×22 cm Immobilon-Ny+ filters (Millipore Corporate, Billerica, Massachusetts). On each filter, 27 648 to 41 472 unique clones were spotted in duplicate, and clones were grown at 37°C for 17 h. Filters were then processed as follows: (1) denaturation on Whatman paper soaked with a solution of 0.5 M NaOH and 1.5 M NaCl for 4 min at room temperature, and for 10 min at 100°C, (2) neutralization on Whatman paper soaked with 1 M TrisHCl pH 7.4, and 1.5 M NaCl for 10 min, incubation in a solution of 0.25 mg/mL proteinase K (Sigma Aldrich, St. Louis, Missouri) for 45 min at 37°C, baking for 45 min at 80°C, and (3) fixation by UV on a Biolink 254 nm crosslinker (Thermo Fischer Scientific, Waltham, Massachusetts) with an energy of 120,000 µJoules. Radiolabelling of probes and hybridization of the filters were performed as described in [93]. Hybridized filters were imaged with a Storm 860 PhosphorImager (GE Healthcare, Little Chalfont, UK), and analyses were performed using the HDFR software (Incogen, Williamsburg, Virginia). Positive BAC clones detected by hybridization were validated individually by PCR amplification using the primer pairs used for probes synthesis (Table S3), and visualisation of PCR products after agarose gel electrophoresis.
Sequencing
A total of fourteen BACs covering the S-locus region of 11 S-haplotypes were sequenced in this study (two partially overlapping BACs were needed for haplotypes Al28, Ah15 and Ah28) : eleven BACs were sequenced at Genoscope; two BACs (containing haplotypes Al39 and Ah43) were sequenced at CNRGV; and a last one (haplotype Al14) was sequenced by Beckman Coulter 485 Genomics.. All clones were sequenced using a 454 multiplexing technology on Titanium sequencer (www.roche.com). De-novo assembly was performed by Newbler (www.roche.com) for each S-haplotype and only contigs representing the extremities of the BACs were organized at this step.
Sequence finishing
BAC sequences covering the 11 S-haplotypes were obtained in two to nine contigs. Suggestion of orientation was provided with assembly for some sequences, but in most cases, only the first and last contigs were oriented. The relative order and orientation of other contigs were therefore unknown. When exons of SCR or SRK were in two different contigs (i.e. haplotypes Al01 and Ah15 for SRK, Ah03 and Al39 for SCR), primers were defined with Primer3 [95] on both contigs. Because of the presence of repeated sequences including transposable elements, long-range (using TaKaRa LA Taq Polymerase) rather than classical PCR were performed in order to confirm the contiguity of the contigs.
Sequence annotation
Annotation of BAC sequences was performed using two gene structure prediction programs with Arabidopsis parameters, FGENESH [96] and GENSCAN [97]. FGENESH has the advantage of being more accurate in detecting Arabidopsis genes but GENSCAN is more sensitive. Detected ORFs were blasted using BLASTX [98] and the obtained proteins were then aligned on BAC sequences with SPALN [99] and FGENESH+ [96] softwares. Because of its high nucleotide diversity, SCR was rarely detected by these two programs. Known SCR proteins were therefore aligned on BAC sequences using the semiglobal alignment procedure implemented on ALN [83], which is more sensitive than SPALN and FGENESH+. The results were then examined by eye in order to find the SCR gene and the cysteine residues that characterize this protein. Transposable elements were annotated with CENSOR [72] using the A. thaliana repetitive elements [v16.02] database of Repbase Update [100]. The results were then filtered and defragmented with PLOTREP [73], using a minimum coverage of merged fragments of 10%.
Comparison of sequences and phylogenetic analysis
The full BAC sequences were aligned and compared using the “glocal” alignment procedure [101] implemented in VISTA [64]. This kind of alignment is able to detect rearrangements and inversions in sequences, and is particularly appropriate for divergent regions like the S-locus. Protein sequences of genes were aligned with CLUSTALW [102]. Alignments were then manually adjusted and phylogenetic trees were constructed using MEGA version 5 [103], according to a Minimum Evolution (ME) analysis with the maximum composite likelihood method. The congruence between topologies of SCR and SRK trees was tested by computing an index of congruence, based on the size of their maximum agreement subtree, and comparing its value to a null-hypothesis distribution obtained by simulation of random trees [69].
Analysis of the transposable elements content
A PERL script was developed to compare TE density between the twelve S haplotypes and the A. lyrata genome. CENSOR [72] was used in local on BAC sequences, excluding the S-locus flanking regions, and on non-overlapping windows of 100 kb along the eight chromosomal sequences of the A. lyrata genome version Araly1 (http://genome.jgi-psf.org/Araly1/Araly1.download.html [59]). Densities were thus calculated for each transposable element family in the A. lyrata genome and in the S-locus, according to the classification in [104].
Supporting Information
Sequence conservation at the S-locus boundaries between Al13 (the reference A. lyrata genome) and each of the other haplotypes. Sequences not available for the U-box side (Ah03 and Ah43) were not represented. Distance from the homology breakpoint is indicated under each graph.
(PDF)
Synonymous nucleotide diversity (ΠS) at S-locus flanking genes for A. halleri (black) and A. lyrata (gray), estimated using DnaSP [105].
(PDF)
Phylogeny of pseudo-ARK3 sequences, SRK and ARK3. Phylogeny was constructed using a Minimum Evolution analysis.
(PDF)
Separate phylogenies of the S-locus Region genes. Phylogenies were obtained by the Minimum Evolution method, and are based on protein sequences, with the A. thaliana reference sequences (Col-0) as outgroup.
(PDF)
Sequence conservation in the SRK-ARK3 region between Ah28 (Class II) and each of the other haplotypes. Distance from homology breakpoint is indicated under the graph.
(PDF)
Annotation of genes and transposable elements for the 12 S-haplotypes. The S-locus genes are represented in black rectangles, with delimitation of their exons. Other genes are depicted in light gray. Transposable elements are shown in dark gray, and their fragmentation is indicated by white gaps.
(PDF)
TE density along A. lyrata chromosomes 1 to 6 and chromosome 8. Transposable elements contents were calculated using CENSOR [72] for non overlapping windows of 100 kb.
(PDF)
Comparative density in different families of transposable elements for the entire genome of A. lyrata, and the S-locus of A. lyrata and A. halleri. Transposable elements classification refers to Wicker et al. [104].
(PDF)
Description of the different clones. Two clones were necessary to cover the entire S-locus for three haplotypes : Al18, Ah28 and Ah15.
(DOC)
List of the transposable elements detected in the BAC sequences.
(DOC)
Primers pairs used to validate BAC clones by PCR amplification. These primers were defined to amplify SRK (primer Sh04), U-box (primer B80) and ARK3 (primer ARK3) genes.
(DOC)
Acknowledgments
We thank Deborah Charlesworth, Gabriel Marais, Tatiana Giraud, and two anonymous reviewers for helpful comments on the manuscript. We are also grateful to Eric Schmitt and Angélique Bourceaux for taking excellent care of plants in the greenhouse. Yalong Guo kindly shared sequences from haplotypes Al16, Al38, and Al50. Jesper Bechsgaard and Mikkel H. Schierup provided the A. lyrata plant material used in this study. Maude Pupin, Hélène Touzet, Camille Roux, and Clémentine Vitte provided computational advice on the use of software for data analysis.
Footnotes
The authors have declared that no competing interests exist.
This project was funded by Genoscope project AP2006/07-project #13 and by ANR “Jeunes Chercheurs” JSV7 008 01. PMG was supported by a CNRS doctoral grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Billiard S, Lopez-Villavivencio M, Devier B, Hood ME, Fairhead C, et al. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biological reviews. 2011;86:421–442. doi: 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- 2.Bachtrog D. Sex chromosome evolution: Molecular aspects of Y-chromosome degeneration in Drosophila. Genome Research. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Q, Hou S, Hobza R, Feltus FA, Wang X, et al. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Molecular Genetics and Genomics. 2007;278:177–185. doi: 10.1007/s00438-007-0243-z. [DOI] [PubMed] [Google Scholar]
- 5.Marais GAB, Nicolas M, Bergero R, Chambrier P, Kejnovsky E, et al. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Current Biology. 2008;18:545–549. doi: 10.1016/j.cub.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Hasselmann M, Beye M. Pronounced differences of recombination activity at the sex determination locus of the Honeybee, a locus under strong balancing selection. Genetics. 2006;174:1469–1480. doi: 10.1534/genetics.106.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodenough UW, Armbrust EV, Campbell AM, Ferris PJ. Molecular genetics of sexuality in Chlamydomonas. Annual Review of Plant Physiology and Plant Molecular Biology. 1995;46:21–44. [Google Scholar]
- 8.Ferris P, Olson BJSC, De Hoff PL, Douglass S, Casero Diaz-Cano D, et al. Evolution of an expanded sex determining locus in Volvox. Science. 2010;328:351–354. doi: 10.1126/science.1186222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N, Bakkeren G, Wong K, Sherwood JE, Kronstad JW. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Genetics. 1999;96:15026–15031. doi: 10.1073/pnas.96.26.15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood ME, Antonovics J, Koskella B. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms. Genetics. 2004;168:141–146. doi: 10.1534/genetics.104.029900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittle CA, Johannesson H. Evidence of the accumulation of allele-specific non-synonymous substitutions in the young region of recombination suppression within the mating-type chromosomes of Neurospora tetrasperma. Heredity. 2011;107:305–314. doi: 10.1038/hdy.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice WR. Evolution of Y sex chromosome in animals. Bioscience. 1996;46:331–343. [Google Scholar]
- 14.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2005;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 15.Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, et al. Are all sex chromosomes created equal? TRENDS in Genetics in press. 2011 doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 17.Lawson Handley LJ, Ceplitis H, Ellegren H. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 2004;167:367–376. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 19.Bergero R, Charlesworth D, Filatov DA, Moore RC. Defining regions and rearrangements of the Silene latifolia Y chromosome. Genetics. 2008;178:2045–2053. doi: 10.1534/genetics.107.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends in Ecology and Evolution. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 21.De Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin: Springer-Verlag; 2001. [Google Scholar]
- 22.Igic B, Lande R. Loss of self incompatibility and its evolutionary consequences. International Journal of Plant Sciences. 2008;169:93–104. [Google Scholar]
- 23.Takayama S, Isogai A. Self-incompatibility in plants. Annual Review of Plant Biology. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- 24.Rea A, Nasrallah JB. Self-incompatibility systems: barriers to self-fertilization in flowering plants. International Journal of Developmental Biology. 2008;52:627–636. doi: 10.1387/ijdb.072537ar. [DOI] [PubMed] [Google Scholar]
- 25.Kubo K, Entani T, Takara A, Wang N, Fields A, et al. Collaborative non-self recognition system in S-RNase-Based self-incompatibility. Science. 2010;330:796. doi: 10.1126/science.1195243. [DOI] [PubMed] [Google Scholar]
- 26.Franklin-Tong NVE, Franklin FCH. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. TRENDS in Plant Science. 2003;8:598–605. doi: 10.1016/j.tplants.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Hiscock SJ, McInnis SM. Pollen recognition and rejection during the sporophytic self-incompatibility response: Brassica and beyond. TRENDS in Plant Science. 2003;8:606–613. doi: 10.1016/j.tplants.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castric V, Vekemans X. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Molecular Ecology. 2004;13:2873–2889. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- 30.Castric V, Bechsgaard J, Grenier S, Noureddine R, Schierup MH, et al. Molecular evolution within and between self-incompatibility specificities. Molecular Biology and Evolution. 2010;27:11–20. doi: 10.1093/molbev/msp224. [DOI] [PubMed] [Google Scholar]
- 31.Dwyer KG, Balent MA, Nasrallah JB, Nasrallah ME. DNA sequences of self-incompatibility genes from Brassica campestris and B. oleracea: polymorphism predating speciation. Plant Molecular Biology. 1991;16:481–486. doi: 10.1007/BF00024000. [DOI] [PubMed] [Google Scholar]
- 32.Sato K, Nishio T, Kimura R, Kusaba M, Suzuki T, et al. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics. 2002;162:931–940. doi: 10.1093/genetics/162.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casselman AL, Vrebalov J, Conner JA, Singhal A, Giovannoni J, et al. Determining the physical limits of the Brassica S locus by recombinational analysis. Plant Cell. 2000;12:23–34. doi: 10.1105/tpc.12.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawabe A, Hannson B, Forrest A, Hagenblad J, Charlesworth D. Comparative gene mapping in Arabidopsis lyrata chromosomes 6 and 7 and A. thaliana chromosome IV: evolutionary history, rearrangements and local recombination rates. Genetical Research Cambridge. 2006;88:45–46. doi: 10.1017/S0016672306008287. [DOI] [PubMed] [Google Scholar]
- 35.Charlesworth D, Awadalla P. The molecular population genetics of flowering plant self-incompatibility polymorphisms. Heredity. 1998;81:1–9. doi: 10.1038/sj.hdy.6884000. [DOI] [PubMed] [Google Scholar]
- 36.Vieira CP, Charlesworth D, Vieira J. Evidence for rare recombination at the gametophytic self-incompatibility locus. Heredity. 2003;91:262–267. doi: 10.1038/sj.hdy.6800326. [DOI] [PubMed] [Google Scholar]
- 37.Kamau E, Charlesworth B, Charlesworth D. Linkage Disequilibrium and Recombination Rate Estimates in the Self-Incompatibility Region of Arabidopsis lyrata. Genetics. 2007;176:2357–2369. doi: 10.1534/genetics.107.072231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kusaba M, Tung C, Nasrallah ME, Nasrallah JB. Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiology. 2002;128:17–20. [PMC free article] [PubMed] [Google Scholar]
- 39.Prigoda NL, Nassuth A, Mable BK. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Molecular Biology and Evolution. 2005;22:1609–1620. doi: 10.1093/molbev/msi153. [DOI] [PubMed] [Google Scholar]
- 40.Uyenoyama MK. A generalized least-squares estimate for the origin of self-incompatibility. Genetics. 1995;139:975–992. doi: 10.1093/genetics/139.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatakeyama K, Watanabe M, Takasaki T, Ojima K, Hinata K. Dominance relationships between S-alleles in self-incompatible Brassica campestris L. Heredity. 1998;80:241–247. [Google Scholar]
- 42.Schierup MH, Vekemans X, Christiansen FB. Evolutionary dynamics of sporophytic self-incompatibility alleles in plants. Genetics. 1997;147:835–846. doi: 10.1093/genetics/147.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billiard S, Castric V, Vekemans X. A general model to explore complex dominance patterns in plant sporophytic self-incompatibility systems. Genetics. 2007;175:1351–1369. doi: 10.1534/genetics.105.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schierup MH, Mikkelsen AM, Hein J. Recombination, balancing selection and phylogenies in MHC and self-incompatibility genes. Genetics. 2001;159:1833–1844. doi: 10.1093/genetics/159.4.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Entani T, Iwano M, Shiba H, Che FS, Isogai A, et al. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells. 2003;8:203–213. doi: 10.1046/j.1365-2443.2003.00626.x. [DOI] [PubMed] [Google Scholar]
- 46.Shiba H, Kenmochi M, Sugihara M, Iwano M, Kawasaki S, et al. Genomic organization of the S-locus region of Brassica. Bioscience Biotechnology and Biochemistry. 2003;67:622–626. doi: 10.1271/bbb.67.622. [DOI] [PubMed] [Google Scholar]
- 47.Wheeler MJ, Armstrong SA, Franklin-Tong VE, Franklin FCH. Genomic organization of the Papaver rhoeas self-incompatibility S1 locus. Journal of Experimental Botany. 2003;54:131–139. doi: 10.1093/jxb/erg006. [DOI] [PubMed] [Google Scholar]
- 48.Tomita RN, Suzuki G, Yoshida K, Yano Y, Tsuchiya T, et al. Molecular characterization of a 313-kb genomic region containing the self-incompatibility locus of Ipomoea trifida, a diploid relative of sweet potato. Breeding Science. 2004;54:165–175. [Google Scholar]
- 49.Tang C, Toomajian C, Sherman-Broyles S, Plagnol V, Guo Y, et al. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 50.Cui Y, Brugière N, Jackman L, Bi Y, Rothstein SJ. Structural and transcriptional comparative analysis of the S locus regions in two self-incompatible Brassica napus lines. The Plant Cell. 1999;11:2217–2231. doi: 10.1105/tpc.11.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki G, Kai N, Hirose T, Fukui K, Nishio T, et al. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics. 1999;153:391–400. doi: 10.1093/genetics/153.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura R, Sato K, Fujimoto R, Nishio T. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleraceae and Brassica rapa. The Plant Journal. 2002;29:215–223. doi: 10.1046/j.1365-313x.2002.01208.x. [DOI] [PubMed] [Google Scholar]
- 53.Fukai E, Fujimoto R, Nishio T. Genomic organization of the S core region and the S flanking regions of a class-II S haplotype in Brassica rapa. Molecular Genetics and Genomics. 2003;269:361–369. doi: 10.1007/s00438-003-0844-0. [DOI] [PubMed] [Google Scholar]
- 54.Fujimoto R, Okazaki K, Fukai E, Kusaba M, Nishio T. Comparison of the genome structure of the self-incompatibility (S) locus in interspecific pairs of S haplotypes. Genetics. 2006;173:1157–1167. doi: 10.1534/genetics.104.037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takuno S, Fujimoto R, Sugimura T, Sato K, Okamoto S, et al. Effects of recombination on hittchhiking diversity in the Brassica self-incompatibility locus complex. Genetics. 2007;177:949–958. doi: 10.1534/genetics.107.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 57.Sherman-Broyles S, Boggs N, Farkas A, Liu P, Vrebalov J, et al. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. The Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Molecular Biology and Evolution. 2006;23:1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- 59.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, et al. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. The Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- 61.Boggs NA, Dwyer KG, Shah P, McCulloch AA, Bechsgaard J, et al. Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics. 2009;182:1313–1321. doi: 10.1534/genetics.109.102442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo Y, Zhao X, Lanz C, Weigel D. Evolution of the S-locus region in Arabidopsis thaliana relatives. Plant Physiology. 2011 doi: 10.1104/pp.111.174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Llaurens V, Billiard S, Castric V, Vekemans X. Evolution of dominance in sporophytic self-incompatibility systems: I. genetic load and coevolution of levels of dominance in pollen and pistil. Evolution. 2009;63:2427–2437. doi: 10.1111/j.1558-5646.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 64.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 65.Ruggiero MV, Jacquemin B, Castric V, Vekemans X. Hitch-hiking to a locus under balancing selection : high sequence diversity and low population subdivision at the S-locus genomic region in Arabidopsis halleri. Genetical Research. 2007;89:1–13. doi: 10.1017/S0016672307008932. [DOI] [PubMed] [Google Scholar]
- 66.Ross-Ibarra J, Wright SI, Foxe JP, Kawabe A, DeRose-Wilson L, et al. Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoS ONE. 2008;3:e2411. doi: 10.1371/journal.pone.0002411. doi: 10.1371/journal.pone.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagenblad J, Bechsgaard J, Charlesworth D. Linkage Disequilibrium Between Incompatibility Locus Region Genes in the Plant Arabidopsis lyrata. Genetics. 2006;173:1057–1073. doi: 10.1534/genetics.106.055780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vekemans X, Slatkin M. Gene and allelic genealogies at the gametophytic self-incompatibility locus. Genetics. 1994;137:1157–1165. doi: 10.1093/genetics/137.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Vienne DM, Giraud T, Martin OC. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- 70.Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, et al. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature. 2010;464:1342–1346. doi: 10.1038/nature08927. [DOI] [PubMed] [Google Scholar]
- 71.Charlesworth D, Bartolomé C, Schierup MH, Mable BK. Haplotype Structure of the Stigmatic Self-Incompatibility Gene in Natural Populations of Arabidopsis lyrata. Molecular Biology and Evolution. 2003;20:1741–1753. doi: 10.1093/molbev/msg170. [DOI] [PubMed] [Google Scholar]
- 72.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;25:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toth G, Deak G, Barta E, Kiss G. PLOTREP: a web tool for defragmentation and visual analysis of dispersed genomic repeats. Nucleic Acids Research. 2006;34:W708–W713. doi: 10.1093/nar/gkl263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sampson DR. Equilibrium frequencies of sporophytic self-incompatibility alleles. Canadian Journal of Genetics and Cytology. 1974;16:611–618. [Google Scholar]
- 75.Mable BK, Schierup MH, Charlesworth D. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity. 2003;90:422–431. doi: 10.1038/sj.hdy.6800261. [DOI] [PubMed] [Google Scholar]
- 76.Schierup MH, Bechsgaard J, Christiansen FB. Selection at work in self-incompatible Arabidopsis lyrata. II. spatial distribution of S haplotypes in Iceland. Genetics. 2008;180:1051–1059. doi: 10.1534/genetics.108.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rahman MH, Suwabe K, Kohori J, Tomita RN, Kakeda K, et al. Physical size of the S locus region defined by genetic recombination and genome sequencing in Ipomoea trifida, Convolvulaceae. Sexual Plant Reproduction. 2007;20:63–72. [Google Scholar]
- 78.Ushijima K, Sassa H, Tamura M, Kusaba M, Tao R, et al. Characterization of the S-locus region of almond (Prunus dulcis): analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics. 2001;158:379–386. doi: 10.1093/genetics/158.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, Wang F, Ma W, Zhang Y, Han B, et al. Structural and transcriptional analysis of S-locus F-box genes in Antirrhinum. Sexual Plant Reproduction. 2003;16:165–177. [Google Scholar]
- 80.Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, et al. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryotic Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferris PJ, Armbrust EV, Goodenough UW. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics. 2002;160:181–200. doi: 10.1093/genetics/160.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boggs NA, Nasrallah JB, Nasrallah ME. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000426. doi: 10.1371/journal.pgen.1000426. doi: 10.1371/journal.pgen.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gotoh O. An improved algorithm for matching biological sequences. Journal of Molecular Biology. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 84.Edh K, Widen B, Ceplitis A. The evolution and diversification of S-locus haplotypes in the Brassicaceae family. Genetics. 2009;181:977–984. doi: 10.1534/genetics.108.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hall C, Welch J, Kowbel DJ, Glass NL. Evolution and diversity of a fungal self/nonself recognition locus. PLoS ONE. 2010;5:e14055. doi: 10.1371/journal.pone.0014055. doi: 10.1371/journal.pone.0014055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fobis-Loisy I, Miege C, Gaude T. Molecular evolution of the S locus controlling mating in the Brassicaceae. Plant Biology. 2004;6:109–118. doi: 10.1055/s-2004-817804. [DOI] [PubMed] [Google Scholar]
- 87.Lemaitre C, Braga MDV, Gautier C, Sagot MF, Tannier E, et al. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biology and Evolution. 2009;1:56–66. doi: 10.1093/gbe/evp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bakkeren G, Jiang G, Warren RL, Butterfield Y, Shin H, et al. Mating factor linkage and genome evolution in basidiomycetes pathogens of cereals. Fungal Genetics and Biology. 2006;4:655–666. doi: 10.1016/j.fgb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 89.Wright SI, Agrawal N, Bureau TE. Effects of recombinaison rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Research. 2003;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schierup MH, Vekemans X. Genomic consequences of selection on self-incompatibility genes. Current Opinion in Plant Biology. 2008;11:116–122. doi: 10.1016/j.pbi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 91.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 92.Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH. Construction of plant bacterial artificial chromosome (BAC) libraries: An illustrated guide. Journal of Agricultural Genomics. 2000;5 [Google Scholar]
- 93.Gonthier L, Bellec A, Blassiau C, Prat E, Helmstetter N, et al. Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae). BMC Research Notes. 2010;3:225. doi: 10.1186/1756-0500-3-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chalhoub B, Belcram H, Caboche M. Efficient cloning of plant genomes into bacterial artificial chromosome (BAC) libraries with larger and more uniform insert size. Plant Biotechnology Journal. 2004;2:181–188. doi: 10.1111/j.1467-7652.2004.00065.x. [DOI] [PubMed] [Google Scholar]
- 95.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz SMS, editor. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. [DOI] [PubMed] [Google Scholar]
- 96.Salamov A, Solovyev V. Ab initio gene finding in Drosophila genomic DNA. Genome Research. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. Journal of Molecular Biology. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 98.Gish W, States DJ. Identification of protein coding regions by database similarity search. Nature Genetics. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 99.Gotoh O. Direct mapping and alignment of protein sequences onto genomic sequence. Bioinformatics. 2008;24:2438–2444. doi: 10.1093/bioinformatics/btn460. [DOI] [PubMed] [Google Scholar]
- 100.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and Genome Research. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 101.Brudno M, Poliakov A, Do CB, Dubchak I, Batzoglou S. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2003;19S1:i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- 102.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using likelihood, distance, and parsimony methods. Molecular Biology and Evolution. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 105.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence conservation at the S-locus boundaries between Al13 (the reference A. lyrata genome) and each of the other haplotypes. Sequences not available for the U-box side (Ah03 and Ah43) were not represented. Distance from the homology breakpoint is indicated under each graph.
(PDF)
Synonymous nucleotide diversity (ΠS) at S-locus flanking genes for A. halleri (black) and A. lyrata (gray), estimated using DnaSP [105].
(PDF)
Phylogeny of pseudo-ARK3 sequences, SRK and ARK3. Phylogeny was constructed using a Minimum Evolution analysis.
(PDF)
Separate phylogenies of the S-locus Region genes. Phylogenies were obtained by the Minimum Evolution method, and are based on protein sequences, with the A. thaliana reference sequences (Col-0) as outgroup.
(PDF)
Sequence conservation in the SRK-ARK3 region between Ah28 (Class II) and each of the other haplotypes. Distance from homology breakpoint is indicated under the graph.
(PDF)
Annotation of genes and transposable elements for the 12 S-haplotypes. The S-locus genes are represented in black rectangles, with delimitation of their exons. Other genes are depicted in light gray. Transposable elements are shown in dark gray, and their fragmentation is indicated by white gaps.
(PDF)
TE density along A. lyrata chromosomes 1 to 6 and chromosome 8. Transposable elements contents were calculated using CENSOR [72] for non overlapping windows of 100 kb.
(PDF)
Comparative density in different families of transposable elements for the entire genome of A. lyrata, and the S-locus of A. lyrata and A. halleri. Transposable elements classification refers to Wicker et al. [104].
(PDF)
Description of the different clones. Two clones were necessary to cover the entire S-locus for three haplotypes : Al18, Ah28 and Ah15.
(DOC)
List of the transposable elements detected in the BAC sequences.
(DOC)
Primers pairs used to validate BAC clones by PCR amplification. These primers were defined to amplify SRK (primer Sh04), U-box (primer B80) and ARK3 (primer ARK3) genes.
(DOC)