Abstract
The solution structure of human U1 snRNA was investigated by using base-specific chemical probes (dimethylsulfate, carbodiimide, diethylpyrocarbonate) and RNase V1. Chemical reagents were employed under various conditions of salt and temperature and allowed information at the Watson-Crick base-pairing positions to be obtained for 66% of the U1 snRNA bases. Double-stranded or stacked regions were examined with RNase V1. The dat gained from these experiments extend and support the previous 2D model for U1snRNA. However, to elucidate some aspects of the solution data that could not be accounted for by the secondary structure model, the information gathered from structure probing was used to provide the experimental basis required to construct and to test a tertiary structure model by computer graphics modeling. As a result, U1 snRNA is shown to adopt an asymmetrical X-shape that is formed by two helical domains, each one being generated by coaxial stacking of helices at the U1 snRNA cruciform. Chemical reactivities and model building show that a few nucleotides, previously proposed to be unpaired, can form A.G and U.U non Watson-Crick base-pairs, notably in stem-loop B. The structural model we propose for regions G12 to A124 integrates stereochemical constraints and is based both on solution structure data and sequence comparisons between U1 snRNAs.
Full text
PDF


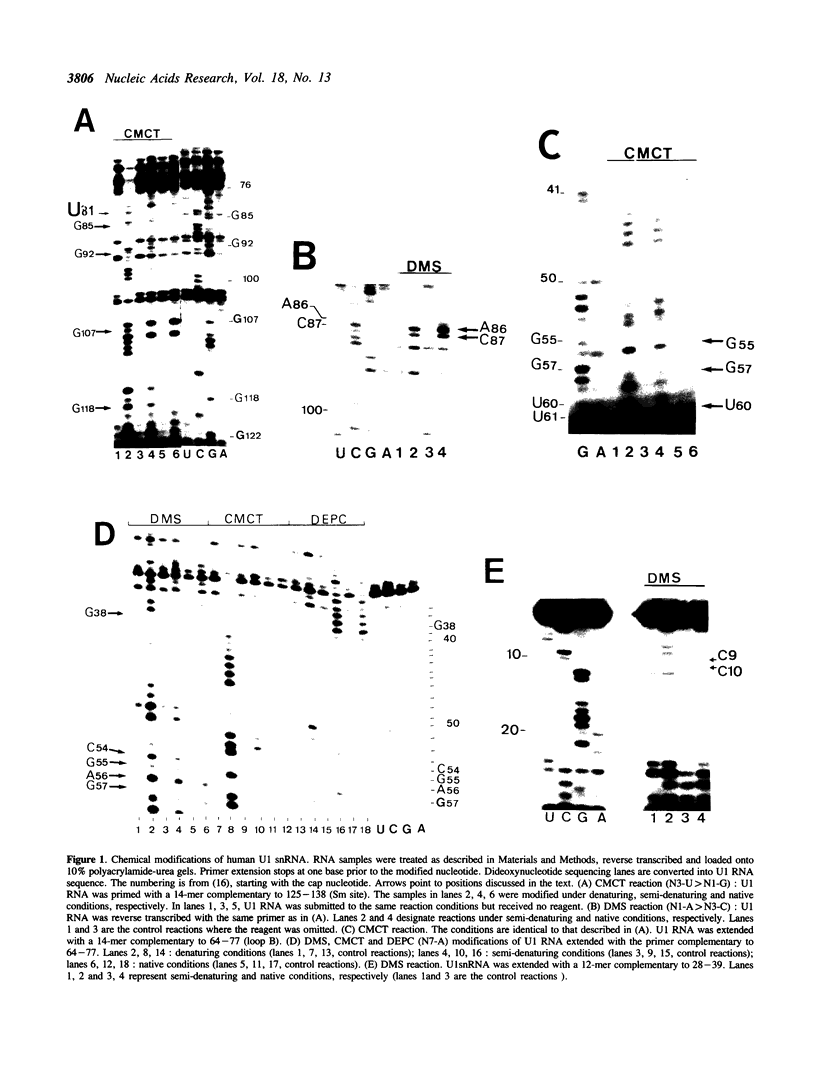

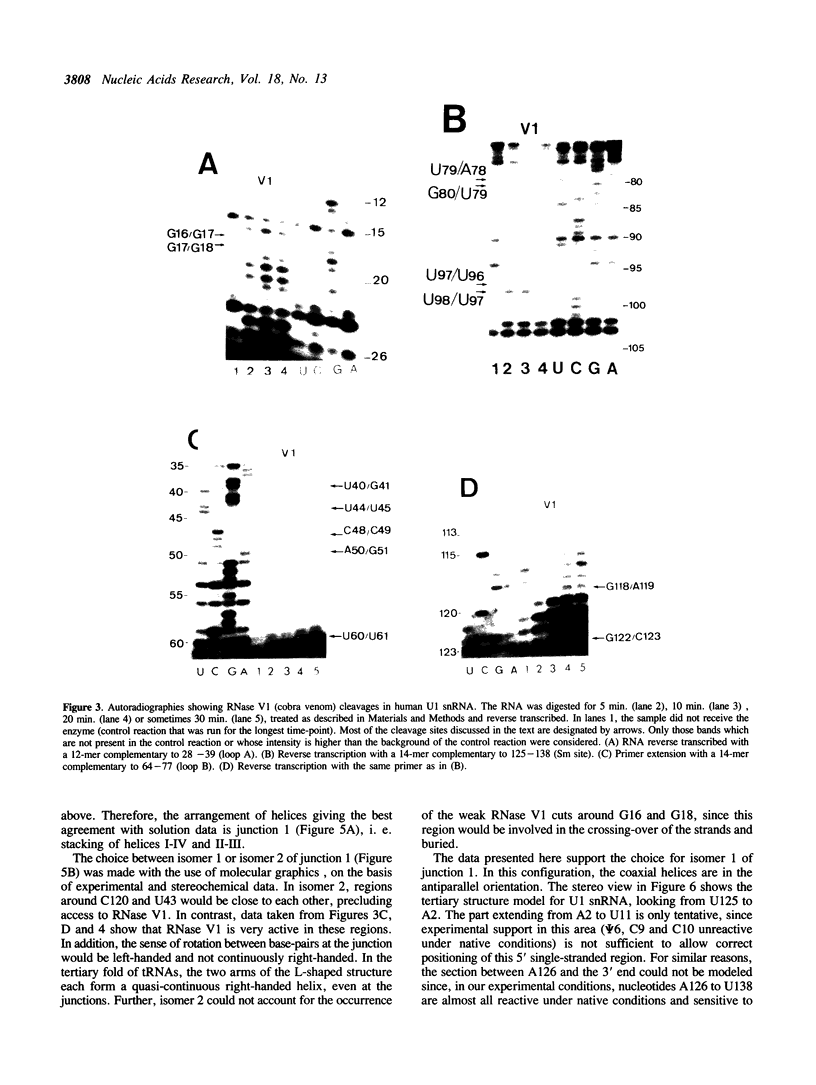
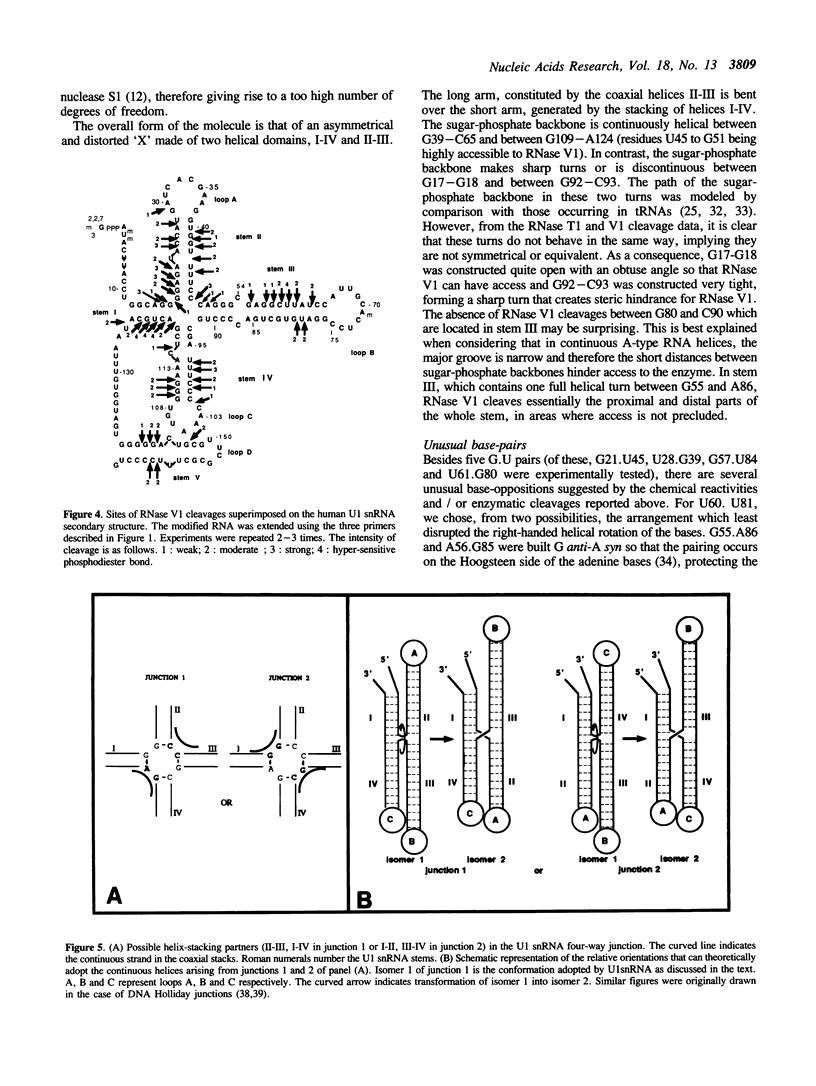
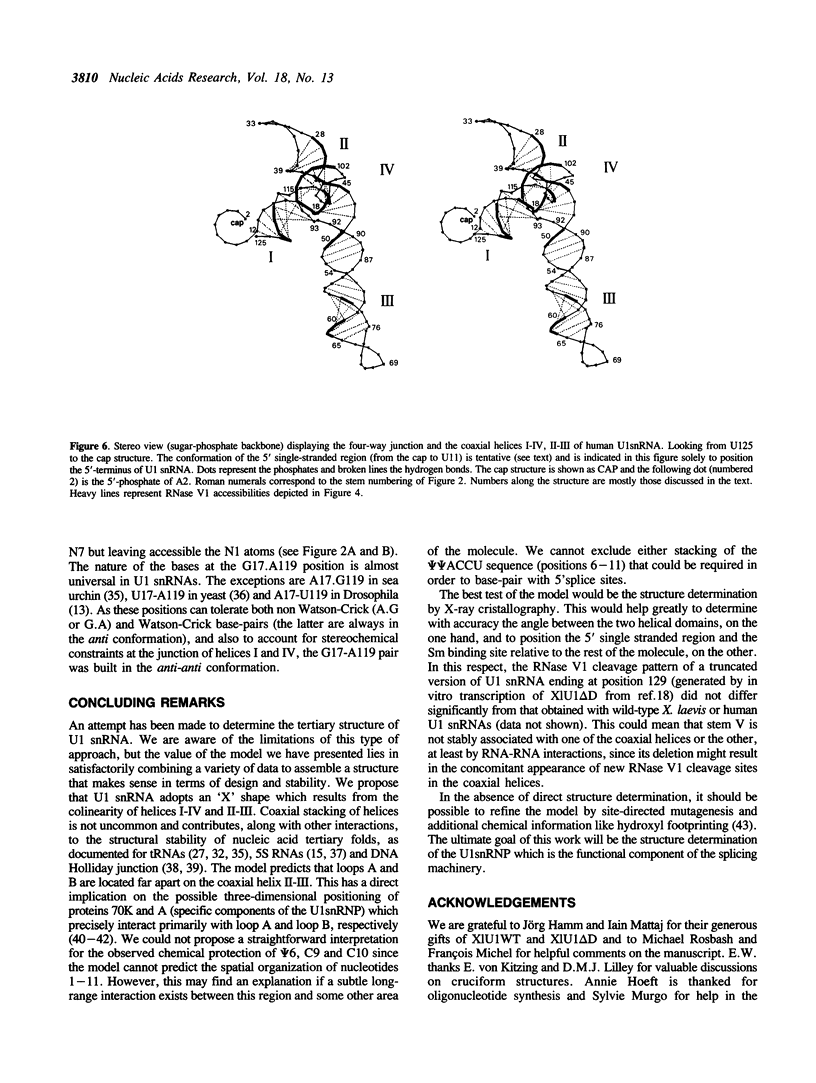

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auron P. E., Weber L. D., Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982 Sep 14;21(19):4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- Bach M., Krol A., Lührmann R. Structure-probing of U1 snRNPs gradually depleted of the U1-specific proteins A, C and 70k. Evidence that A interacts differentially with developmentally regulated mouse U1 snRNA variants. Nucleic Acids Res. 1990 Feb 11;18(3):449–457. doi: 10.1093/nar/18.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochnig P., Reuter R., Bringmann P., Lührmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987 Oct 15;168(2):461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Ebel J. P., Gallinaro H., Lazar E., Jacob M. The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res. 1981 Feb 25;9(4):841–858. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990 Feb 13;29(6):1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- Christiansen J., Brown R. S., Sproat B. S., Garrett R. A. Xenopus transcription factor IIIA binds primarily at junctions between double helical stems and internal loops in oocyte 5S RNA. EMBO J. 1987 Feb;6(2):453–460. doi: 10.1002/j.1460-2075.1987.tb04775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Westhof E., Giegé R., Moras D. Solution structure of a tRNA with a large variable region: yeast tRNASer. J Mol Biol. 1989 Apr 20;206(4):707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hamm J., Kazmaier M., Mattaj I. W. In vitro assembly of U1 snRNPs. EMBO J. 1987 Nov;6(11):3479–3485. doi: 10.1002/j.1460-2075.1987.tb02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Lauble H., Frank R., Blöcker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987 Nov 25;15(22):9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Kim S. H. Correlation between chemical modification and surface accessibility in yeast phenylalanine transfer RNA. Biopolymers. 1983 Apr;22(4):1145–1166. doi: 10.1002/bip.360220410. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Kastner B., Lührmann R. Electron microscopy of U1 small nuclear ribonucleoprotein particles: shape of the particle and position of the 5' RNA terminus. EMBO J. 1989 Jan;8(1):277–286. doi: 10.1002/j.1460-2075.1989.tb03374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Krol A., Rosbash M. Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains. Proc Natl Acad Sci U S A. 1990 Jan;87(2):851–855. doi: 10.1073/pnas.87.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A., Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Lepeschkin E., Jones J. L., Rush S., Jones R. E. Local potential gradients as a unifying measure for thresholds of stimulation, standstill, tachyarrhythmia and fibrillation appearing after strong capacitor discharges. Adv Cardiol. 1978;21:268–278. doi: 10.1159/000400463. [DOI] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983 Jun;33(2):509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Steitz J. A. Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981 Dec 11;9(23):6351–6368. doi: 10.1093/nar/9.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Richmond T. J. Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect. J Mol Biol. 1984 Sep 5;178(1):63–89. doi: 10.1016/0022-2836(84)90231-6. [DOI] [PubMed] [Google Scholar]
- Scherly D., Boelens W., van Venrooij W. J., Dathan N. A., Hamm J., Mattaj I. W. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J. 1989 Dec 20;8(13):4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Sinha N. D., Biernat J., McManus J., Köster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984 Jun 11;12(11):4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub W., Sussman J. L. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res. 1982 Apr 24;10(8):2701–2708. doi: 10.1093/nar/10.8.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Yu J. C., Nash M. A., Santiago C., Marzluff W. F. Structure and expression of a second sea urchin U1 RNA gene repeat. Nucleic Acids Res. 1986 Dec 22;14(24):9977–9988. doi: 10.1093/nar/14.24.9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapp M. L., Berget S. M. Evidence for nuclear factors involved in recognition of 5' splice sites. Nucleic Acids Res. 1989 Apr 11;17(7):2655–2674. doi: 10.1093/nar/17.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]




