Abstract
Sucrose is known to repress the translation of Arabidopsis thaliana AtbZIP11 transcript which encodes a protein belonging to the group of S (S - stands for small) basic region-leucine zipper (bZIP)-type transcription factor. This repression is called sucrose-induced repression of translation (SIRT). It is mediated through the sucrose-controlled upstream open reading frame (SC-uORF) found in the AtbZIP11 transcript. The SIRT is reported for 4 other genes belonging to the group of S bZIP in Arabidopsis. Tobacco tbz17 is phylogenetically closely related to AtbZIP11 and carries a putative SC-uORF in its 5′-leader region. Here we demonstrate that tbz17 exhibits SIRT mediated by its SC-uORF in a manner similar to genes belonging to the S bZIP group of the Arabidopsis genus. Furthermore, constitutive transgenic expression of tbz17 lacking its 5′-leader region containing the SC-uORF leads to production of tobacco plants with thicker leaves composed of enlarged cells with 3–4 times higher sucrose content compared to wild type plants. Our finding provides a novel strategy to generate plants with high sucrose content.
Introduction
Growing world population in combination with climate change demand higher productivity and alternative resources for the production of food and biofuels in the future. To meet these needs agriculturally two approaches have been developed: i) extensive yield oriented selection of important traits; ii) molecular breeding for production of stress-resistant phenotypes to abiotic and biotic factors [1]. Yet another way to gain more exploitable biomass that can be used to obtain ethanol by fermentation, might be by maximizing the conversion efficiency of solar energy to soluble sugars such as sucrose [2].
Sucrose plays a crucial role not only in carbon and energy metabolism as one of the primary end products of photosynthesis but also as a signaling molecule [3], [4], [5], [6]. Smeekens and his colleagues [7] reported that sucrose negatively controls the translation of AtbZIP11 main open reading frame (ORF) which encodes a basic leucine zipper (bZIP) type-transcription factor belonging to the group of S (S - stands for small) family in Arabidopsis thaliana [8]. This post-transcriptional control is termed sucrose-induced repression of translation (SIRT) [9]. The SIRT is mediated through one of the upstream ORFs (uORFs) found in the unusually long 5′-leader region of AtbZIP11 transcript. This uORF exhibits high identity to uORFs found in 4 other genes (AtbZIP1, -2, -44 and -53) of the Arabidopsis S bZIP group [10], thus it is called the sucrose controlled upstream open reading frame (SC-uORF).
We have been studying bZIP homologues in various plant species and termed them as lip19 subfamily of the bZIP gene family. The lip19 subfamily members are phylogenetically closely related to Arabidopsis group S bZIP members [11]. They are upregulated in various abiotic stress conditions: i.e., rice lip19 and maize mlip15 in low temperature stress [12], [13], [14], and Nicotiana tabacum tbz17 responds to low temperature and salt stresses [15]. Upregulation of tbz17 in senescing leaves was also reported [16]. In these conditions, energy and hence sucrose levels decrease abruptly to limited levels, which might lead to energy deprivation condition. In Arabidopsis, it has been shown that low energy condition triggers changes in the expression of various genes via activation of KIN10/KIN11 kinases followed by the activation of group S-bZIP transcription factor genes including AtbZIP11 and AtbZIP53 [17]. AtbZIP11 and AtbZIP53 were well studied, and asparagine (Asn) synthetase gene {ASN1, also known as dark-inducible 6 (DIN6) [18]} and/or proline (Pro) dehydrogenase (PDH) were identified as their targets [19], [20], [21]. However, the relationship between the group S-bZIP transcription factors and endogenous sucrose has not been studied yet.
In this study we selected N. tabacum tbz17 along with Arabidopsis AtbZIP53 genes and, showed SIRT in tbz17 for the first time. Additionally, SIRT in AtbZIP53 was also confirmed by our experiments (Figure S1). Then we revealed the crucial roles of TBZ17 and AtBZIP53 to control endogenous sucrose through the generation of SIRT-insensitive tbz17- and AtbZIP53- overexpressing plants. Based on the results, we proposed a novel strategy to generate plants with enhanced endogenous sucrose levels.
Results
SIRT in Tobacco tbz17
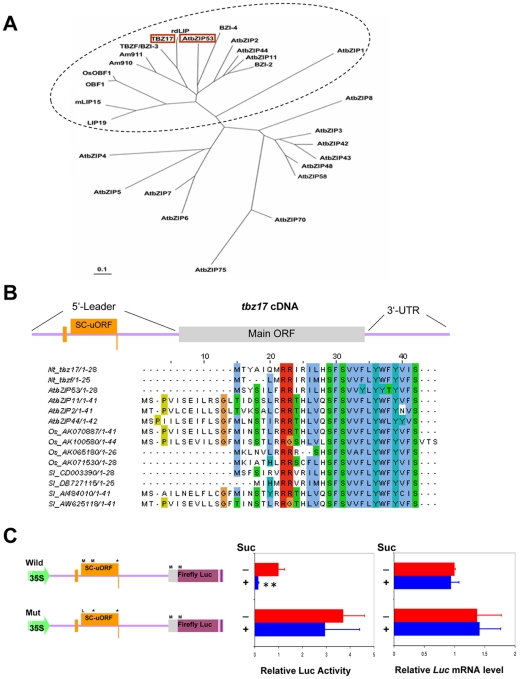
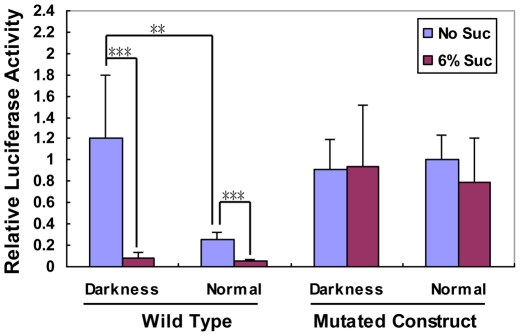
The lip19 subfamily members which include tobacco tbz17, are phylogenetically closely related to the Arabidopsis group S of bZIP genes carrying SC-uORFs (Figure 1A) [11]. tbz17 cDNA contains 3 uORFs in its 5′-leader of which the second uORF shows high identity to the SC-uORF [15] (Figure 1B, Figure S2). To address the question of whether tbz17 has a SIRT mechanism, we firstly performed a transient assay using Arabidopsis mature rosette leaves and a luciferase (LUC) reporter gene construct. In the wild type (Wild) construct, the LUC activity was profoundly inhibited by the presence of 6% sucrose (Figure 1C), whereas, in the mutated construct (Mut), in which the start codon and the second Met codon of the SC-uORF were changed to Leu codon (TTG) and stop codon (TAA), respectively, the LUC activity was not repressed by sucrose (Figure 1C). The result indicated that SIRT mediated by the SC-uORF exists in tbz17. Next we assayed the relative LUC activity in Arabidopsis rosette leaves that have been exposed to different light conditions; one was 4 h-light in a 16 h light/8 h dark photocycle (normal condition) and the other was complete darkness for 44 h (dark condition, see Materials and Methods). In dark condition, the LUC activity with Wild construct was almost comparable to the one with Mut, while, in normal light condition, the LUC activity with Wild construct was ca. 75% lower compared to that with Mut construct (Figure 2). It suggests that the translation of tbz17 main ORF is controlled by endogenous sucrose levels. SIRT in tbz17 was further confirmed by a transgenic approach. We generated transgenic tobacco plants which carried the CaMV 35S promoter-driven tbz17 intact 5′-leader sequence translationally-fused to a GUS reporter gene. Histochemical GUS staining in two independent transgenic lines (#1–1 and #3–1) was specifically repressed by sucrose in a dose-dependent manner but not by glucose or by fructose (Figure S3B). In those seedlings, the GUS transcripts were detected at the similar levels whatever sugars were present, while the GUS protein levels were decreased in the presence of sucrose (Figure S3C, D). The evidence indicates that SIRT functions in tobacco tbz17 through the SC-uORF, suggesting that SIRT is a common phenomenon in plants.
Figure 1. SIRT is found in tobacco tbz17 gene.
A, Phylogenetic relationship between 17 bZIP proteins of Arabidopsis class S [7] and LIP19 subfamily members including tobacco TBZ17. LIP19 subfamily is indicated by dotted-circle line and TBZ17 and AtbZIP53 are highlighted. The amino acid sequence alignment was constructed by the ClustalW program and the relationship was visualized by TREEVIEW program [39]. AtbZIP1 (At5g49450), AtbZIP2 (GBF5, At2g18160), AtbZIP11 (ATB2, At4g34590), AtbZIP44 (At1g75390), AtbZIP53 (At3g62420), Am910 (Y13675), Am911 (Y13676), BZI-2 (AY045570), BZI-4 (AY045572), LIP19 (X57325), mLIP15 (D26563), OBF1 (X62745), rdLIP (AB015187), TBZ17 (D63951), TBZF (identical to BZI-3, AB032478), OsOBF1 (AB185280). B, Structure of tobacco tbz17 cDNA and its evolutionary conserved uORF. The first, second and third uORFs in the 5′-leader region are positioned in the second-, first- and third-frames of tbz17 cDNA, respectively. The second uORF, here called SC-uORF, is highly conserved. The predicted amino acid sequence encoded by tbz17 SC-uORF is aligned with those of the other group S bZIP-encoded cDNAs. Nt, Nicotiana tabacum; At, Arabidopsis thaliana; Os, Oryza sativa; Sl, Solanum lycopersicum. C, SIRT found in tbz17 is mediated by its SC-uORF. The constructs used are as follows; Wild: the intact tbz17 5′-leader sequence (+1 to +358 of tbz17 cDNA) was inserted between the CaMV 35S promoter and the firefly luciferase (LUC) gene. Mut: the start codon (ATG) of SC-uORF is mutated to Leu codon (TTG) and the second Met codon (ATG) is replaced by stop codon (TAA) by site-directed mutagenesis. The constructs were delivered into Arabidopsis thaliana rosette leaves (collected from 3 week-old-seedlings). The bombarded leaves were incubated in half-strength MS media for 20 h with or without 6% sucrose. The relative LUC mRNA levels and LUC activities were analyzed. The error bar represents the SD. Asterisk indicates significant differences that were observed due to sucrose treatment (Student’s t-test: **P<0.01).
Figure 2. Wild type SC-uORF supresses the activity of Luc reporter in leaves kept in normal light condition but not in the dark condition.
The mutated SC-uORF lost such regulation ability. Leaves of intact Arabidopsis plants that were exposed to normal light regime or kept in darkness for 24 h, respectively, were detached and bombarded with the corresponding SC-uORF luciferase constructs, WT construct and mutated one (shown in Fig. 1C). The detached leaves were incubated in half-strength MS solution with or without containing 6% sucrose for 20 h in darkness and then the Luc activity was assayed. The error bar indicates the SD. Asterisk indicates significant difference (Student’s t-test: **P<0.01; ***P<0.001).
Generation of N. tabacum plants overexpressing SIRT-insensitive tbz17
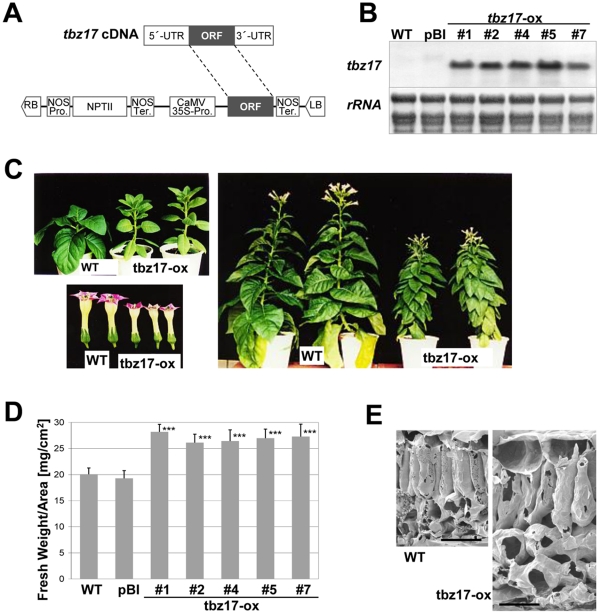
In general, the primary role of highly conserved uORFs seems to be translational regulation of the downstream main ORFs mediated through specific metabolites in a feedback manner: arginine [22], polyamine [23], [24], [25] and choline [26]. These facts inspired us to examine whether TBZ17 is involved in sucrose metabolism. For that purpose, we generated transgenic tobacco plants constitutively expressing the tbz17 ORF lacking the 5′-leader containing the SC-uORF under the control of the CaMV 35S promoter (Figure 3A). Translation of the transgene-derived tbz17 may be SIRT-insensitive in these transgenic plants which are referred as tbz17-ox lines. The constitutive expression of transgene-derived tbz17 was confirmed in 5 independent lines by RNA blot hybridization (Figure 3B). The tbz17-ox plants showed a clear phenotype of smaller and pale-green leaves in younger stage and their vegetative growth was slightly slower compared to wild type (WT)- and the control transgenic (pBI) plants. The sizes of flowers of the tbz17-ox plants were also smaller compared to WT plants (Figure 3C); however, they set fertile seeds. Another prominent feature of the tbz17-ox plants was increased fresh weight per leaf area (cm2) (Figure 3D), suggesting that tbz17-ox plants have thicker leaves compared to WT and pBI. This postulation was based on the evidence from cryo-scanning electron microscopy method (Figure 3E). Leaves of tbz17-ox plants were about 1.5-fold thicker than WT and pBI. Both mesophyll- and parenchyma-cells were enlarged in tbz17-ox plant leaves.
Figure 3. Generation of SIRT-insensitive tbz17-ox tobacco plants.
A, Structure of the binary vector construct lacking the 5′-leader region required for translational repression by sucrose. tbz17 main ORF is inserted into pBI121 vector (see Materials and Methods). B, Expression of tbz17 in five independent transgenic tobacco lines. Total leaf RNA was separated on agarose gels, transferred onto nylon membrane and hybridized with the 32P-labelled tbz17-ORF cDNA-fragment. rRNA stained by methylene blue solution is shown as a loading control. WT, non transgenic tobacco plants; pBI, control transgenic tobacco plants; tbz17-ox, transgenic tobacco plants transformed by the construct shown in A. C, Representative growth phenotypes of tobacco plants overexpressing tbz17. Left upper image, young seedlings; left bottom image, flowers; right image, mature stage of tobacco plants. D, tbz17-ox plants have thicker leaves in relative to WT and the control transgenic plants. Discs from leaves of similar growth stage of WT, pBI and tbz17-ox plants were punched out with a cork borer and their fresh weights were measured. The error bar represents the SD. The differences between WT/pBI and tbz17-ox lines were highly significant as calculated by Students t-test (***P<0.001). E, Vertically dissected leaf images of WT and tbz17-ox plants. Leaf sections were observed by cryo-SEM. Bar = 50 µm.
Sucrose accumulation in tbz17-ox plants
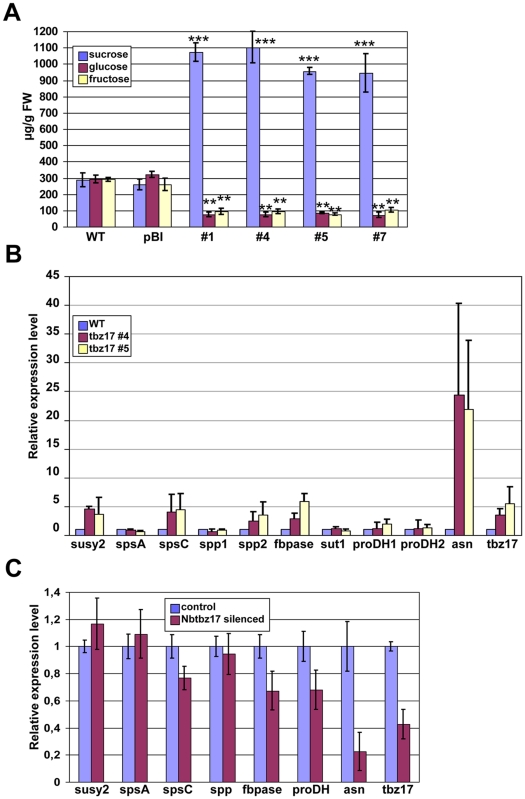
We hypothesized that one possible reason to have enlarged cells was the constitutive accumulation of some compatible solutes and, in this particular case, accumulation of sucrose. Thus, we measured the contents of sucrose, glucose and fructose, in 4 independent tbz17-ox plants and those of WT and pBI. All the tbz17-ox lines contained about 3- to 4-fold higher sucrose content relative to WT and pBI while glucose and fructose contents were reduced in tbz17-ox plants compared to WT and pBI (Figure 4A). Sucrose biosynthesis in higher plants is catalyzed by the sequential reaction of sucrose phosphate synthase (SPS) and sucrose-6′-phosphate phosphatase (SPP). The activity of SPS, a key enzyme of the pathway, is regulated at multiple levels; i.e., feedback regulation by positive or negative allosteric effectors and by phosphorylation [3], [27], [28]. In spite of the fact that sucrose synthesis is highly regulated at the post-translational level, we investigated the expression of genes which are involved in sucrose-metabolism considering that tbz17 encodes a transcription factor. As mentioned earlier, target genes of AtbZIP11 have been identified as ASN1 and PDH2 [19]. One of the target genes of AtbZIP53 is PDH2 [20]. In this context, we included N. tabacum ASN gene (accession number AY061820) and two PDH genes (accession numbers AY639145 & AY639145) in the qRT-PCR analysis of tbz17-ox and wild type plants. As seen in Figure 4B, ASN gene was highly upregulated in the tbz17-ox plants, whereas the expression of two PDH genes was not much changed compared to WT. We could tentatively conclude that one of the TBZ17-target genes is, in fact, ASN. It should be noted that tbz17 expression is senescence-associated [16] and the expression of DIN6, identified as ASN, is darkness-induced and sucrose-repressed [18], [29]. Interestingly, transcripts for fructose 1, 6-bisphosphatase (FBPase) gene, class C sucrose phosphate synthase gene (SPSC) [30], sucrose phosphate phosphatase 2 gene (SPP2) and sucrose synthase 2 gene (SuSy2) accumulated 4- to 6-fold in both lines, #4 and #5, of the tbz17-ox plants compared to those of WT (Figure 4B). To further confirm the correlation between the levels of tbz17 expression and ASN, PDH and sucrose-synthesizing genes, a virus-induced gene-silencing (VIGS) approach was taken. N. benthamiana plants in which Nbtbz17 (tbz17 ortholog of N. benthamiana) was silenced using VIGS method showed about 80% reduced expression of the ASN gene, and ca. 20–30% reduced expression of PDH, FBPase and SPSC genes, respectively (Figure 4C). The results indicated that the transcripts’ levels of ASN and sucrose synthesizing genes, especially FBPase and SPSC genes, positively and tightly correlated with the levels of tbz17 transcripts. In parallel, we generated transgenic Arabidopsis plants overexpressing AtbZIP53 ORF lacking its 5′-leader region (Methods S1). The transgenic Arabidopsis plants generated with this construct are referred as AtbZIP53-ox. Independent lines (#10, #12 and #22) were further analyzed (Figure S4A-C). Sucrose contents in AtbZIP53-ox plants are 1.5- to 2.5- fold higher compared to WT plants and, in contrast, glucose and fructose contents decreased in AtbZIP53-ox (Figure S4D). In relation to this, two SPS genes, class A of SPS1 (At5g20280) and class C of SPS4 (At4g10120), were upregulated in all AtbZIP53-ox plants (Figures S3E and S4). It should be noted that Arabidopsis SPS4 belongs to the same clade to which tobacco SPSC belongs (Figure S5). Chen et al. [30] reported that N. tabacum SPSA and SPSB are expressed in whole plant body and in reproductive organs (anther and ovary), respectively, while SPSC is specifically expressed in source leaves under physiological condition. Therefore, the upregulation of this class C SPS gene may contribute to higher sucrose content. Next question was whether TBZ17 transactivated SPSC gene directly or indirectly. To address this issue, we took an indirect approach and tested whether AtbZIP53 directly transactivates 4 kinds of SPS genes. The results showed that AtbZIP53 transactivates the ASN gene but not all SPS genes (Figure S6), suggesting that upregulation of tobacco SPSC in tbz17-ox plants is a secondary effect.
Figure 4. Sucrose accumulation in tbz17-ox plants and correlation analysis between tbz17 and sucrose synthesis genes.
A, Sugar contents in WT, control transgenic (pBI) and tbz17-ox tobacco plants. The differences in sucrose, glucose and fructose contents between WT/pBI and tbz17-ox lines were highly significant as calculated by Students t-test B, Quanitative real-time RT-PCR analysis on sugar-metabolizing genes in tbz17-ox tobacco plants. C, Quanitative real-time RT-PCR analysis on sugar-metabolizing genes in Nbtbz17-silencing Nicotiana benthamiana. The error bar represents the SD. *P<0.05; **P<0.01; ***P<0.001.
Discussion
We have shown that a tobacco bZIP gene, tbz17, retains SIRT mechanism mediated by a conserved SC-uORF (Figure 1). This is a first report on SIRT beyond Arabidopsis genus, suggesting that SIRT is a common phenomenon in higher plants. Furthermore, the translation of tbz17 main ORF was decreased to ca. one-forth in the leaves exposed to normal light condition compared to the one of the leaves placed under complete darkness (Figure 2). This difference was not observed with the SC-uORF-mutated construct (Figure 2). Thus, the reasons for the difference in translation might be due to elevated endogenous sucrose levels causing SIRT, suggesting that sucrose is a physiological effector to control the translation of tbz17 main ORF.
Hanfrey et al. [25] revealed that the sequence conserved uORFs control the translation of the main ORF encoding S-adenosylmethionine decarboxylase (SAMDC) in response to cellular polyamine contents in Arabidopsis. In this post-transcriptional regulation system, the uORFs are polyamine-sensors and the system contributes to polyamine homeostasis in the cells. Similarly to this feedback mechanism, it is likely that the SC-uORF functions as a sucrose-sensor and that SIRT contributes to sucrose homeostasis. To assess this hypothesis, we introduced the SIRT-insensitive tbz17 construct, deleting its 5′-leader region containing SC-uORF, into tobacco plants. The resulting tobacco plants had thicker leaves composed of enlarged cells (Figure 3). In de-regulated condition of SIRT (at least for the introduced ‘tbz17’ gene), the transgenic tobacco plants contained 3- to 4-fold higher sucrose in the cells (Figure 4A). Taking a similar strategy for AtbZIP53, the transgenic Arabidopsis contained 1.5- to 2.5-fold higher sucrose (Figure S4). These combined results support our hypothesis.
Arabidopsis SnRK1 (SNF1-related protein kinase 1) -like kinases, KIN10 and KIN11, function as central signal integrators for adapting to low energy condition such as darkness, low sugar and stress conditions [17]. The KIN10 kinase-signal pathway was mediated by a specific subset of group S bZIPs including bZIP53, and activate DIN6 ( = ASN1) transcription. ASN1 transcriptional activation was blocked by sugars [18]; i.e., sucrose and glucose because those sugars inhibited KIN10/KIN11 activation [17]. To superimpose the KIN10 kinase signal pathway to tobacco plant, the order of signaling is predicted to be: KIN10-like SnRK1 kinase(s) – TBZ17 – ASN and/or PDH. In fact, the transcription of ASN but not PDH was positively correlated with the levels of tbz17 transcripts in tbz17-ox plants and in Nbtbz17-silenced N. benthamiana plants (Figures 4B, C). In addition, the transcription of FBPase and class C SPS genes was correlated with the levels of tbz17 transcripts. Even in AtbZIP53-ox plants, classes A and C SPS genes were upregulated (Figures S4, S5). We predict that the enhanced transcription of the class C member of SPS gene family contributes to sucrose accumulation. The transcription assay shows that AtbZIP53 transactivates ASN but not all of SPS genes (Figure S6), suggesting that the transcript accumulation of classes A and C SPS genes in AtbZIP53-ox plants occurred in an indirect manner. Analysis of the global gene expression regulated by KIN10 showed that it controls divergent metabolic reprogramming in promoting catabolic processes and suppressing anabolic processes [17]. Recently, Dietrich et al. [31] showed that the heterodimer composed of AtbZIP1 and AtbZIP53 directly binds to the G-box-like sequence of the promoters of ASN1 and PDH genes and causes the changes in Pro, Asn and branched-chain amino acid metabolism to adapt to low energy stress. Taking into account all the information, we propose a model for explaining sucrose accumulation in tbz17-ox plants (Figure 5); in wild tobacco plant cells, TBZ17 transactivates ASN and induces metabolic reprogramming, which turns into activation of the ‘sucrose synthesis pathway’, and if the end product sucrose reaches a upper threshold, SIRT is operated and suppresses the translation of TBZ17 in a feedback regulation. This sucrose-sensing circuit regulates a certain range of sucrose content in the cells. In contrast, in SIRT-insensitive tbz17-ox plants, even if sucrose contents reach an upper level, SIRT does not operate and thus the TBZ17 translation continues. Notable modification of SPS is reversible phosphorylation that inactivates the enzyme [27], [28]. In spinach, serine-158 of SPS is phosphorylated by SnRK1 kinase(s). It was shown that KIN10 is able to phosphorylate SPS at serine-158 in vitro, while KIN10 was inhibited by high concentrations of sucrose [17]. So once cellular sucrose reaches higher levels, KIN10-like SnRK1 kinase(s) may be inactivated and SPS may stay active because the dephosphorylated form is major. A similar trend of sucrose accumulation was observed in AtbZIP53-ox plants, but the level of sucrose accumulation was smaller compared to that of tbz17-ox plants. The upper threshold for sucrose content in Arabidopsis may be lower compared to that of tobacco [32]. We could argue that the introduction of SIRT-insensitive group S bZIP genes leads to sucrose accumulation in the host plant cells.
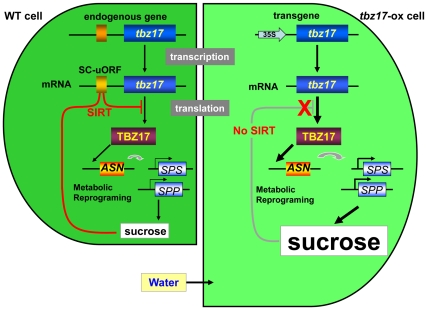
Figure 5. A proposed model to explain how tbz17-ox plant cell accumulates higher sucrose and becomes enlarged.
In WT tobacco cells, when the endogeneous tbz17 gene is transcribed and its transcript contained SC-uORF. The gene product, TBZ17, transactivates NtASN gene and then indirectly activates SPS and SPP genes through metabolic reprogramming, which turns on the sucrose accumulation. If the SC-uORF of tbz17 transcript senses higher sucrose concentration, SIRT is induced, therefore, cellular sucrose concentration is maintained in a certain range. tbz17-ox plant cells (1.5-fold enlarged cell) contain not only the endogenous tbz17 gene but also the transgene `tbz17’ which transcript does not contain SC-uORF. In the latter cells, the gene product TBZ17 directs the transactivation of ASN gene and indirectly upregulates `SPS and SPP’ genes. Increased sucrose concentration represses the translation of endogenous transcript carrying SC-uORF but not the transgene-derived transcript, thus sucrose concentration increased more compared to WT cells. To adjust the high osmotic pressure due to high sucrose concentration, the cell adsorbs more water and become enlarged.
In relation with this, it is worthy to note that transcriptomic analysis using sugarcane (Saccharum officinarum) revealed that the expression of genes encoding sucrose transporter, ornithine aminotransferase (OAT) and ASN is positively correlated with sucrose content [33]. OAT and ASN are involved in Pro and Asn metabolism, respectively. Dehydrin, LEA and PDH transcript levels were also higher in the higher sucrose-contained sugarcane culms. Iskander et al. also used several genotypes of sugarcanes differing in sucrose contents and drew a similar conclusion [33]. While the adaptation mechanism to high sucrose content in sugarcane is still unclear, it should be emphasized that most of the related genes were also upregulated in high sucrose-contained tobacco plants.
To conclude, our finding provides a novel strategy to generate high-content sucrose plants. Genes coding for the type of hypothetical sucrose-sensitive bZIP transcription factors investigated in this work are present in at least 3–4 copies per plant (Figure S2). Theoretically, our proposed strategy might be applicable to the genes belonging to the group of S bZIP genes. Specifically, the similar engineering of rice, lip19, is of interest. Plants transformed with the engineered bZIP gene might be useful as a novel source of biofuel production. Finally, the same strategy of transgenic expression of the bZIP homologue lacking the regulatory 5′-leader but under control of a fruit-specific promoter might result in a production of fruits with enhanced sucrose content. Such an approach is currently under way with tomato plants.
Materials and Methods
Plant materials and growth condition
N. tabacum cv. Xanthi nc and N. benthamiana were grown in soil at 25°C or at 23°C under a 16 h light/8 h dark photocycle, respectively. A. thaliana (ecotype Columbia Col-0) was grown in soil at 22°C under a 16 h light/8 h dark photocycle.
Transient ‘SIRT’ assay
The basal vectors pUC19K and pUC19K-Luc were constructed as follows: Cauliflower mosaic virus (CaMV) 35S promoter- and Nos terminator- fragments derived from pBI221 (Clontech) were subcloned into HindIII-SmaI sites and SacI-EcoRI sites of pUC19, respectively, resulting in pUC19K. Firefly luciferase (LUC) gene sandwiched with SmaI and SacI sites was inserted into the respective sites of pUC19K, yielding pUC19K-Luc. Then, intact 5′- leader sequence of tbz17 cDNA [15] in size of 361 bp was amplified by using a primer pair (forward, 5′-AGCTCTAGATGATCTTTTTTGTTAATACCT-3′ and reverse, 5′-TGACCCGGGAGCCATGTCGATTGATA-3′, the underlined XbaI and SmaI sequences were incorporated for cloning purpose, respectively). The sequence-verified fragment was digested with XbaI and SmaI, and inserted into the corresponding sites of pUC19K-Luc. The resulting plasmid was used as ‘wild type’ reporter construct of 5′-leader of tbz17 (see Figure 1C). The mutant construct was made by inserting point mutations through PCR in which the start codon (ATG) of the 2nd uORF of tbz17 5′-leader sequence was changed to a Leu codon (TTG) and the 2nd methionine codon of the 2nd uORF was changed to a stop codon (TAG). The two reporter constructs were used for transient assay experiments. Gold particles were coated with the reporter plasmid and the reference plasmid pPTRL [34], in which Renilla luciferase (RLUC) gene is placed under the control of the CaMV 35S promoter. Mature rosette leaves of A. thaliana were bombarded with the coated gold particles using a PSD-1000/He Particle Delivery System (Bio-Rad). Transformed leaves were kept on half strength Murashige-Skoog (MS) media supplemented with or without 6% sucrose for 20 h under darkness to avoid fluorescence quenching. Darkness treatment of Arabidopsis leaves was applied as follows: leaves were kept in darkness 24 h prior to bombardment and 20 h after bombardment, then LUC activity was measured. To normalize the efficiency of particle bombardment, relative luciferase (LUC/RLUC) activities were measured according to the protocol of the Dual-Luciferase Reporter Assay System (Promega) by using a luminescence reader (Lumat LB9507, Berthold Japan, Tokyo). The experiment was repeated three times with three replicates per time.
Generation of transgenic tobacco plants overexpressing tbz17
A fragment of the binary vector pBI121 (Clontech), encompassing GUS coding region and Nos terminator, was eliminated by restriction with BamHI and EcoRI and replaced with the BamHI-EcoRI Nos terminator fragment derived from pCaMV-neo (provided by Dr. Virgina Walbot), yielding pBI001. The coding region of tbz17 cDNA [15] was amplified by PCR with a primer pair (tbz17-F, 5′- GCGGATCCATGGCTTCCACTCAGCAAGC-3′ and tbz17-R, 5′-CGGGATCCTCAAAACAGCAACATATCAGAAG-3′), where BamHI sites are shown as underlined. The BamHI-digested fragment was inserted into pBI001, and the recombinant with sense orientation of tbz17-main ORF was named pBI-tbz17. tbz17-overexpressing tobacco plants were generated by infection with Agrobacterium tumefaciens strain LBA4404 [35] carrying pBI-tbz17.
Northern blot analysis
Total RNAs were isolated according to the method of Nagy et al. [36]. Aliquots (20 µg each) were separated by electrophoresis on formaldehyde-1.0% (w/v) agarose gels and blotted onto Hybond N+ membranes (GE Healthcare) in 20×SSC. The 32P labelled-fragment covering the main ORF of tbz17 cDNA or AtbZIP53 cDNA, respectively, was used as a probe. Hybridization was performed as described previously [37].
Cryo-scanning electron microscopy
For scanning-electron microscopy (SEM) leaves of tobacco plants were frozen in liquid nitrogen and subsequently freeze-dried. Sections of ca. 2 mm edge length were cut, placed on a carbon grid and sputtered for 5 min with gold. Examinations were performed with a Hitachi S-4500 scanning electron microscope.
Determination of sugar contents in plants
The contents of sucrose, glucose and fructose were determined by enzyme-coupled reactions, based on the measurement of NADPH absorption at 340 nm, using the Sucrose/D-Glucose/D-Fructose kit (r-biopharm, Darmstadt, Germany), as described by the manufacturer. Fresh leaf material was ground with a mortar and a pestle under liquid nitrogen to fine powder and 200 mg of the powder was weighed into a microcentrifuge tube, briefly homogenized with 600 µl of distilled water and immediately boiled for 10 min in a water bath. After centrifugation (20,000 × g, 10 min at 4°C), 100 µl of the supernatant was used in the assay with a spectrophotometer (Hitachi U-2900).
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) analysis
Quantitative real-time RT-PCR was performed with FastStart Universal SYBR Green Master (ROX) (Roche Applied Science, Mannheim, Germany). First-strand cDNA was synthesized with Rever Tra Ace (Toyobo Co. Ltd., Osaka, Japan) and oligo-dT primers. The subsequent quantitative PCR was performed in a StepOne real-time PCR system (Life Technologies Japan, Tokyo, Japan) using the appropriate primer pairs (see Table S1). The steady state levels of the transcripts were determined in relative to the levels of the transcripts of a housekeeping gene encoding ribosomal protein L-25 as an internal control. All quantitative RT-PCR experiments were performed with biologically independent samples at least three times.
Virus-induced gene silencing (VIGS) in N. benthamiana
For VIGS, the method described was employed [38]. The fragment of Nbtbz17 cDNA (tbz17 ortholog of N. benthamiana) was amplified with the primer pair (forward, 5′-ATGAGCTCCGTATTCTGCACTCTTTCTCAGTA-3′ and reverse, 5′-TTTCTAGAGTTCAATGAATTCAAACGTTCAGT-3′, SacI and XbaI sites were underlined, respectively). The resulting 500-bp-fragment was cloned into the pTRV2 vector. Agrobacterium cultures containing pTRV1, and pTRV2 or its derivative plasmid were similarly mixed in a 1∶1 ratio, and were infiltrated into the lower leaf of 4-leaf stage plants using a needle-less syringe.
Statistical analysis
The data analysis was performed using the statistical tools (Student’s t test) of Microsoft Excel software.
Supporting Information
Confirmation of SIRT in Arabidopsis AtbZIP53 cDNA. A, Schematic drawing of the cloned fragment in binary vector construct for transformation. AtbZIP53 genome DNA fragment, spanning from -919 to +552, was inserted into PstI and BamHI-digested pBI101 vector (Clontech), yielding pAtZIP53G. The recombinant was a GUS-translational fusion construct. B, Histochemical staining of the AtbZIP53 promoter-GUS transgenic Arabidopsis seedlings. Among the transgenics, two independent lines were used for assay. 5-day-old Arabidopsis seedlings were incubated with or without 20 mM or 100 mM sucrose for 2 days, then rinsed with distilled water twice, and subjected to histochemical staining according to the procedure described by Jefferson (1987). C and D, GUS transcript levels (C) and GUS protein levels (D) in the transgenics incubated with or without 20 mM or 100 mM sucrose. Methylene blue-stained rRNA (C) and CBB-stained large subunit (LSU) of RuBisCO (D) were used for loading controls. GUS protein was detected with anti-GUS antibody (abcam, UK).
(TIF)
SC-uORF is highly conserved in higher plants. Almost all members of plants harbor 3–5 members of the group S bZIP genes carrying the highly conserved SC-uORFs per organism. Redundant and non-redundant databases were screened by using tblastn and later analyzed manually. Sequence alignment was carried out by multiple sequence alignment software ClustalW in default parameters and then edited with Jalview editor (http://www.jalview.org/training.html). Abbreviation: Ac - Allium cepa, At - Arabidopsis thaliana, Am - Artemisia amuna, Ah - Arachis hypogaea, Aa - Artemisia amuna, Ao - Asparagus officinalis, Bv - Beta vulgaris, Bn - Brassica napus, Bo - Brassica oleracea, Br - Brassica rapa, Cc - Citrus clementina, Cp - Carica papaya, Ci - Cichorium intybus, Cic - Citrus clementina, Cr - Citrus reticulate, Cs - Citrus sinensis, Coc - Coffea canephora, Et - Eragrostis tef, Ee - Euphorbia escula, Fa - Festuca arundinacea, Fv - Fragaria vesca, Gm - Glycine max, Ga - Gossypium arboretum, Gh - Gossypium hirsutum, Gr - Gossypium raimondi, Ha - Helianthus annuus, Ht - Helianthus tuberosus, Hv - Hordeum vulgare, In - Ipomoea nil, Ls - Lactuca sativa, Lj - Lotus japonicus, Md - Malus x domestica, Me - Manihot esculenta, Mt - Medicago truncatula, Mc - Mesembryanthemum crystallinum, Nt - Nicotiana tabacum, Os - Oryza sativa, Pv - Panicum virgatum, Pc - Phaseolus coccineus, Phv - Phaseolus vulgaris, Pt - Poncirus trifoliate, Pn - Populus nigra, Pot - Populus tremula x Populus tremuloides, Potri - Populus trichocarpa, Pa- Prunus armeniaca, Pd - Prunus dulcis, Pp - Prunus persica, Rr - Raphanus raphanistrum, Rs - Raphanus sativus, Rc - Ricinus communis, So - Saccharum officinarum, Sc - Secale cereale, Sl - Solanum lycopersicum, St - Solanum tuberosum, Sb - Sorghum bicolor, Tc - Theobroma cacao, Tp - Triphysaria pusilla, Tv - Triphysaria versicolor, Ta - Triticum aestivum, Tt - Triticum turgidum, Vu - Vigna unduiculata, Vv - Vitis vinifera, Zm - Zea mays.
(TIF)
Confirmation of SIRT in tobacco tbz17 cDNA. A, Schematic drawing of the cloned fragment in binary vector construct. The tbz17 cDNA fragment (+1 to +358) was placed under the control of CaMV-35S promoter and translationally fused to GUS gene. B, Histochemical staining of the tbz17 5′-leader GUS transgenic plants. Among the tobacco transgenic plants, two independent lines (#1–1 and #3–1) were used for the assay. Two-week-old tobacco seedlings were incubated with or without sucrose, glucose and fructose for 2 days, then rinsed with distilled water twice, and subjected to histochemical staining according to the procedure described by Jefferson [4]. C and D, relative GUS transcript levels (C) and GUS protein levels (D) in the transgenics incubated with or without sugars. PCR amplification of EF-1α cDNA (C) and CBB staining of large subunit (LSU) of RuBisCO (D), respectively, were used for loading controls. GUS protein was detected with anti-GUS antibody (abcam, UK).
(TIF)
Generation of transgenic Arabidopsis plants overexpressing AtbZIP53 . A, AtbZIP53 cDNA and construction of the binary vector, pBI-AtbZIP53. B, Growth phenotype of 6-day-old representative Arabidopsis seedlings of wild-type (WT) and transgenic plants overexpressing AtbZIP53 (AtbZIP53-ox). C, RNA blot hybridization of AtbZIP53 in WT and AtbZIP53-ox plants. AtbZIP53 endogenous-transcripts and the transgene-derived transcripts were indicated by e and t, respectively. The open reading frame (ORF) of AtbZIP53 was used as a hybridization probe. D, Sugar contents in leaves of wild-type plants (WT) and 3 transgenic lines. The error bar represents the SD. The differences in sucrose contents between WT, pBI and AtbZIP53-ox lines were highly significant as calculated by Students t-test (*P<0.05; **P<0.01). E, RT-PCR analysis on 4 kinds of sucrose phosphate synthase (SPS) genes (At5g20280, At5g11110, At1g04920, At4g10120) and FBPase (At1g43670) genes. Tubulin cDNA was amplified as a control.
(TIF)
Phylogenetic analysis of N. tabacum and Arabidopsis SPS protein sequences. An unrooted neighbor-joining tree was constructed. Accession numbers of N. tabacum SPS cDNAs and AGI codes for Arabidopsis sequences are as follows: NtSPSA (AF194022), NtSPSB (DQ213015), NtSPSC (DQ213014), AtSPS1F (At5g20280), AtSPS2F (At5g11110), AtSPS3F (At1g04920), AtSPS4F (At4g10120).
(TIF)
AtbZIP53 transactivates ASN1 gene but not 4 kinds of SPS genes. A, Schematic drawing of two effectors and 5 reporter plasmids. Those constructs were generated using the primers shown in Table S2. Horizontal short bars inside the 700-bp promoter fragments indicate the transcriptional start sites. B, Transactivation activity assays of AtbZIP53. Effector, reporter and Renilla LUC reference plasmids were co-bombarded into Arabidopsis mature rosette leaves by a particle delivery system (PDS-1000 He, Bio-Rad, Hercules, CA). After 18 h of incubation at 23oC under darkness, relative luciferase (LUC/RLUC) activities were determined. The values obtained from three independent experiments in duplicate assays were calculated with the means+SD. **P<0.01.
(TIF)
The primers used in this study.
(RTF)
The primers used for transactivation assay.
(DOC)
Supplemental Methods. Generation of transgenic Arabidopsis plants overexpressing AtbZIP53.
(RTF)
Acknowledgments
We thank S.P. Dinesh-Kumar, D. Baulcombe and V. Walbot for providing plasmids and bacterial strain. The initial stage of this work was performed by Kazuyuki Sugawara. We thank Anthony J. Michael and Elena Herzog for critically reading the manuscript. S.K.T. is a recipient of MEXT fellowship.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by Grant-in-Aids from the Japan Society for the Promotion of Science to TK (19658039), the research funding programme “LOEWE - Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz” of Hesse’s Ministry of Higher Education, Research, and the Arts to TB, and Next-Generation BioGreen 21 Program (No. ADFC2011), Rural Development Administration, Republic of Korea to SHY. No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 2.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 3.Winter H, Huber SC, Sharkey T. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Cri Rev Biochem Mol Biol. 2000;35:253–289. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- 4.Koch K. Sucrose Metabolism: regulatory mechanism and pivotal roles in sugar sensing and plant development. Curr Opi Plant Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Fernie AR, Willmitzer L, Trethewey RN. Sucrose to starch: a transition in molecular plant physiology. Trends Plant Sci. 2002;7:35–41. doi: 10.1016/s1360-1385(01)02183-5. [DOI] [PubMed] [Google Scholar]
- 6.Rolland F, Gonzalez EB, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Ann Rev Plant Biol. 2006;57:676–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 7.Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, et al. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998;15:253–263. doi: 10.1046/j.1365-313x.1998.00205.x. [DOI] [PubMed] [Google Scholar]
- 8.Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 9.Wiese A, Elzinga N, Wobbes B, Smeekens S. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell. 2004;16:1717–1729. doi: 10.1105/tpc.019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiese A, Elzinga N, Wobbes B, Smeekens S. Sucrose-induced translational repression of plant bZIP-type transcription factors. Biochem Soc Trans. 2005;33:272–275. doi: 10.1042/BST0330272. [DOI] [PubMed] [Google Scholar]
- 11.Lee SS, Yang SH, Berberich T, Miyazaki A, Kusano T. Characterization of AtbZIP2, AtbZIP11 and AtbZIP53 from the group S basic region-leucine zipper family in Arabidopsis thaliana. Plant Biotechnol. 2006;23:249–258. [Google Scholar]
- 12.Aguan K, Sugawara K, Suzuki N, Kusano T. Low-temperature-dependent expression of a rice gene encoding a protein with a leucine-zipper motif. Mol Gen Genet. 1993;240:1–8. doi: 10.1007/BF00276876. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu H, Sato K, Berberich T, Ozaki R, Miyazaki A, et al. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol. 2005;46:1623–1634. doi: 10.1093/pcp/pci178. [DOI] [PubMed] [Google Scholar]
- 14.Kusano T, Berberich T, Harada M, Suzuki N, Sugawara K. A maize DNA binding factor with bZIP motif is induced by low temperature. Mol Gen Genet. 1995;248:507–517. doi: 10.1007/BF02423445. [DOI] [PubMed] [Google Scholar]
- 15.Kusano T, Sugawara K, Harada M, Berberich T. Molecular cloning and partial characterization of a tobacco cDNA encoding a small bZIP protein. Biochim Biophys Acta. 1998;1395:171–175. doi: 10.1016/s0167-4781(97)00161-9. [DOI] [PubMed] [Google Scholar]
- 16.Yang SH, Berberich T, Sano H, Kusano T. Specific association of transcripts of tbzF and tbz17, tobacco genes encoding basic region leucine zipper-type transcriptional activators, with guard cells of senescing leaves and/or flowers. Plant Physiol. 2001;127:23–32. doi: 10.1104/pp.127.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–943. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 18.Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, et al. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant. 2001;111:345–352. doi: 10.1034/j.1399-3054.2001.1110312.x. [DOI] [PubMed] [Google Scholar]
- 19.Hanson J, Hanseen M, Weise A, Hendriks MMWB, Smeekens S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of asparagine synthetase1 and proline dehydrogenase2. Plant J. 2008;53:935–949. doi: 10.1111/j.1365-313X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- 20.Satoh R, Fujita Y, Nakashima K, Shinozaki K, Shinozaki KY. A novel subgroup of bZIP proteins functions as transcriptional activaters in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 2004;45:309–317. doi: 10.1093/pcp/pch036. [DOI] [PubMed] [Google Scholar]
- 21.Alonso R, Onate-Sanchez L, Weltmeier F, Ehlert A, Diaz I, et al. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell. 2009;21:1747–1761. doi: 10.1105/tpc.108.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Z, Sachs MS. Role of upstream open reading frame in mediating arginine-specific translation control in Neurospora crassa. J Bacteriol. 1996;178:2172–2177. doi: 10.1128/jb.178.8.2172-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raney A, Baron AC, Mize GJ, Law GL, Morris DR. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J Biol Chem. 2000;275:24444–24450. doi: 10.1074/jbc.M003364200. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov IP, Loughram G, Atkins JF. uOFR with unusual translation start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Nat Acad Sci USA. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, et al. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J Biol Chem. 2005;277:44131–44139. doi: 10.1074/jbc.M509340200. [DOI] [PubMed] [Google Scholar]
- 26.Tabuchi T, Okada T, Azuma T, Nanmori T, Yasuda T. Posttranscriptional regulation by the upstream open reading frame of the phophoetanolamine n-methyltransferase gene. Biosci Biotechnol Biochem. 2006;70:2330–2334. doi: 10.1271/bbb.60309. [DOI] [PubMed] [Google Scholar]
- 27.Sugden C, Donaghy PG, Halford NG, Hardie G. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang JZ, Huber SC. Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase. Influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol. 2001;42:1079–1087. doi: 10.1093/pcp/pce137. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida S. Molecular regulation of leaf senescence. Current Opinion Plant Biol. 2003;6:79–84. doi: 10.1016/s1369526602000092. [DOI] [PubMed] [Google Scholar]
- 30.Chen S, Hajirezaei M, Börnke F. Differential expression of sucrose-phosphate synthase isoenzymes in tobacco reflects their functional specialization during dark-governed starch mobilization in source leaves. Plant Physiol. 2005;139:1163–1174. doi: 10.1104/pp.105.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, et al. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell. 2011;23:381–395. doi: 10.1105/tpc.110.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stitt M, Lunn J, Usadel B. Arabidopsis and primary photosynthetic metabolism: more than the icing on the cake. Plant J. 2010;61:1067–1091. doi: 10.1111/j.1365-313X.2010.04142.x. [DOI] [PubMed] [Google Scholar]
- 33.Iskander HM, Casu RE, Fletcher AT, Schmidt S, Xu J, et al. Identification of drought-response genes and study of their expression during sucrose accumulation and water deficit in sugarcane. BMC Plant Biology. 2011;11:12. doi: 10.1186/1471-2229-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horsch RB, Fry JE, Hoffman NL, Eichholtz D, Roger SG, et al. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 36.Nagy F, Kay SA, Chua NH. Analysis of gene expression in transgenic plants. In: Gelvin, S.B. Schilperoort, R.A. ‘(eds) Plant Molecular Biology Manual, B4. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 1988;1–29 [Google Scholar]
- 37.Berberich T, Sano H, Kusano T. Involvement of a MAP kinase, ZmMPK5, in senescence and recovery from low-temperature-stress in maize. Mol. Gen. Genet. 1999;262:534–542. doi: 10.1007/s004380051115. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 39.Page RDM. Treeview: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of SIRT in Arabidopsis AtbZIP53 cDNA. A, Schematic drawing of the cloned fragment in binary vector construct for transformation. AtbZIP53 genome DNA fragment, spanning from -919 to +552, was inserted into PstI and BamHI-digested pBI101 vector (Clontech), yielding pAtZIP53G. The recombinant was a GUS-translational fusion construct. B, Histochemical staining of the AtbZIP53 promoter-GUS transgenic Arabidopsis seedlings. Among the transgenics, two independent lines were used for assay. 5-day-old Arabidopsis seedlings were incubated with or without 20 mM or 100 mM sucrose for 2 days, then rinsed with distilled water twice, and subjected to histochemical staining according to the procedure described by Jefferson (1987). C and D, GUS transcript levels (C) and GUS protein levels (D) in the transgenics incubated with or without 20 mM or 100 mM sucrose. Methylene blue-stained rRNA (C) and CBB-stained large subunit (LSU) of RuBisCO (D) were used for loading controls. GUS protein was detected with anti-GUS antibody (abcam, UK).
(TIF)
SC-uORF is highly conserved in higher plants. Almost all members of plants harbor 3–5 members of the group S bZIP genes carrying the highly conserved SC-uORFs per organism. Redundant and non-redundant databases were screened by using tblastn and later analyzed manually. Sequence alignment was carried out by multiple sequence alignment software ClustalW in default parameters and then edited with Jalview editor (http://www.jalview.org/training.html). Abbreviation: Ac - Allium cepa, At - Arabidopsis thaliana, Am - Artemisia amuna, Ah - Arachis hypogaea, Aa - Artemisia amuna, Ao - Asparagus officinalis, Bv - Beta vulgaris, Bn - Brassica napus, Bo - Brassica oleracea, Br - Brassica rapa, Cc - Citrus clementina, Cp - Carica papaya, Ci - Cichorium intybus, Cic - Citrus clementina, Cr - Citrus reticulate, Cs - Citrus sinensis, Coc - Coffea canephora, Et - Eragrostis tef, Ee - Euphorbia escula, Fa - Festuca arundinacea, Fv - Fragaria vesca, Gm - Glycine max, Ga - Gossypium arboretum, Gh - Gossypium hirsutum, Gr - Gossypium raimondi, Ha - Helianthus annuus, Ht - Helianthus tuberosus, Hv - Hordeum vulgare, In - Ipomoea nil, Ls - Lactuca sativa, Lj - Lotus japonicus, Md - Malus x domestica, Me - Manihot esculenta, Mt - Medicago truncatula, Mc - Mesembryanthemum crystallinum, Nt - Nicotiana tabacum, Os - Oryza sativa, Pv - Panicum virgatum, Pc - Phaseolus coccineus, Phv - Phaseolus vulgaris, Pt - Poncirus trifoliate, Pn - Populus nigra, Pot - Populus tremula x Populus tremuloides, Potri - Populus trichocarpa, Pa- Prunus armeniaca, Pd - Prunus dulcis, Pp - Prunus persica, Rr - Raphanus raphanistrum, Rs - Raphanus sativus, Rc - Ricinus communis, So - Saccharum officinarum, Sc - Secale cereale, Sl - Solanum lycopersicum, St - Solanum tuberosum, Sb - Sorghum bicolor, Tc - Theobroma cacao, Tp - Triphysaria pusilla, Tv - Triphysaria versicolor, Ta - Triticum aestivum, Tt - Triticum turgidum, Vu - Vigna unduiculata, Vv - Vitis vinifera, Zm - Zea mays.
(TIF)
Confirmation of SIRT in tobacco tbz17 cDNA. A, Schematic drawing of the cloned fragment in binary vector construct. The tbz17 cDNA fragment (+1 to +358) was placed under the control of CaMV-35S promoter and translationally fused to GUS gene. B, Histochemical staining of the tbz17 5′-leader GUS transgenic plants. Among the tobacco transgenic plants, two independent lines (#1–1 and #3–1) were used for the assay. Two-week-old tobacco seedlings were incubated with or without sucrose, glucose and fructose for 2 days, then rinsed with distilled water twice, and subjected to histochemical staining according to the procedure described by Jefferson [4]. C and D, relative GUS transcript levels (C) and GUS protein levels (D) in the transgenics incubated with or without sugars. PCR amplification of EF-1α cDNA (C) and CBB staining of large subunit (LSU) of RuBisCO (D), respectively, were used for loading controls. GUS protein was detected with anti-GUS antibody (abcam, UK).
(TIF)
Generation of transgenic Arabidopsis plants overexpressing AtbZIP53 . A, AtbZIP53 cDNA and construction of the binary vector, pBI-AtbZIP53. B, Growth phenotype of 6-day-old representative Arabidopsis seedlings of wild-type (WT) and transgenic plants overexpressing AtbZIP53 (AtbZIP53-ox). C, RNA blot hybridization of AtbZIP53 in WT and AtbZIP53-ox plants. AtbZIP53 endogenous-transcripts and the transgene-derived transcripts were indicated by e and t, respectively. The open reading frame (ORF) of AtbZIP53 was used as a hybridization probe. D, Sugar contents in leaves of wild-type plants (WT) and 3 transgenic lines. The error bar represents the SD. The differences in sucrose contents between WT, pBI and AtbZIP53-ox lines were highly significant as calculated by Students t-test (*P<0.05; **P<0.01). E, RT-PCR analysis on 4 kinds of sucrose phosphate synthase (SPS) genes (At5g20280, At5g11110, At1g04920, At4g10120) and FBPase (At1g43670) genes. Tubulin cDNA was amplified as a control.
(TIF)
Phylogenetic analysis of N. tabacum and Arabidopsis SPS protein sequences. An unrooted neighbor-joining tree was constructed. Accession numbers of N. tabacum SPS cDNAs and AGI codes for Arabidopsis sequences are as follows: NtSPSA (AF194022), NtSPSB (DQ213015), NtSPSC (DQ213014), AtSPS1F (At5g20280), AtSPS2F (At5g11110), AtSPS3F (At1g04920), AtSPS4F (At4g10120).
(TIF)
AtbZIP53 transactivates ASN1 gene but not 4 kinds of SPS genes. A, Schematic drawing of two effectors and 5 reporter plasmids. Those constructs were generated using the primers shown in Table S2. Horizontal short bars inside the 700-bp promoter fragments indicate the transcriptional start sites. B, Transactivation activity assays of AtbZIP53. Effector, reporter and Renilla LUC reference plasmids were co-bombarded into Arabidopsis mature rosette leaves by a particle delivery system (PDS-1000 He, Bio-Rad, Hercules, CA). After 18 h of incubation at 23oC under darkness, relative luciferase (LUC/RLUC) activities were determined. The values obtained from three independent experiments in duplicate assays were calculated with the means+SD. **P<0.01.
(TIF)
The primers used in this study.
(RTF)
The primers used for transactivation assay.
(DOC)
Supplemental Methods. Generation of transgenic Arabidopsis plants overexpressing AtbZIP53.
(RTF)