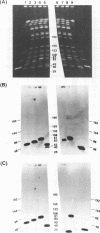
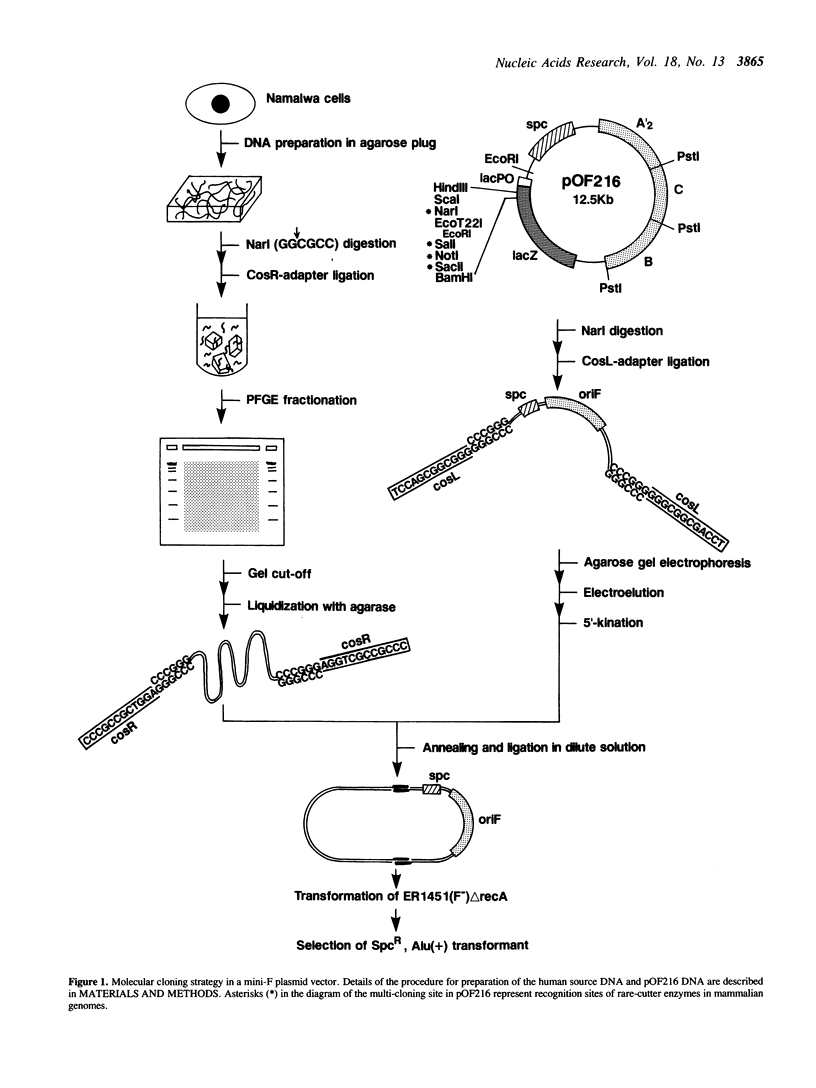
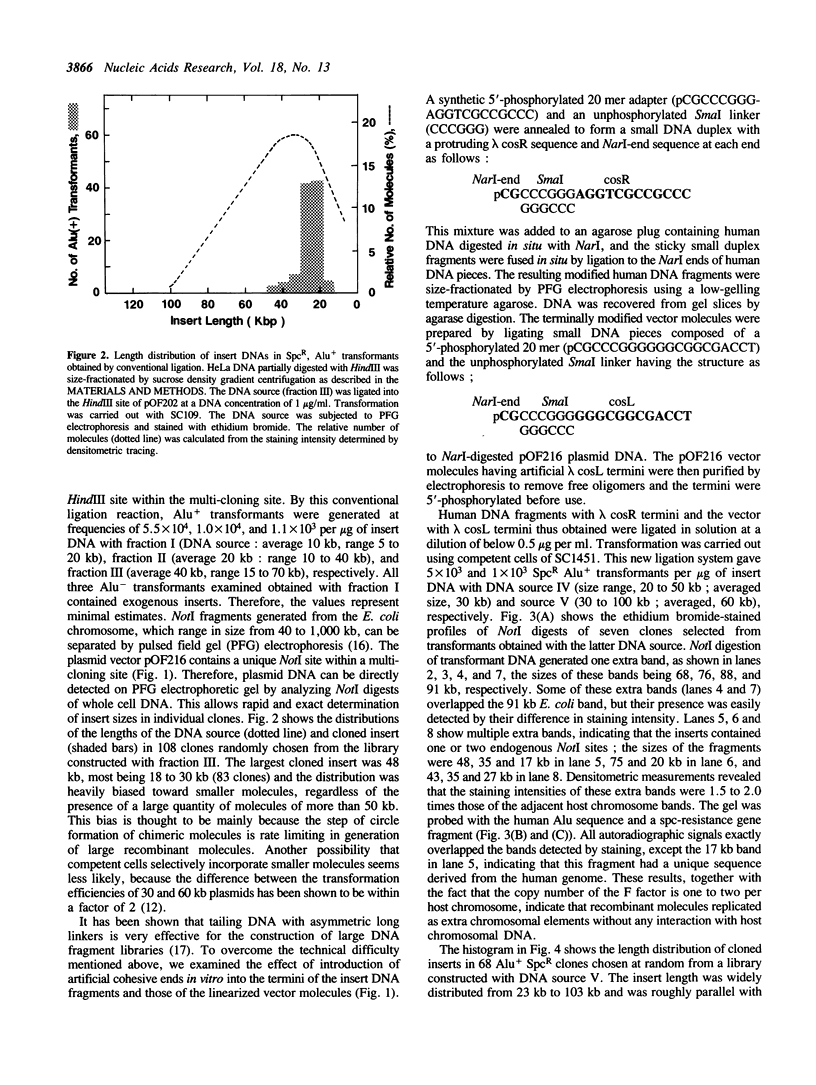
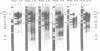Abstract
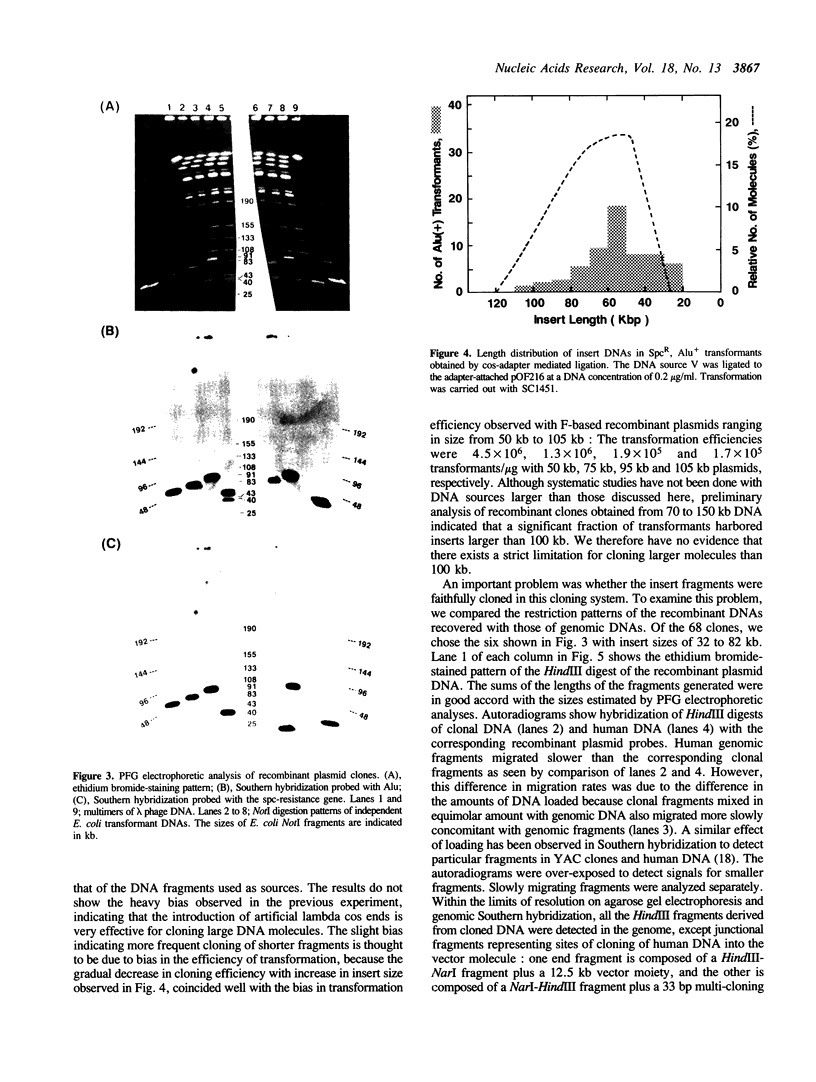
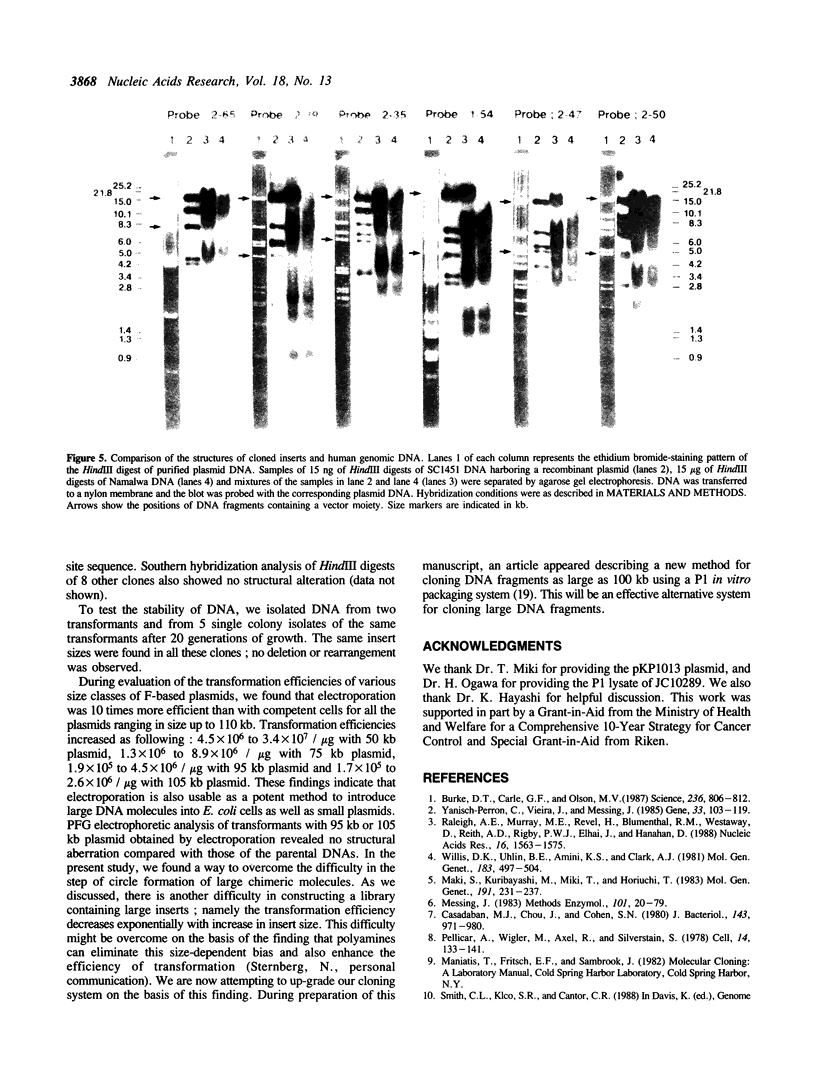
An effective technique using an Escherichia coli plasmid system was developed to clone fragments of exogenous DNA of as large as 100 kilobase pairs. The characteristic features of this technique are the use of a low copy number (one to two) mini-F based plasmid vector and the introduction of artificial lambda cosR ends into the termini of DNA sources and then of the cosL ends into those of linearized vector molecules. This terminal modification greatly facilitated the formation of active large recombinant molecules, which was rarely achieved when the modification was omitted. The efficiency with which large recombinant clones can be generated is high enough to allow construction of a comprehensive library of higher organisms. All analyses of the plasmids recovered have revealed that the inserts were faithful replicas of the human DNAs used as sources.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brownstein B. H., Silverman G. A., Little R. D., Burke D. T., Korsmeyer S. J., Schlessinger D., Olson M. V. Isolation of single-copy human genes from a library of yeast artificial chromosome clones. Science. 1989 Jun 16;244(4910):1348–1351. doi: 10.1126/science.2544027. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Haymerle H., Herz J., Bressan G. M., Frank R., Stanley K. K. Efficient construction of cDNA libraries in plasmid expression vectors using an adaptor strategy. Nucleic Acids Res. 1986 Nov 11;14(21):8615–8624. doi: 10.1093/nar/14.21.8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S., Kuribayashi M., Miki T., Horiuchi T. An amber replication mutant of F plasmid mapped in the minimal replication region. Mol Gen Genet. 1983;191(2):231–237. doi: 10.1007/BF00334819. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mori H., Kondo A., Ohshima A., Ogura T., Hiraga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986 Nov 5;192(1):1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Matsubara K., Sugisaki H., Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981 Nov;15(2-3):257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- Pellicer A., Wigler M., Axel R., Silverstein S. The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell. 1978 May;14(1):133–141. doi: 10.1016/0092-8674(78)90308-2. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C. M., Houck C. M., Deininger P. L., Friedmann T., Schmid C. W. Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature. 1980 Mar 27;284(5754):372–374. doi: 10.1038/284372a0. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(1):103–107. doi: 10.1073/pnas.87.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. K., Uhlin B. E., Amini K. S., Clark A. J. Physical mapping of the srl recA region of Escherichia coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet. 1981;183(3):497–504. doi: 10.1007/BF00268771. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]