Abstract
We have developed a multifaceted highly specific reporter for multimodal in vivo imaging and applied it for detection of brain tumors. A metabolically biotinylated, membrane-bound form of Gaussia luciferase was synthesized, termed mbGluc-biotin. We engineered glioma cells to express this reporter and showed that brain tumor formation can be temporally imaged by bioluminescence following systemic administration of coelenterazine. Brain tumors expressing this reporter had high sensitivity for detection by magnetic resonance and fluorescence tomographic imaging upon injection of streptavidin conjugated to magnetic nanoparticles or fluorophore, respectively. Moreover, single photon emission computed tomography showed enhanced imaging of these tumors upon injection with streptavidin complexed to 111In-DTPA-biotin. This work shows for the first time a single small reporter ( 40 kDa) which can be monitored with most available molecular imaging modalities and can be extended for single cell imaging using intravital microscopy, allowing real-time tracking of any cell expressing it in vivo.
Introduction
Advances in bioengineering, chemistry and imaging techniques have enabled tracking of gene expression, stem cells tracking, tumor growth and regression, molecular events in the tumor as well as gene therapy1–4. The most commonly used reporters are the green fluorescent protein (GFP), the red fluorescent protein (RFP) and more recently, near infra-red fluorescent proteins which are used in combination with fluorescence imaging5,6. Luciferases from the American firefly Photinus pyralis, the sea pansy Renilla reniformis or the marine copepod Gaussia Princeps are used as reporters for bioluminescence imaging (BLI)4,7. Thymidine kinase (TK) from the herpes simplex virus (HSV) type-1 is commonly used for positron emission tomographic (PET) imaging8. Other reporters include transferrin receptors overexpression to increase the uptake of magnetic resonance (MR) imaging agents9 and sodium–iodine symporters (NIS) for PET and single photon computed molecular tomographic (SPECT) imaging10. All these reporter genes and the corresponding imaging technique have unique benefits as well as specific limitations, making them better suited for some applications over others1. For instance, some optical imaging methodologies are well suited for in vitro studies, frequent small animal imaging, and are cost-effective as well as time efficient. However, they can be limited by depth of light penetration and scatter and in the case of bioluminescence, do not yet provide tomographic information; PET and SPECT are highly sensitive translational tools but are costly and not suited for frequent imaging due to the use of radionuclides; MR on the other hand provides a high degree of spatial resolution and is well suited for tumor phenotyping and anatomical reconstruction but can lack molecular detail. Many of the limitations of each of these techniques can be overcome by the use of a flexible multimodal reporter platform which allows the selection of the imaging modality to be used, thereby facilitating the validation of the study in animal models and its translation into humans11. As a general approach, fusion proteins consisting of two or more reporters are generated through recombinant DNA constructs the expression of which can be imaged with different modalities in both cells and living organisms12,13. These reporters have limitations such as loss of activity, large fusion size and the best described reporter thus far allowed imaging with up to three modalities.
The strong interaction of biotin with streptavidin have been used for decades for different applications ranging from protein and nucleic acid analysis in vitro, protein tagging and purification in cultured cells to tumor targeting in vivo14–17. On the other hand, metabolic biotinylation of a biotin acceptor peptide (BAP) in which the enzyme biotin ligase catalyses the addition of a single biotin moiety to a specific lysine residue within its sequence has been used for DNA detection in vitro18, to monitor gene expression19; purify and target viral vectors20; monitor of cell and tumor distribution in real time in vivo21; and tag proteins for purification, localization and trafficking22,23.
In this paper, we took advantage of the natural secretion feature of Gaussia luciferase24–26 and engineered a membrane-bound variant fused to BAP. We show that both Gluc and biotin are displayed on the cell surface in an active form (mbGluc-biotin). More importantly, we show that brain tumors expressing this reporter can be imaged with different imaging modalities including BLI, fluorescence, MR, as well as radionuclide imaging such as SPECT using either coelenterazine, the substrate for Gluc, or different labeled streptavidin moieties. This is the first report of a single small reporter (~40 kDa) which can be monitored with most available molecular imaging modalities and is successfully used to localize tumors in deep tissues.
Experimental Section
Cell culture
Gli36 human glioma cells [obtained from Dr. Anthony Capagnoni (UCLA, Los Angeles, CA)] were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO), 100 U penicillin and 0.1 mg of streptomycin (Sigma) per milliliter at 37 °C and 5% CO2 in a humidified atmosphere.
mbGluc-biotin reporter construct
The sequence for Gluc, codon optimized for mammalian gene expression25, was amplified by polymerase chain reaction (PCR) using specific primers introducing EcoRI and BglII restriction sites at the 5′ and 3′ end respectively. The plasmid pDisplay-BAP-TM21 carring the Igk signal sequence fused to the hemagglutinin (HA) antigenic epitope followed by BAP of the prokaryotic Propionibacterium shermanii 1.3S transcarboxylase domain (PSTCD) and the transmembrane domain (TM) of the platelets-derived growth factor receptor (PDGFR) was digested with similar restriction enzymes removing the Igk signal sequence and HA-tag. Gluc PCR product was then digested and cloned in frame with BAP-TM producing pmbGluc-biotin. The mbGluc-biotin reporter sequence was then amplified by PCR using primers which introduce an NheI site at both sides and cloned in similarly digested CSCW-IG26, a self-inactivating lentivirus vector plasmid which expresses both mbGluc-biotin as well as GFP separated by an internal ribosome entry site (IRES) element under the control of cytomegalovirus (CMV) promoter (CSCW-mbGluc-biotin-IG). CSCW-IG which expresses GFP alone was used as a control. Lentivirus vectors were produced and titered as transducing units (TU)/mL as previously described27. Initially Gli36 cells were engineered by lentivirus vector to overexpress a secreted form of the bacterial biotin ligase, codon-optimized for mammalian gene expression (sshBirA) in order to enhance the biotinylation efficiency of the PSTCD BAP in mammalian cells as we previously described28. These cells, named Gli36-sshBirA, were used throughout this study. Gli36 glioma cells are highly transduced with this lentivirus vector (>95% efficiency)26 and therefore transduced cells were used in all experiment directly without any sorting.
Western blot analysis
Gli36-sshBirA cells (2 × 105) were infected with the lentivirus vector expressing either mbGluc-biotin or GFP control. Forty-eight hours later, cells were collected and lysed in 50 μl RIPA buffer (150mM NaCl, 1% NP40, 0.5% deoxycholic acid, 0.1% SDS in 50 mM TRIS-HCl (pH 8.0) with 0.5 μl protease inhibitor (PI Complete: Boehringer Mannheim). The protein concentration was measured using a Bradford reagent (Bio-Rad). The lysates were eletrophoresed on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). The blots were stained with ponceau to verify even protein loading. Then conjugated streptavadin-horse radish peroxidase (streptavidin-HRP; Amersham Pharmacia Biotech, Piscataway, NJ) was diluted (1:5000) in Tris-buffered saline with Tween-20 (TBS-T) and the membrane was incubated for thirty minutes followed by washing. The same blot was also detected with monoclonal Gluc antibody (Massachusetts General Hospital, antibody production core; 1:500 in TBS-T)29 for one hour, washed, incubated with HRP-conjugated secondary anti-mouse antibody (1:10,000; Amersham Pharmacia Biotech). Blots were developed using SuperSignal West Pico Chemiluminescent Substrate™ (Pierce, Rockford, IL). Blots were finally detected with a combination of anti-beta-tubulin antibody and secondary-HRP conjugate to show equal protein loading.
mbGluc expression
Gli36 cells (1 × 104 cells/well) expressing mbGluc-biotin or the wild-type naturally secreted Gluc were plated in a white 96-well plate and forty-eight hours later cells were washed with PBS. Gluc activity was assayed by adding 50 μl 40 μM coelenterazine (Nanolight, Pinetop, AZ) diluted in PBS directly to the cells, and photon counts were acquired for ten seconds using a plate luminometer (Dynex, Richfield, MN).
FACS analysis
Gli36 cells (2.5 × 105) expressing mbGluc-biotin or control Gli36 cells expressing GFP were plated in a 6-well plate and twenty-four hours later, cells were resuspended in 100 μl of of anti-biotin-APC (Miltenyi Biotec, Auburn, CA) diluted 10x in PBS pH 7.2, 0.5% bovine serum albumin (BSA) and 2 mM EDTA and incubated for five minutes in the dark. Cells were then washed using the same buffer and resuspended in 200 μl PBS followed by analysis using a FACS-Calibur cytometer (Becton Dickinson, San Jose, CA).
Cell-surface staining
Gli36 cells were infected with lentivirus vectors expressing either mbGluc-biotin and GFP or GFP alone as control. These cells were then plated on coverslips in a 12-well plate (1.2×105 cells/well). The next day, cells were washed two times with PBS and incubated with either streptavidin-Alexa647 (Molecular probes 1:100) for ten minutes (to stain for biotin on the cell surface) or with monoclonal Gluc antibody (1:100 dilution in PBS) for ten minutes at room temperature followed by secondary anti-mouse antibody conjugated to Alexa647 (Molecular probes; 1:400 dilution) for thirty minutes at room temperature. Cells were then washed and fixed in 4% PFA for ten minutes at room temperature. Finally the coverslips were mounted onto microscope slides using fluorescent mounting medium. Images were captured using an inverted fluorescent microscope (Nikon TE 200-U) coupled to a digital camera.
In vivo studies
All animal studies were approved by the institutional animal care committee and were performed in accordance to their guidelines and regulations. The serum level of biotin in mice was reduced to make the level comparable to that in humans30,31 by placing the mice on a biotin-deficient diet (Purina Biotin-deficient diet 5839; Purina Test Diet) 3 – 5 days before imaging. Mice were anesthetized with i.p. injection of ketamine (100 mg/kg) and xylazine (5 mg/kg). Gli36 human glioma cells (2×105) expressing either mbGluc-biotin or GFP control were washed and resuspended in 2 μL PBS and implanted in the left midstriatum of anesthetized nude mice (n=6 / group) using the following coordinates from bregma in mm: anterior-posterior +1, medio-lateral +2, dorso-ventral −2.5. Tumor cell injection was performed using a Micro 4 Microsyringe Pump Controller (World Precision Instruments, Sarasota, FL) attached to a Hamilton syringe with a 33-gauge needle (Hamilton, Rena, NV) at a rate of 0.4 μl/min. Bioluminescence, FMT and SPECT imaging were performed on these groups around three weeks post-injection. Later, these mice were imaged with MRI around five weeks post-injection.
Bioluminescence imaging
Mice were anesthetized as above and i.v. injected with 150 μl coelenterazine (4 mg/kg body weight) diluted in PBS. Immediately, photon counts were recorded over five minutes using a cooled CCD camera with no illumination25. A light surface image of the animal was taken in the chamber using dim polychromatic illumination. Processing of images, quantification and analysis were performed using CMIR-image, a program developed by the Center of Molecular Imaging Research using image display and analysis suite developed in IDL (Research Systems Inc., Boulder, CO). Tumors were defined using an automatic intensity contour procedure to identify bioluminescence signals with intensities significantly greater than the background. The Gluc-activity was quantified by calculating photon counts after subtraction of the background noise.
FMT
Quantitative fluorescent tomographic imaging was carried out on a commercial imaging system (FMT2500, Perkin Elmer, Waltham MA). Prior to imaging, mice received an intravenous injection of Streptavidin-Alexa750 at a concentration of 70 nmol/kg body weight (with respect to fluorophore). Twenty-four hours after injection, mice were non-invasively imaged under general isoflurane anesthesia (1–1.5% at 2l/min). Paired absorption and fluorescent data sets were collected using a scanning laser, and 3-dimensional reconstructions were generated utilizing the TrueQuant FMT software.
Magnetic resonance imaging
Streptavidin-MNP conjugate was prepared as we previously described21. Mice were injected i.v. with 5 mg/kg body weight of streptavidin-MNP and imaged with MR before and 24 hours post-injection. MR imaging was performed using a 7T small animal MR scanner using a dedicated head coil and respiratory monitoring (Bruker BioSpin, Billerica, MA). The imaging protocol for mice consists of axial T1-weighted fast spin echo (pre-injection) and T2-weighted multi-slice multi-echo (MSME) pulse sequences (pre- and post-injection). The standard T1-weighted sequence had the following parameters: rapid acquisition with relaxation enhancement sequence (RARE), 873/12.4 msec (TR/TE), 4 averages, matrix size 256×192, FOV 2.5 × 2.5 cm and slice thickness 1 mm. The parameters for the T2-mapping sequence were the following: MSME 3200 ms (TR/TE), 4 averages, echo spacing 8.8 ms (8.8 ms to 141 ms). Sixteen slices with a slice thickness of 0.7 mm, field of view of 2.5 × 2.5 cm, and matrix of 128 × 128 were acquired. Image analysis was performed using ImageJ 1.26T and OsiriX 3.5 software (Image J: National Institutes of Health, Bethesda, MD; OsiriX32). T2 values were computed by fitting selected regions of interest in images acquired at varying TE of 8.8 to 141 ms to a standard exponential transverse relaxation model (Moexp(-TE/T2) using OsiriX and a Macintosh computer equipped with an Intel dual core processor.
Biotin-DTPA conjugate synthesis
DTPA-Lys(Biotin)-NH2 was synthesized by standard Fmoc/tBu solid phase peptide synthesis following procedures we previously described33,34. The (CO2t-Bu)4DTPA-Lys(ivDde) sequence was elongated on a Rink Amide MBHA resin. After selective removal of the ivDde protecting group, biotin was coupled using PyBOP activation. The compound was fully deprotected and cleaved from the resin using a cocktail of TFA/H2O/TIS (95:2.5:2.5). After ether precipitation, the DTPA-Lys(Biotin)-NH2 compound was solubilized in water/acetonitrile (5:2) and purified by RP-HPLC (UV monitoring at 220 nm, 0–60% acetonitrile in 30 minutes gradient, 21 mL.min−1 flow rate). The fraction collected (tR=11 minutes) was lyophilized. DTPA-Lys(Biotin)-NH2 (44.1 mg, 59.05 μmol) was obtained as a white powder. Liquid chromatography-mass spectrometry (LC-MS) was used to purify and analyze the conjugate: C30H50N8O12S; MW: 746.83 g.mol−1; calculated exact mass: 746.33; Electrospray Ionisation (ESI)-MS found m/z: [M+H]+=747.7, [M+2H]2+=374.3.
Biotin-DTPA was then used to chelate 111In followed by complexation to streptavidin as follows: 111InCl3 (2.6 mCi) was diluted with 50 μL ammonium acetate (0.4 M, pH 5) and added to 150 μL of a 2.5 μM solution of DTPA-Lys(Biotin)-NH2 (280 ng, 0.4 nmol) in ammonium acetate (0.4 M, pH 5). The reaction mixture was incubated for 1 h at 70 °C, diluted with water (5 mL), and purified on a SepPak tC18 cartridge preconditioned with ethanol and deionized water. Unreacted 111InCl3 was removed by elution with water (5 mL) while the pure radiolabeled probe was released by elution with ethanol (1 mL). The solvent was evaporated by microwave heating at 65 °C under a flow of argon gas for 5 min to give 2.4 mCi 111 In-DTPA-Lys(Biotin)-NH2. 111In-DTPA-Lys(Biotin)-NH2 was dissolved in 0.25 mL DPBS and to this was added 100 μl of 6.5 μM (0.7 nmol) streptavidin (Sigma) solution in DPBS. After incubation for 1 h at 37 °C, the reaction was loaded on to a NAP-10 column for size-exclusion purification. Concentration of the 111In-DTPA-Lys(Biotin)-NH2/Streptavidin conjugate positive fractions provided 2.26 mCi.
SPECT/CT
Mice were imaged twenty-four hours post injection of 111In-DTPA-streptavidin complex (500 μCi) using a high resolution X-SPECT/CT system (Gamma Medica) equipped with dual gamma cameras and a medium energy pinhole collimator. The images were acquired over 64 projections (128 total) at 45 seconds per projection. Images were reconstructed using the ordered subset-expectation maximization (OSEM) iterative reconstruction algorithm as previously described35,36. A CT scan of the mouse was performed using the Siemens Inveon system with a 80 kVp 500 mA tube over 256 projections. The CT scans were reconstructed using a modified Feldkamp cone beam reconstruction algorithm (COBRA). The SPECT-CT images were fused using the normalized mutual information algorithm in Amira software for brain segmentation and tumor analysis.
Results
mbGluc is metabolically biotinylated in mammalian cells
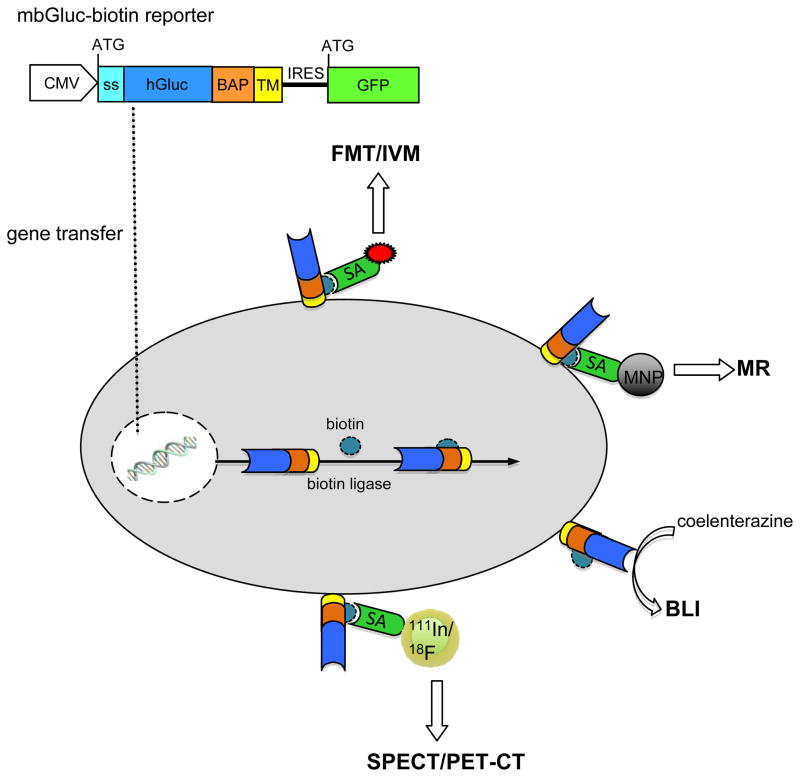
A lentivirus vector construct bearing the naturally secreted Gaussia luciferase cDNA, including its signal sequence (ss; 185 amino acids) fused to the transmembrane domain of the PDGFR (mbGluc), separated by the PSTCD BAP sequence (129 amino acids) was generated (the fusion is around 1kb and 40 kDa). This expression construct also contains the sequence of the enhanced green fluorescent protein (GFP), separated from the reporter by an internal ribosome entry site (IRES), driven by a cytomegalovirus (CMV) promoter (Fig. 1). Upon gene transfer, GFP is expressed. Further, this reporter is biotinylated by biotin ligase with both biotin and Gluc displayed on the cell surface (mbGluc-biotin). Upon injection of coelenterazine, the Gluc substrate, this reporter can be imaged by BLI. When streptavidin conjugated to magnetic nanoparticles, fluorophore, or radionuclide are injected, the reporter can be imaged with MR, fluorescence/intravital microscopy (IVM) or SPECT/PET respectively. Natively, cells can be visualized via GFP, which can also be imaged with IVM and tracked with fluorescence analysis cell sorting (FACS) and histology.
Figure 1. mbGuc-biotin reporter.
Schematic overview of multimodal imaging of the mbGluc-biotin reporter system. Upon vector-mediated delivery and expression, GFP is expressed. Biotin ligase will metabolically tag the BAP sequence within the reporter with a single biotin moiety followed by processing through the secretory pathway leading to displaying both Gluc and biotin on the cell surface. The biotin serves as a “molecular beacon” which can attract any imaging agent conjugated to streptavidin suited for fluorescence-mediated tomography (FMT), intravital microscopy (IVM), magnetic resonance (MR) and radionuclide imaging such as single emission photon computed tomography (SPECT) or positron emission tomography (PET). Further, the presence of Gluc on the cell surface allows bioluminescence imaging of this reporter.
Tracking of mbGluc-biotin in mammalian cells
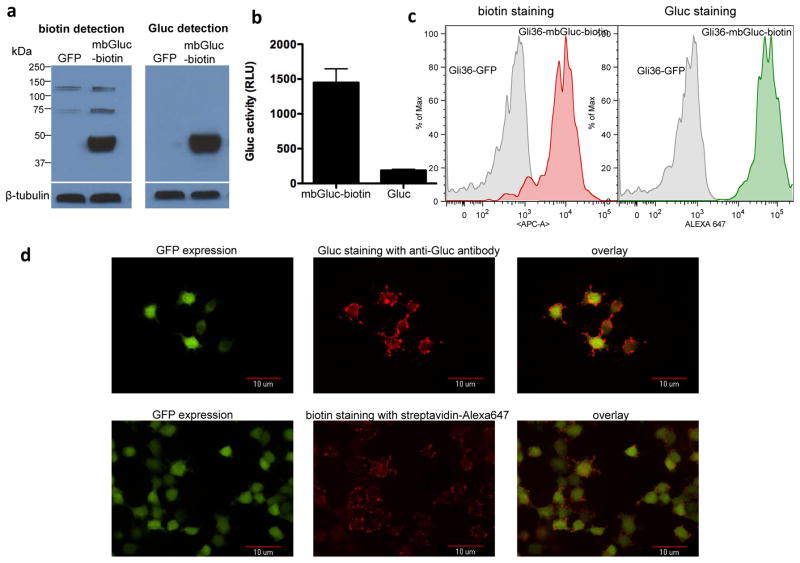
Recently, we have shown that overexpression of the bacterial biotin ligase, codon-optimized for mammalian gene expression, and directed to the same cellular compartment as the BAP fusion protein (sshBirA; 1kb) enhances the metabolic biotinylation of the PSTCD BAP28. Gli36 glioma cell line used in this study was therefore engineered by lentivirus vector to stably express a secreted of the bacterial biotin ligase (Gli36-sshBirA) as described28. In order to measure the expression of the mbGluc-biotin in mammalian cells, Gli36-sshBirA cells were transduced with the lentivirus vector expressing mbGluc-biotin or GFP control. Forty-eight hours later, Western blot analysis was performed on cell lysates using either streptavidin-horseradish peroxidase (HRP) conjugate (to detect biotinylation efficiency) or Gluc specific antibody showing that this reporter is expressed and metabolically biotinylated (Fig. 2a). To confirm the expression of Gluc on the cell surface in an active form, Gli36 cells expressing mbGluc-biotin or native secreted Gluc were plated in a 96-well plate. Forty-eight hours later, cells were washed and the cellular-associated Gluc signal was measured using a luminometer after addition of coelenterazine. Cells expressing mbGluc-biotin showed much higher Gluc signal (>7 fold) as compared to native Gluc confirming the expression and full activity of Gluc on the cell surface (Fig. 2b). To localize Gluc and biotin to the cell surface, we labeled viable Gli36-sshBirA cells expressing mbGluc-biotin reporter or GFP control with either an anti-biotin antibody conjugated to allophycocyanin (APC) or anti-Gluc antibody followed by a secondary-Alexa647 conjugate. Fluorescence-activated cell sorting (FACS) analysis confirmed that the mbGluc-biotin reporter is metabolically biotinylated and showed that both Gluc as well as biotin are displayed on the cell surface (Fig. 2c). To visualize the expression of Gluc and biotin on the cell surface, these cells were plated on coverslips and stained with either Gluc antibody followed by secondary-Alexa647 or streptavidin-Alexa647 conjugate (for biotin detection) followed by analysis using fluorescence microscopy which showed a distinct surface staining for both Gluc and biotin, confirming the FACS analysis data (Fig. 2d).
Figure 2. Analysis of cells expressing mbGluc-biotin reporter.
(a) Gli36-sshBirA cells were transduced with the lentivirus vector expressing mbGluc-biotin or GFP control. Forty-eight hours later, cells were lysed and lysates were analyzed by Western blotting for both biotinylation efficiency and Gluc expression using either streptavidin-HRP conjugate or a combination of Gluc-specific antibody followed by secondary-HRP conjugate respectively. Blots were also analyzed for β-tubulin to show equal loading. (b) Gli36 cells expressing either mbGluc-biotin or native secreted Gluc were imaged for Gluc expression on the cell surface by adding coelenterazine directly to viable cells followed by photon count using a luminometer. Data presented as average ± standard deviation (SD). (c) Viable Gli36-sshBirA cells infected with lentivirus vector encoding mbGluc-biotin and GFP or GFP alone were labeled with anti-biotin antibody conjugated to APC or Gluc antibody followed by a secondary-Alexa647 conjugate and analyzed by FACS. (d) Gli36-sshBirA cells expressing mbGluc-biotin and GFP were plated on coverslips and stained with either streptavadin-Alexa647 for biotin presentation on the cell surface (bottom pannel) or with a combination of Gluc antibody followed by secondary-Alexa647 conjugate (top pannel). Cells were then fixed using 4% paraformaldehyde and analyzed by fluorescence microscopy for both GFP expression and biotin or Gluc staining.
Optical imaging of brain tumors with mbGluc-biotin reporter
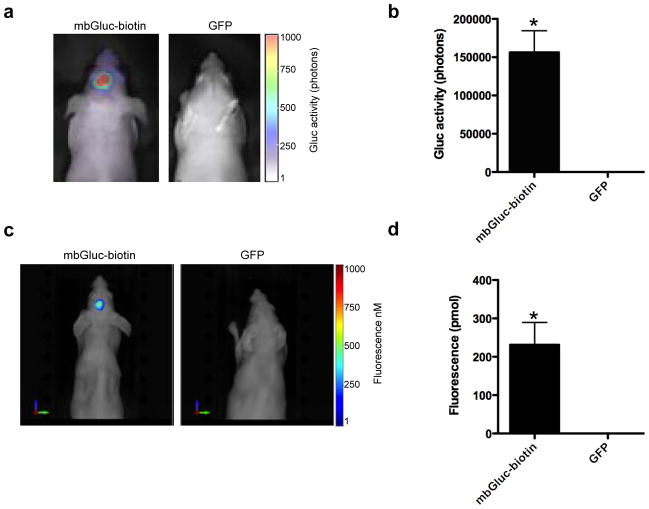
We first engineered Gli36-sshBirA human glioma cells by a lentivirus vector to stably express the mbGluc-biotin reporter or GFP as a control (n=5 per group). We injected either of these cells intracranially into nude mice. Two weeks later, we first imaged tumors with bioluminescence imaging by injecting coelenterazine intravenously (i.v.)26, the substrate for Gluc. A signal was detectable only in the mbGluc-biotin-expressing cells and not the control cells showing that this reporter is successfully detected with BLI (Fig. 3a and b). We next determined whether brain tumors could be detected with fluorescence imaging. The same mice were injected with streptavidin-Alexa750 conjugate and imaged with fluorescence-mediated tomography (FMT) twenty-four hours post-injection. This waiting period allows the free streptavidin-conjugate to cleared from circulation and was previously found to be the optimum time point for imaging tumor-associated conjugate30. An accumulation of Alexa750 fluorophore was observed in brain tumors expressing the mbGluc-biotin reporter and not GFP, showing that brain tumors can be tracked with fluorescence imaging (Fig. 3c and d and Supplementary figure 1).
Figure 3. Optical imaging of mice bearing intracranial tumors.
Gli36 cells expressing either mbGluc-biotin or GFP control (n=5/group) were implanted intracranially in nude mice. (a) Three weeks later, mice were i.v. injected with coelenterazine and imaged by bioluminescence using a cooled CCD camera. (b) Quantification of tumor-associated bioluminescence signal in (a). (c) The same mice from (a) were i.v. injected with streptavadin-Alexa750 conjugate and imaged twenty-four hours later with fluorescence-mediated tomography (FMT). (d) Quantification of tumor associated fluorochrome levels in (c). A representative mouse from each group is shown in (a) and (c). Data shown in (b) and (d) as mean ± SD (n=5; *p<0.001 as calculated by student’s t-test).
Radionuclide imaging of brain tumors expressing mbGluc-biotin
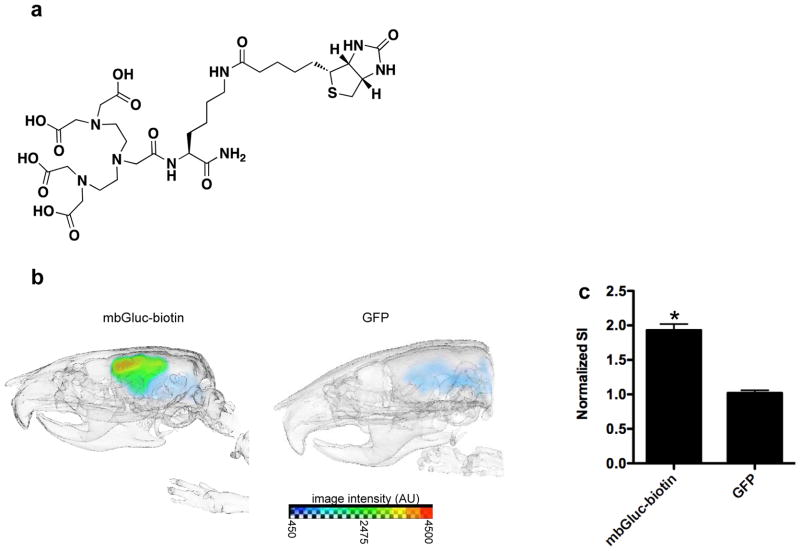
We first synthesized biotin-DTPA conjugate using traditional Fmoc/t-Bu solid phase peptide synthesis (Fig. 4a and Supplementary figure 2). The biotin-DTPA conjugate was purified by RP-HPLC and analyzed by LC-MS (Supplementary figure 3). The LC trace shows a major peak at 6.32 min corresponding to DTPA-biotin as identified by its mono ([M+H]+=747.7) and doubly charged ([M+2H]2+=374.3) peaks on the ESI mass spectrum (Supplementary figure 3). The DTPA-biotin was then used to chelate 111In (through DTPA) followed by complexation to streptavidin (through biotin). The same mice bearing intracranial brain tumors which were imaged with bioluminescence and FMT (n=4) were i.v. injected with this complex and imaged with SPECT twenty-four hours later. High accumulation of radionuclide probe was observed in the mbGluc-biotin-expressing brain tumors as compared to GFP control tumors, with respect to the normal brain, showing that this reporter can be used to image gliomas with radionuclide imaging techniques including SPECT and can be extended to PET, e.g., using 64Cu or 68Ga that can coordinate with DTPA (Fig. 4b and c and Supplementary figure 4).
Figure 4. In vivo radionuclide imaging of mbGluc-biotin-expressing brain tumors.
(a) biotin-DTPA conjugate structure. (b) biotin-DTPA was used to chelate 111In which was then complexed to streptavidin. Mice bearing glioma tumor expressing either mbGluc-biotin or GFP (n=4/group) were i.v. injected with this agent (500 μCi) and imaged using single photon computed molecular tomography (SPECT). A representative mouse from each group is shown. (c) tumor-associated radionuclide signal intensity (SI) was quantified and normalized to unaffected, normal brain signal from the contralateral side. Data shown as the mean ratio of tumor-accumulated radionuclide versus normal brain ± SD (n=4; *p<0.001).
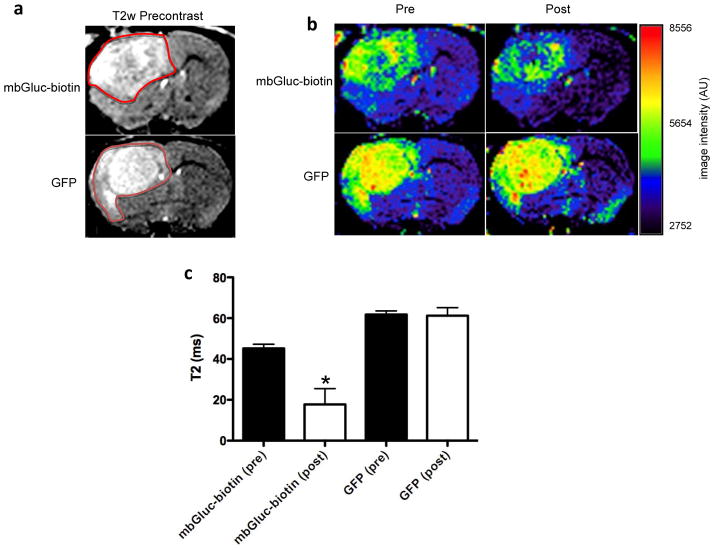
Magnetic resonance imaging of brain tumors expressing mbGluc-biotin
To show the utility of the mbGluc-biotin reporter in imaging brain tumors with magnetic resonance, a T2w precontrast image was first obtained from the same mice from above (n=4) showing equal tumor volume (Fig. 5a). These mice were then i.v. injected with a conjugate composed of streptavidin and magnetic nanoparticle. Twenty-four hours later, a decrease in the T2-weighted image intensity in the mbGluc-biotin-expressing brain tumor was found but not in the GFP control tumor (Fig. 5b and Supplementary figure 5). Correspondingly, a shortening of the spin-spin relaxation time (T2) was observed in tumors expressing mbGluc-biotin as compared to GFP control tumors, proving that this reporter can also be imaged with magnetic resonance (Fig. 5c).
Figure 5. Magnetic resonance imaging of brain tumors expressing mbGluc-biotin.
Mice-bearing glioma tumors expressing either mbGluc-biotin or GFP control (which were imaged with bioluminescence, FMT and SPECT) were i.v. injected with streptavadin-MNP conjugate and imaged with T2-weighted magnetic resonance before and twenty-four hours post-injection. (a) T2w precontrast images demonstrating similar tumor size between the two groups. Red outline indicates the approximate area of the tumors. (b) T2-weighted axial magnetic resonance imaging section through the brain of a typical mouse from each group is shown. In the tumor expressing mbGluc-biotin, there was decreased T2-weighted signal (darker colors) on the post-injection images, reflecting the uptake of the streptavidin-MNP conjugate. This was not observed in the GFP control mouse, in which the T2-weighted signal of the tumor remained the same. (c) Quantification of T2 values from T2-mapping imaging experiments of tumors in (b) shown as mean ± SD (n=4; *p<0.001).
Discussion
Recent advances in non-invasive imaging, including the introduction of genetically encoded reporters into cells and the use of different imaging techniques, had a critical role in understanding normal physiology, disease progression and response to therapy2–4. Imaging modalities are becoming increasingly important in the field of cancer as it can allow a fast quantitative measurement of tumor volume and treatment response, thereby accelerating the development of experimental therapeutic strategies1. Since every imaging modality has its own characteristics and limitations (depth, resolution, cost, throughput), a multimodal reporter provides the option to select the best modality for a particular application, thereby giving insight into the anatomy, physiology and functions of living organisms, facilitating the validation of the study in experimental models and its translation into the clinic11.
In this study, we present a very small multimodal reporter based on a membrane-bound form of the sensitive Gaussia luciferase reporter fused to a biotin acceptor peptide. Upon expression, biotin ligase tags the BAP peptide with a single biotin moiety at a specific lysine residue followed by processing through the secretory pathway and binding on the cell surface. The biotin now serves as a “molecular beacon” to attract any imaging agent coupled to streptavidin. Further, the presence of Gluc on the cell surface can be imaged with bioluminescence imaging upon injection of its substrate, coelenterazine. We showed that brain tumors expressing this reporter were tracked with high sensitivity with most commonly used molecular imaging modalities including bioluminescence, fluorescence, radionuclide (SPECT) and magnetic resonance.
Fusion proteins consisting of two or more reporters, generated through recombinant DNA expression constructs, and which expression can be imaged with different imaging modalities in both individual cells and living organisms have been described. For instance, fusions between a fluorescent protein and a PET reporter protein37,38, a triple fusion protein between luciferase, RFP and thymidine kinase (TK)12 or a hybrid between the sodium iodide symporter (NIS) and eGFP linked with an IRES element13 were developed and applied for multimodal imaging of tumors. Although such fusions or linked proteins have proven to be applicable for experimental purposes, a considerable amount of activity was lost (80% for RFP, and 50% for luciferase) for each reporter due to steric hindrance and/or lower level of expression due to reduced translation of coding sequences following the IRES element39. Thus, the use of these multi-fusions reduces detection sensitivity. Further, the size of these and similar fusion proteins is considerably large (Rluc-mRFP-TK; 2.4 kb; 90 kDa; NIS-IRES-eGFP; 3.7 kb; 140 kDa) making the expression cassettes too large to be carried in some viral vectors such as AAV vectors which are widely used in clinical trials for gene transfer.
The interaction of biotin with streptavidin is one of the strongest non-covalent binding known being orders of magnitude higher than most antibodies40. Naturally, biotin in its physiologically active form is covalently attached at the active site of a class of important metabolic enzymes, biotin carboxylase and decarboxylases. These enzymes are found either in the cytoplasm (acetyl coA carboxylase) or mitochondria (pyruvate carboxylase, propionyl-CoA caboxylase and 3-methylcrotonyl-CoA carboxylase)41 and therefore do not interfere with imaging of the mbGluc-biotin reporter which is expressed on the cell surface. The attachment of biotin to the BAP sequence, through biotin ligase, is a highly selective process and has already been used in different fields ranging from protein/vectors purification to cell tracking19,20,23. Further, the streptavidin-biotin system has been used in clinical trials for targeted tumor therapy as well as for imaging using magnetic nanoparticles and radionuclides42,43,14,17. The mbGluc-biotin reporter described here has several advantages compared to commonly used multimodal reporters: (1) the reporter has a small size (~40 kDa) which makes it a valuable reporter for several delivery systems; (2) This reporter can be imaged with four different imaging modalities including BLI, FMT, MR and SPECT and can be extended for PET and IVM imaging; (3) the very strong binding of biotin to streptavidin makes the reporter very sensitive even in detecting tumors in deep tissues such as the brain. The mbGluc-biotin reporter provides a strong platform for multimodal imaging of tumors and can be extended to many different fields including stem cells tracking for gene and cell therapy.
Supplementary Material
Acknowledgments
This work was supported partly by grants from NIH/NCI P50CA86355 (RW and BAT) and 4R00CA126839 (BAT) as well as NIH/NINDS P30NS045776 (BAT), R01NS070835 (JWC), R01NS072167 (JWC), and K08HL081170 (JWC). We would like to thank Miss Danielle Morse and Jorien Koelen for technical assistance.
References
- 1.Weissleder R, Pittet MJ. Nature. 2008;452:580. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Nat Rev Drug Discov. 2008;7:591. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 3.Tang C, Russell PJ, Martiniello-Wilks R, Rasko JE, Khatri A. Stem Cells. 2010;28:1686. doi: 10.1002/stem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dothager RS, Flentie K, Moss B, Pan MH, Kesarwala A, Piwnica-Worms D. Curr Opin Biotechnol. 2009;20:45. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Nat Biotechnol. 2011;29:757. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 7.Badr CE, Tannous BA. Trends Biotechnol. 2011;29:624. doi: 10.1016/j.tibtech.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaumik S. Curr Pharm Biotechnol. 2011 doi: 10.2174/138920111795163896. [DOI] [PubMed] [Google Scholar]
- 9.Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca EA, Basilion JP. Nat Med. 2000;6:351. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 10.Dadachova E, Carrasco N. Semin Nucl Med. 2004;34:23. doi: 10.1053/j.semnuclmed.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Cherry SR. Semin Nucl Med. 2009;39:348. doi: 10.1053/j.semnuclmed.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Cancer Res. 2004;64:1323. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Che J, Doubrovin M, Serganova I, Ageyeva L, Zanzonico P, Blasberg R. Mol Imaging. 2005;4:128. doi: 10.1162/15353500200504193. [DOI] [PubMed] [Google Scholar]
- 14.Lesch HP, Kaikkonen MU, Pikkarainen JT, Yla-Herttuala S. Expert Opin Drug Deliv. 2010;7:551. doi: 10.1517/17425241003677749. [DOI] [PubMed] [Google Scholar]
- 15.Pardridge WM. Adv Drug Deliv Rev. 2007;59:141. doi: 10.1016/j.addr.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laitinen OH, Nordlund HR, Hytonen VP, Kulomaa MS. Trends Biotechnol. 2007;25:269. doi: 10.1016/j.tibtech.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Gruaz-Guyon A, Raguin O, Barbet J. Curr Med Chem. 2005;12:319. doi: 10.2174/0929867053363225. [DOI] [PubMed] [Google Scholar]
- 18.Verhaegent M, Christopoulos TK. Anal Chem. 2002;74:4378. doi: 10.1021/ac025742k. [DOI] [PubMed] [Google Scholar]
- 19.Parrott MB, Barry MA. Biochem Biophys Res Commun. 2001;281:993. doi: 10.1006/bbrc.2001.4437. [DOI] [PubMed] [Google Scholar]
- 20.Barry MA, Campos SK, Ghosh D, Adams KE, Mok H, Mercier GT, Parrott MB. Expert Opin Biol Ther. 2003;3:925. doi: 10.1517/14712598.3.6.925. [DOI] [PubMed] [Google Scholar]
- 21.Tannous BA, Grimm J, Perry KF, Chen JW, Weissleder R, Breakefield XO. Nat Methods. 2006;3:391. doi: 10.1038/nmeth875. [DOI] [PubMed] [Google Scholar]
- 22.Luque-Garcia JL, Martinez-Torrecuadrada JL, Epifano C, Canamero M, Babel I, Casal JI. Proteomics. 2010;10:940. doi: 10.1002/pmic.200900441. [DOI] [PubMed] [Google Scholar]
- 23.Chen I, Howarth M, Lin W, Ting AY. Nat Methods. 2005;2:99. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 24.Santos EB, Yeh R, Lee J, Nikhamin Y, Punzalan B, La Perle K, Larson SM, Sadelain M, Brentjens RJ. Nat Med. 2009;15:338. doi: 10.1038/nm.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Mol Ther. 2005;11:435. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, Tannous BA. Nat Methods. 2008;5:171. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AWJ. Virol Methods. 2004;122:131. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Niers JM, Chen JW, Weissleder R, Tannous BA. Anal Chem. 2011;83:994. doi: 10.1021/ac102758m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badr CE, Hewett JW, Breakefield XO, Tannous BA. PLoS One. 2007;2:e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axworthy DB, Reno JM, Hylarides MD, Mallett RW, Theodore LJ, Gustavson LM, Su F, Hobson LJ, Beaumier PL, Fritzberg AR. Proc Natl Acad Sci U S A. 2000;97:1802. doi: 10.1073/pnas.97.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusckowski M, Fogarasi M, Fritz B, Hnatowich DJ. Nucl Med Biol. 1997;24:263. doi: 10.1016/s0969-8051(97)00061-9. [DOI] [PubMed] [Google Scholar]
- 32.Rosset A, Spadola L, Ratib OJ. Digit Imaging. 2004;17:205. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garanger E, Aikawa E, Reynolds F, Weissleder R, Josephson L. Chem Commun (Camb) 2008:4792. doi: 10.1039/b809537j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garanger E, Blois J, Hilderbrand SA, Shao F, Josephson LJ. Comb Chem. 2010;12:57. doi: 10.1021/cc900141b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepp LA, Vardi Y. IEEE Trans Med Imaging. 1982;1:113. doi: 10.1109/TMI.1982.4307558. [DOI] [PubMed] [Google Scholar]
- 36.Querol M, Chen JW, Weissleder R, Bogdanov A., Jr Org Lett. 2005;7:1719. doi: 10.1021/ol050208v. [DOI] [PubMed] [Google Scholar]
- 37.Loimas S, Wahlfors J, Janne J. Biotechniques. 1998;24:614. doi: 10.2144/98244st01. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs AH, Winkeler A, Hartung M, Slack M, Dittmar C, Kummer C, Knoess C, Galldiks N, Vollmar S, Wienhard K, Heiss WD. Hum Gene Ther. 2003;14:277. doi: 10.1089/10430340360535823. [DOI] [PubMed] [Google Scholar]
- 39.Finn J, MacLachlan I, Cullis P. FASEB J. 2005;19:608. doi: 10.1096/fj.04-2769fje. [DOI] [PubMed] [Google Scholar]
- 40.Diamandis EP, Christopoulos TK. Clin Chem. 1991;37:625. [PubMed] [Google Scholar]
- 41.Kirkeby S, Moe D, Bog-Hansen TC, van Noorden CJ. Histochemistry. 1993;100:415. doi: 10.1007/BF00267821. [DOI] [PubMed] [Google Scholar]
- 42.Paganelli G, Grana C, Chinol M, Cremonesi M, De Cicco C, De Braud F, Robertson C, Zurrida S, Casadio C, Zoboli S, Siccardi AG, Veronesi U. Eur J Nucl Med. 1999;26:348. doi: 10.1007/s002590050397. [DOI] [PubMed] [Google Scholar]
- 43.Cremonesi M, Ferrari M, Chinol M, Stabin MG, Grana C, Prisco G, Robertson C, Tosi G, Paganelli G. Eur J Nucl Med. 1999;26:110. doi: 10.1007/s002590050366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.