Abstract
Central to synaptic function are protein scaffolds associated with neurotransmitter receptors. α7 neuronal nicotinic acetylcholine receptors (nAChRs) modulate network activity, neuronal survival and cognitive processes in the CNS, but protein scaffolds that interact with these receptors are unknown. Here we show that the PDZ-domain containing protein PICK1 binds to α7 nAChRs and plays a role in their clustering. PICK1 interacted with the α7 cytoplasmic loop in yeast in a PDZ-dependent way, and the interaction was confirmed in recombinant pull-down experiments and by co-precipitation of native proteins. Some α7 and PICK1 clusters were adjacent at the surface of SH-SY5Y cells and GABAergic interneurons in hippocampal cultures. Expression of PICK1 caused decreased α7 clustering on the surface of the interneurons in a PDZ-dependent way. These data show that PICK1 negatively regulates surface clustering of α7 nAChRs on hippocampal interneurons, which may be important in inhibitory functions of α7in the hippocampus.
Keywords: Nicotinic receptor, α7 nAChR, PICK1, Clustering, Hippocampus, GABAergic interneuron
Introduction
Molecular scaffolds organize synaptic structures and downstream signaling processes. Among nAChRs, members of the PSD95 family interact with α3 and β4 subunits in the peripheral nervous system (Conroy et al., 2003; Parker et al., 2004), but no intracellular proteins regulating clustering of nAChRs have been identified in the central nervous system (CNS), yet. α7 nAChRs are prominent nAChRs and constitute α-bungarotoxin-(α-BT)-binding sites widely expressed throughout the CNS (Jones et al., 1999). They are important in learning, attention, nicotine addiction, and involved in neurodegenerative diseases and schizophrenia (Jones et al., 1999; Martin et al., 2004; O’Neill et al., 2002). α7 nAChRs are highly permeable for calcium (Seguela et al., 1993), present at synaptic and extrasynaptic sites (Fabian-Fine et al., 2001; Kawai et al., 2002; Levy and Aoki, 2002; Shoop et al., 1999) and have numerous functions in cell survival and synaptic plasticity (Dajas-Bailador and Wonnacott, 2004), implying specific interaction with appropriate signaling and scaffolding molecules (Berg and Conroy, 2002; Huh and Fuhrer, 2002). Src-family kinases (SFKs) have recently been found to associate with α7 nAChRs, causing α7 phosphorylation and decreased receptor activity (Charpantier et al., 2005). Unlike in the case of the neuromuscular AChR, however (Sadasivam et al., 2005; Willmann et al., 2006), SFKs do not seem to control clustering of α7 nAChRs (Wiesner and Fuhrer, 2006).
In the hippocampus, which receives rich cholinergic innervation from the septal complex, α7 nAChRs are highly expressed in GABAergic interneurons where they form postsynaptic clusters (Kawai et al., 2002), mediate cholinergic synaptic input (Alkondon et al., 1998; Frazier et al., 1998) and regulate inhibition within the hippocampal network (Alkondon et al., 1997; Jones and Yakel, 1997). Activation of these α7 receptors blocks concurrent STP and LTP induction in pyramidal cells (Ji et al., 2001). Inhibition of pyramidal neurons by postsynaptic α7 nAChRs on interneurons also underlies hippocampal auditory gating, suggesting that α7 might play a role in the pathogenesis of schizophrenia (Martin et al., 2004; Ripoll et al., 2004). Neuregulin, neurotrophins and NMDA receptor activity increase interneuronal α7 nAChR levels or clustering in hippocampus (Kawai et al., 2002; Liu et al., 2001) whereas raft-like lipid microdomains are important in α7 clustering in neurons of the ciliary ganglion (Bruses et al., 2001) — but in all these cases the intracellular proteins mediating or modulating α7 clustering remain unknown.
Here we identify PICK1 as a first scaffolding protein that interacts with α7 nAChRs. PICK1 was originally isolated as a binding protein of protein kinase C (PKCα) (Staudinger et al., 1995), and PICK1 is important in synaptic targeting and clustering of other neurotransmitter receptors. Presynaptically, PICK1 binds to the C-terminus of mGluR7a and causes receptor clustering and phosphorylation by PKC (Boudin et al., 2000; Dev et al., 2000). Postsynaptically, PICK1 binds to and clusters kainate receptors through its PDZ domain (Hirbec et al., 2003). GluR2-containing AMPA receptors are clustered by PICK1 in heterologous cells (Xia et al., 1999). Furthermore, in neurons PICK1 influences glutamate receptor transport processes suggesting a role of PICK1 in the release of AMPA receptors from synaptic anchors and in receptor transport from the synaptic membrane towards endocytotic pathways (Perez et al., 2001; Steinberg et al., 2006; Terashima et al., 2004).
We find that the α7–PICK1 interaction involves the PDZ domain of PICK1 and a segment of the α7 intracellular loop. Interaction is shown in the yeast two-hybrid system and is confirmed in precipitation assays using recombinant and native proteins. Interestingly, PICK1 negatively regulates clustering of α7 receptors in hippocampal GABAergic interneurons, suggesting that PICK1 may play a specific role in α7-mediated inhibition of the hippocampal network.
Results
Identification of PICK1 as an α7 interaction partner using the yeast two-hybrid system
To search for intracellular molecules that interact with α7 nAChRs, we used the cytoplasmic loop of α7 as bait to screen a rat brain cDNA library using the yeast two-hybrid (YTH) technique (Fields and Song, 1989) (bait 1, aa 332–467, Fig. 1A). This loop is situated between transmembrane domains 3 and 4 and comprises most of the cytoplasmic portion of the α7 receptor. Positive candidates were verified by cotransformation of bait and prey clones into yeast and repeated lift filter assays. We classified binding results as positive or negative (+ or −; see Fig. 1A), in accordance to Staudinger et al. (1997), Xia et al. (1999) and Boudin et al. (2000). Among others, we identified two clones that encode full-length PICK1 (aa 1–417, Fig. 1A), showing that the α7 loop interacts with PICK1 in yeast.
Fig. 1.
Interaction between α7 nAChR and PICK1 in yeast. (A) Yeast strain AH109 was cotransformed with plasmids encoding the GAL4 DNA-binding domain fused to different sequences of the cytoplasmic loop of rat α7 nAChR (or rat α4or β2 nAChR, as indicated) and the GAL4 activation domain fused to different PICK1 sequences. Protein–protein interaction was assayed by growing the yeast on selective medium and by galactosidase assays. The specificity of this interaction was tested using control plasmids; + indicates interaction, – no interaction. n.d., not done. (B) PICK1 prey constructs used. CC, coiled coil domain; AR, acidic region. (C) Sequence alignment of the bait 9 region of α7 with other nAChR subunits and with the C-terminus of other proteins known to bind PICK1. A separate alignment of α7 with Arf1 and Arf3 is shown at the bottom. Note the two putative PDZ-binding motifs, EVRYand ESEV. (D) Mutation of the putative PDZ-binding motifs (EVRY and ESEV) in α7 nAChR bait 1 and bait 9. The interaction with PICK1 prey vectors was not affected. Polarity colors mark the residues according to the polarity of amino acids (www.clcbio.com).
Besides the homopentameric α7 nAChRs that form α-BT binding sites, heteropentameric α4/β2 nAChRs are abundant in brain (Lindstrom et al., 1995). We characterized the specificity of the α7 nAChR-PICK1 interaction by examining the binding of PICK1 to the cytoplasmic loop sequence of the α4 and β2 nAChR in the YTH system. PICK1 did not interact with α4or β2 nAChR subunits, illustrating the specificity of the PICK1–α7 loop interaction in yeast (Fig. 1A).
A C-terminal segment of the α7 loop and the PDZ-domain of PICK1 mediate binding
To map the site of interaction between α7 nAChR and PICK1, various bait constructs of the α7 cytoplasmic loop were tested for interaction with full-length PICK1 (Fig. 1A). Deleting the C-terminal region of the α7 loop bait eliminated the interaction (baits 7, 8), while baits containing this region still interacted with PICK1 (baits 9, 10). These data show that a C-terminal segment (aa 429– 467; bait 9) close to the TM4 domain of α7 nAChR is necessary and sufficient to bind to PICK1 in yeast.
PICK1 comprises three major structural domains important for protein interactions, an N-terminal PDZ domain (aa 20–110), a coiled-coil domain (aa 139–166) and a C-terminal acidic region (aa 380–390) (Staudinger et al., 1997; Xia et al., 1999) (Fig. 1B). Previous studies indicated that these domains are important for clustering and synaptic localization of PICK1 (Boudin and Craig, 2001), the PDZ domain being necessary for interactions with various neurotransmitter receptors (Boudin et al., 2000; Hirbec et al., 2003; Xia et al., 1999). To determine whether the PICK1 PDZ domain also mediates binding to α7 nAChRs, we used two additional PICK1 prey constructs, one containing the PDZ domain only (aa 1–126), the other lacking it (aa 126–417, Fig. 1B). None of the α7 nAChR baits interacted with PICK1 lacking its PDZ domain (Fig. 1A). Baits 1, 9 and 10, which interacted with the full-length PICK1, also interacted with the short prey containing PICK1’s PDZ domain only (Fig. 1A). This shows that the PDZ domain of PICK1 is both necessary and sufficient for interaction with the α7 nAChR loop.
Protein interactions mediated by PDZ-domains are of great versatility, as PDZ domains bind to small C-terminal peptides (through class I, II and III binding motifs), internal protein segments, other PDZ domains or even lipids (Nourry et al., 2003). We analyzed the sequence of the α7 cytoplasmic loop for potential class I, II, and III PDZ-binding motifs and identified two putative motifs in the shortest PICK1-interacting bait (bait 9, Fig. 1C). Sequence comparison revealed that one of these motifs (EVRY) is partially conserved between nAChR subunits, whereas the other (ESEV) only occurs in α7 (Fig. 1C; Nourry et al., 2003). Alignments of nAChR subunits with the C-termini of proteins known to bind to PICK1 resulted in a low degree of conservation with no particular signs of homology (Fig. 1C). These proteins were: Arf1 and Arf3 (Takeya et al., 2000), EphB2 (Cowan et al., 2000), GluR2 and GluR3 (Xia et al., 1999), GluR52b and GluR6 (Hirbec et al., 2003), mGluR7a (Boudin et al., 2000) and PKCα (Staudinger et al., 1997). Given the absence of binding between α4or β2 nAChR subunits with PICK, the α7-specific sequence ESEV appeared as a candidate to mediate binding to PICK1.
We point-mutated the sequences EVRY and ESEV singly or together in bait 1 and 9 (Fig. 1D). Mutation was done at the 2nd and 4th amino acids of the consensus by replacement with alanine, to inactivate the motif (Nourry et al., 2003). We found that all mutated baits still bound normally to full-length PICK1 through its PDZ domain (Fig. 1C). Thus the α7 nAChR-PICK1 interaction reported here does not depend on α7 sequences similar to class I, II and III PDZ-binding motifs. Similarly, the C-termini of Arf1 and Arf3 bind to PICK1 but lack such binding motifs (Fig. 1C) (Takeya et al., 2000). In a specific comparison between α7 and these two proteins, no obvious homologies were observed (Fig. 1C).
We can conclude the following: most PICK1-interacting proteins known so far bind the PDZ domain of PICK1 through class I or II motifs. Exceptions are Arf1 and Arf3, which do use their C-termini to bind PICK1, but this binding does not occur via consensus sequences. Another exception is α7, which uses neither the C-terminus nor consensus motifs to bind PICK1. Instead, this binding occurs via a segment of the internal α7 loop close to transmembrane domain 4. Further characterization of this binding region will require systematic deletions and amino acid replacements.
Interaction of recombinant α7 and PICK1 in heterologous cells
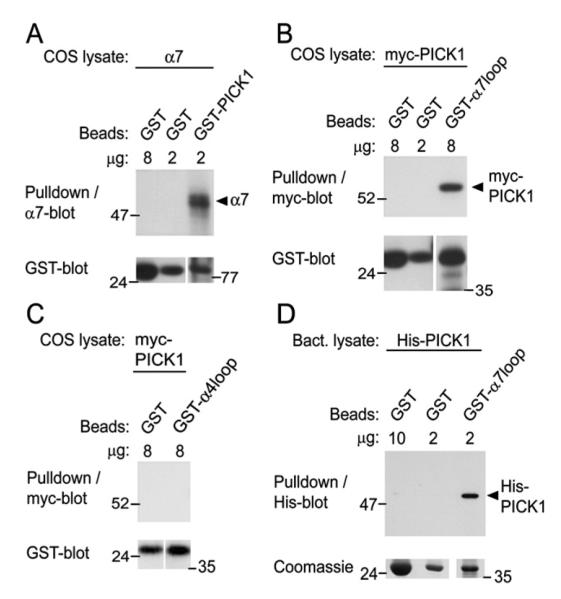
The interaction of α7 nAChR and PICK1 was further examined by recombinant protein pull-down experiments and immunoblotting. COS cells or bacteria were transfected with either full-length α7 nAChR or tagged (myc, His) PICK1 expression vectors. Cell lysates were incubated with glutathione-S-transferase (GST) fusion proteins immobilized to glutathione-Sepharose beads (Fig. 2). These fusion proteins contained either full-length PICK1 (GST-PICK1) or the cytoplasmic loop of α7 nAChR (GST-α7loop), or the cytoplasmic loop of α4 nAChR (GST-α4loop) as a control. These assays showed that GST-PICK1 beads precipitated α7 nAChRs from COS cells (Fig. 2A), while GST-α7loop beads pulled down myc-PICK1 from COS lysates (Fig. 2B) and also His-PICK1 from bacteria (Fig. 2D). In contrast, GST-α4loop beads did not pull down myc-PICK1 from COS cells indicating the specificity of the interaction between recombinant α7 nAChR and PICK1. These results confirm the YTH data and demonstrate the interaction of the α7 loop and PICK1. Our data also show, using both YTH and COS cell experiments, that the α4 subunit of nAChRs does not interact with PICK1.
Fig. 2.
Interaction of recombinant α7 and PICK1. (A–C) COS cells were transfected with full-length α7 or myc-PICK1 expression constructs, lysed and incubated with the indicated amounts of GST proteins immobilized on beads. Bead pellets were analysed by α7- or myc-immunoblotting, and blots were reprobed for GST, showing that GST-PICK1 precipitates α7 from the COS lysate (A), while GST-α7loop pulls down myc-PICK1 (B), and GST-α4loop does not pull down myc-PICK1 (C). As a control, non-transfected COS cells produced no immunoblot signals (not shown). Panels of GST-blots show GST, GST-PICK1, GST-α7loop or GST-α4loop proteins at their respective molecular weights. To probe α7 nAChR, the following antibodies were used for immunoblots: polyclonal anti-α7 (Santa Cruz; shown) and mAb306 (not shown), with identical results. (D) Bacteria expressing His-PICK1 were lysed, incubated with the indicated GST beads, and precipitates were analyzed by His-immunoblotting, showing that GST-α7 loop pulls down His-PICK1. Parallel samples were Coomassie-stained to reveal GST and GST-α7loop proteins, shown at their respective molecular weight.
Association of native PICK1 with α7 nAChRs in brain
We next performed co-precipitation experiments to test for interaction between the native PICK1 and α7 nAChR proteins in rat brain. From synaptosome preparations of adult rat hippocampus, α7 nAChRs were first precipitated with α-BT coupled to sepharose beads according to established protocols (Drisdel and Green, 2000; Fuhrer and Hall, 1996). Samples were analyzed by PICK1 immunoblotting, revealing the presence of PICK1 in the α7 precipitates (Fig. 3A). The presence of α7 nAChR after α-BT-precipitation was verified using anti-α7 antibodies (Fig. 3A). Pre-incubation with free excess toxin abolished the α7 signal and strongly decreased levels of PICK1 signal, demonstrating specific α7–PICK1 association of native proteins in brain (Fig. 3A). In addition, the specificity of the α-BT-precipitation and the presence of PICK1 in the α7 precipitates were demonstrated by nicotine-competition, which eliminated the α7 nAChR signal and strongly reduced the PICK1 signal in the corresponding Western blots (Fig. 3B). The weak PICK1-signal in the control lanes (+T, +Nic) originates from non-specific binding of PICK1 to the α-BT-sepharose resin. We also precipitated α7 nAChRs from synaptosomes using anti-α7 antibodies and again detected associated PICK1 by immunoblotting (Fig. 3C, left). Omitting antibodies or synaptosomes from the precipitation eliminated the PICK1 signal (Fig. 3C, left). α7-precipitation from synaptosomal preparations of cerebellum or cerebral cortex showed reduced signals compared to hippocampus (Fig. 3C, left) as expected from the high relative abundance of α7 in hippocampus (Seguela et al., 1993). To further assess the specificity of the α7 immunoprecipitation we used non-immune IgG as a control and found no associated PICK1 signal (Fig. 3C, right). In all controls, the molecular weight range of PICK1 was free of signal, with the anti-α7 antibody band and the non-immune IgG band running above the PICK1 range (Fig. 3C).
Fig. 3.
Interaction of endogenous α7 nAChRs and PICK1 in adult rat brain. (A) Synaptosomes were prepared from dissected hippocampi of adult rats, and α7 nAChRs were precipitated with α-BT-Sepharose beads (Tox-P). As controls for specificity, excess free α-BT (+T) was added. A fraction of the total synaptosomal lysate was loaded as a control (Tot). PICK1 immunoblotting reveals specific association with α7 nAChRs, which themselves are visualized in an α7-blot using mAb306 (shown) or mAb319 (not shown; identical results). (B) From adult rat brain lysates, α7 nAChRs were precipitated with α-BT-Sepharose beads (Tox-P) and analyzed by PICK1- or α7-immunoblotting (anti-α7 from Abcam). Nicotine-competition eliminated the α7 nAChR signal and strongly reduced the PICK1 signal, demonstrating specific α7–PICK1-association. (C) Synaptosomes (left) or total hippocampal tissue (right) were prepared from hippocampus (Hip), cerebellum (Cer) or cortex (Cor), lysed, and α7 precipitated using mAb319. As controls, mAb319 was omitted (Ab), brain tissue was left out, or a fraction of total hippocampal synaptosomes was loaded without precipitation (Tot). α7-associated PICK1 was visualized by immunoblotting and mostly detected in hippocampus. Levels of α7 were highest in hippocampus, as revealed by α7-immunoblotting (not shown). Nonimmune IgG was used as a control (right). *Indicates the non-immune IgG antibody band, and **denotes the α7-antibody band. (D) Hippocampal synaptosomes were processed as in panel C, but antibodies against the PSD95-family or GluR2 were used for immunoblotting. PSD95-family proteins and GluR2 AMPAR subunits were present in hippocampal synaptosomes but not associated with α7.
To further illustrate the specificity of the α7–PICK1 interaction, we probed the same α7 immunoprecipitates from hippocampal synaptosomes as used in Fig. 3C for the presence of other synaptic proteins, GluR2 (an AMPA receptor subunit) and members of the PSD95 family (using pan-PSD95 antibodies). Neither GluR2 nor PSD95-family members were associated with α7, but were clearly visible in the starting synaptosomal preparation (Fig. 3D). Taken together, the co-precipitation experiments demonstrate that native α7 nAChRs are specifically associated with PICK1 in the hippocampus. The experiments involve two independent methods – precipitation of α7 receptors with α-BT or with antibodies – and thus represent solid and specific evidence for in vivo interaction of α7nAChRs and PICK1.
PICK1 partially colocalizes with α7 in heterologous cells but does not induce α7 clustering
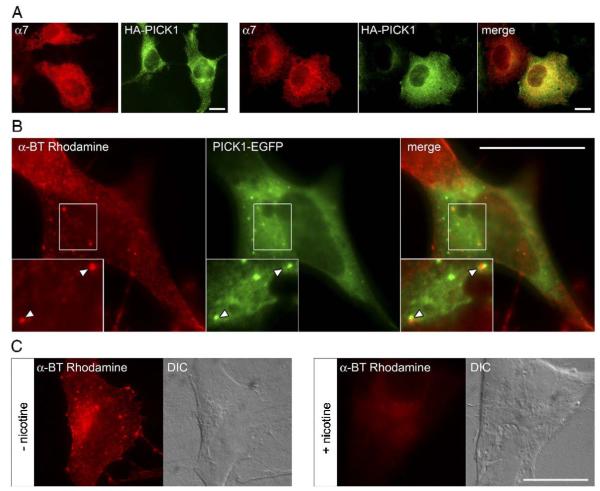
PICK1 has been shown to cluster AMPARs (Dev et al., 1999; Xia et al., 1999) and mGluR7a (Boudin and Craig, 2001; Boudin et al., 2000) in heterologous expression systems. To examine if PICK1 could induce α7 nAChR clustering, we transfected PICK1 and α7 into COS cells, HEK 293T cells and the human neuroblastoma SH-SY5Y cell line, and analyzed α7 and PICK1 distribution by immunofluorescence staining and α-BT labeling (Fig. 4). In transfected COS and HEK cells, α7 nAChRs remain in an immature conformation and are mostly intracellular (Cooper and Millar, 1997). In contrast, in SH-SY5Y cells recombinant α7 nAChRs can form functional channels at the surface and bind α-BT (Charpantier et al., 2005; Cooper and Millar, 1997; Peng et al., 1994). Thus, our experiments allowed determining whether immature intracellular α7 and mature surface α7 nAChRs colocalize with PICK1.
Fig. 4.
Partial colocalization of α7 and PICK1 in transfected heterologous cells. (A) COS cells were transfected either with α7 expression vector or with HA-tagged PICK1 expression construct (left). Alternatively, they were transfected with both plasmids (right). Cells were permeabilized, stained for α7 using anti-α7 antibodies (red), HA-tag (green), or both, and analyzed by conventional fluorescence microscopy. Anti-α7 antibodies were from Abcam (shown) or mAb306 (not shown), with identical results. In all cases, α7 and PICK1 signals are diffuse and around the nucleus. Coexpression does not affect this and reveals partial overlay in the perinuclear area (yellow). Untransfected COS cells produced no signal (not shown). Scale bar, 20 μm. (B) SH-SY5Y cells stably expressing α7 were transfected with PICK1-EGFP expression vector using magnetofection. They were subjected to surface staining of α7 (using α-BT-rhodamine, red) and analyzed by fluorescence microscopy. A cell expressing α7 clusters is shown, with PICK1-EGFP expression in green. This EGFP signal is diffuse and in clusters; the clusters partially overlap and are adjacent to the α7 clusters (note the arrowheads in the white box, which was rotated 90° clockwise to produce the higher magnification insert). In cells not transfected with PICK1-EGFP (not shown), α7 clusters are very similar. Scale bar, 20 μm. (C) As control for specificity, SH-SY5Y cells were stained with α-BT-rhodamine in the presence or absence of nicotine. Differential interference contrast (DIC) shows the cells present. Nicotine-competition (1 mM nicotine added 10 min before α-BT-rhodamine) caused a strong reduction in α-BT surface staining, demonstrating the specificity of the α-BT signal for α7 receptors. Scale bar, 20 μm.
In COS cells transfected with either α7 nAChR or HA-tagged PICK1, we observed largely diffuse intracellular immunofluorescence for these proteins, most intense in the perinuclear area, and occasionally some HA-PICK1 clusters (Fig. 4A left, and data not shown). The same result was seen when HA-PICK1 and α7nAChR were expressed together (Fig. 4A, right). Thus, although no re-distribution was seen upon co-transfection, α7 and PICK1 are perfectly positioned to interact with each other (Fig. 4A, yellow in overlay). Likewise, in HEK 293T cells, α7 immunofluorescence was diffusely distributed and did not reveal clusters (data not shown) whether or not HA-PICK1 was co-expressed. PICK1 formed clusters in HEK 293T cells, also when α7 was not co-expressed, and the PICK1 clusters did not overlap with clusters of α7 (data not shown).
We transfected SH-SY5Y cells stably expressing α7 (Charpantier et al., 2005) with a PICK-EGFP fusion construct to visualize both markers in intact cells. α-BT-rhodamine staining revealed α7 nAChR clusters at the cell surface, besides some diffuse signal (Fig. 4B). PICK1-EGFP also formed clusters, which often were adjacent, or even apposed, to the α7 clusters (Fig. 4B merge). Here again, the distribution and appearance of α7 nAChR clusters was identical in cells not transfected with PICK1-EGFP. The specificity of the α7 nAChR signal on SH-SY5Y cells was demonstrated by displacing α-BT-rhodamine with nicotine, resulting in a drastic reduction of α-BT-rhodamine staining (Fig. 4C).
Taken together, these data indicate that in heterologous cells PICK1 does not induce or affect clustering of α7 nAChRs, although PICK1 itself, in agreement with previous studies (Xia et al., 1999), can form clusters in such cells. Immature intracellular α7 protein is positioned to interact with PICK1 in the perinuclear area in COS cells, whereas some clusters of PICK1 and mature α7 nAChRs are adjacent and partially overlapping at the surface of SH-SY5Y cells. These data are consistent with those of Figs. 1, 2 and 3 showing interaction between PICK1 and α7 nAChRs.
Clusters of PICK1 are adjacent to α7 nAChR clusters at the surface of rat hippocampal GABAergic interneurons
We next assessed the subcellular distribution of α7 receptors and PICK1 in neurons using immunofluorescence microscopy. Primary cultures of rat hippocampal neurons were stained during the second and third week in vitro (Fig. 5). Fluorescent α-BT, added to intact cells, specifically labeled surface α7 clusters along membranes (Fig. 5), confirming the pattern demonstrated previously (Kawai et al., 2002). The labeling was specific as it was blocked by adding excess unlabeled α-BT or nicotine (not shown; but see Kawai et al., 2002). α7 clusters were easily detectable on dendritic processes proximal and distal to the soma, and often appeared grouped into larger aggregates on the cell soma (Fig. 5). At higher magnification, individual clusters were seen on dendrites (box in Fig. 5A).
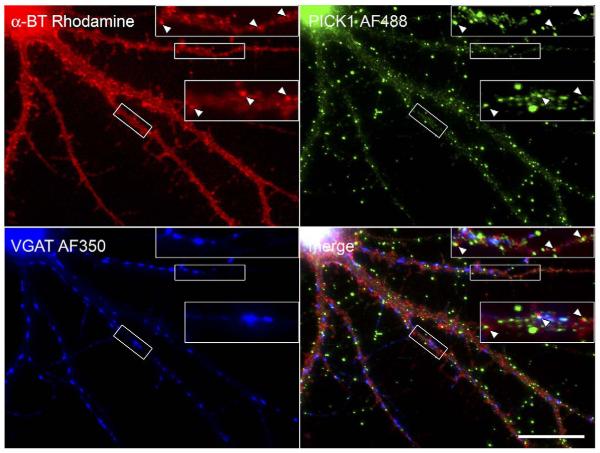
Fig. 5.
α7 nAChRs are clustered at the surface of GABAergic hippocampal interneurons. Cultured dissociated hippocampal neurons were labeled with fluorescent α-BT, permeabilized, and incubated with antibodies against VGAT or GAD followed by fluorescent secondary antibodies. Image overlays reveal that α7-expressing cells are VGAT- and GAD-positive. α7 clusters occur on the soma, often grouped into larger aggregates, and more individually along dendrites, and show some overlap with VGAT or GAD. Panels show a maximal projection of confocal stacks. Note the individual dendrite-associated clusters of α7 nAChRs in the higher magnification (white box). Scale bars: 20 μm.
α7 nAChR clusters labeled with α-BT were seen on only a subset of neurons. To determine their identity, we double-labeled cells with α-BT and antibodies recognizing GAD (glutamic acid decarboxylase) or VGAT (vesicular GABA transporter) (Fig. 5). 5–10% of all neurons were GAD- or VGAT-positive, revealing thus a low density of GABAergic interneurons. Consistent with a previous report (Kawai et al., 2002), α7 nAChR clusters were only present on GAD- or VGAT-positive neurons and labeled most of these cells (Fig. 5). Furthermore, α7 nAChR clusters showed some overlap with VGAT- or GAD-immunoreactivity (IR) (Fig. 5) and also with GABAA receptor α1 subunit-IR (data not shown), suggesting that some of these α7 clusters are located close to GABAergic synapses. Average density of α-BT-clusters in dendrites was 15.0 clusters per 100 μm segment (averaged from 145 dendrite segments of 100 μm length from 40 cells of three independent cultures). This value is similar to published GABAA receptor α2 subunit clusters apposed to GAD boutons (14.7 per 100 μm segment) (Brunig et al., 2002a,b).
The synaptic localization of many interneuronal α7 clusters remains unclear in cultured hippocampal cells (Kawai et al., 2002) although some overlap with synaptotagmin label has been reported (Zarei et al., 1999). Extra- and perisynaptic α7 receptors are found in hippocampal and ciliary ganglion tissue sections (Fabian-Fine et al., 2001; Shoop et al., 1999). We performed pair-wise double-labels with α-BT and antibodies against bassoon, gephyrin and PSD-95 in our hippocampal cultures. The overlap of α-BT signal with these markers was very low (K. Baer and C. Fuhrer, unpublished observations). Since cultured hippocampal cells lack cholinergic neurons, it is thus possible that many of our α7 clusters represent extrasynaptic receptors that, in vivo, may be recruited postsynaptically by cholinergic nerve terminals.
Immunofluorescence labeling of neurons for endogenous PICK1 revealed a clustered pattern outlining the soma and dendrites (Fig. 6), in good agreement with previous studies (Torres et al., 2001; Xia et al., 1999). Double-labeling for α7 nAChR with α-BT-rhodamine showed a punctate distribution for both proteins in interneurons and indicated few colocalizing spots. Rather, as seen before in SH-SY5Y cells (Fig. 4), some α7 clusters were adjacent or even apposed to PICK1 clusters in interneurons (Fig. 6 — note the examples pointed out by arrowheads in the white boxes at higher magnification). Collectively, our data demonstrate that α7 nAChR clusters are found mainly on GABAergic interneurons and tend to closely associate with PICK1 clusters.
Fig. 6.
Some clusters of α7 and PICK1 are adjacent and partially overlapping at the surface of hippocampal interneurons. Cultured hippocampal neurons were labeled with α-BT-rhodamine, permeabilized, and incubated with PICK1 and VGAT antibodies, followed by secondary AlexaFluor 488- and 350-coupled antibodies and fluorescence microscopical analysis. The panel demonstrates one interneuron (VGAT marker in blue) with strong α-BT-rhodamine (red) and endogenous PICK1 immunoreactivity (green). Two dendritic areas are shown enlarged with arrowheads highlighting discrete punctae immunoreactive for α7 and PICK1 clusters. The merged image shows the partial overlap of adjacent α7 nAChRs and PICK1 clusters. Scale bars: 20 μm.
PICK1 reduces α7 nAChR surface clustering in interneurons
To determine whether PICK1 controls clustering of α7 nAChRs at the surface of hippocampal interneurons, we expressed a bicistronic EGFP-PICK1 construct (Terashima et al., 2004) in neuronal cultures using Sindbis virus, enabling the identification of infected neurons by EGFP fluorescence. For comparison, we used Sindbis virus expressing a mutant PICK1 (AA) containing two point mutations in the PICK1 PDZ domain that eliminate PDZ-dependent interactions (Terashima et al., 2004; Xia et al., 1999). As a control, Sindbis virus expressing only EGFP was used.
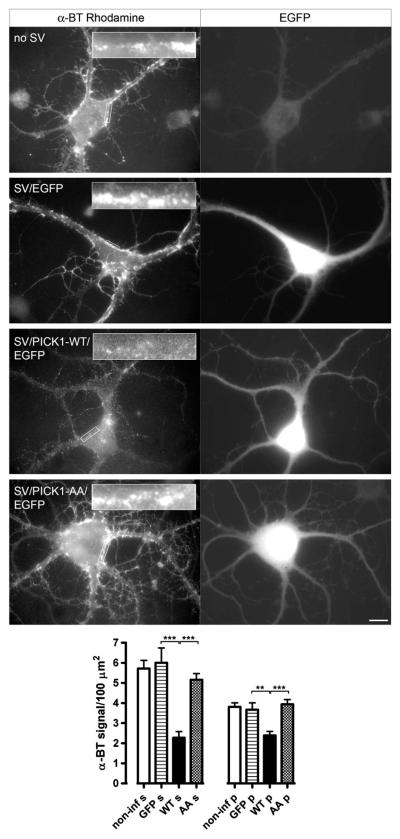
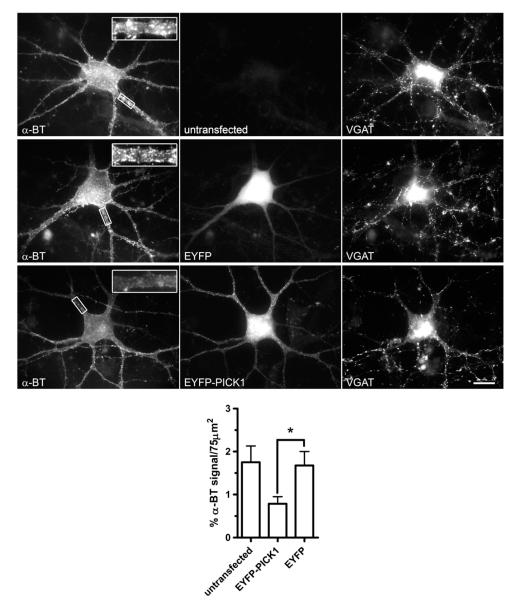
To analyze the effects of PICK1 on α7 nAChR surface clusters, we examined α-BT-rhodamine labeling following virus infection in 14-day-old hippocampal neurons. Infected interneurons were readily detected by EGFP fluorescence in their somata and dendrites. A reduction in α-BT labeling of α7 nAChR at the surface was evident in interneurons infected with EGFP-PICK1 virus compared to non-infected cells (Fig. 7). Furthermore, overexpressing EGFP only or the mutant PICK1-AA protein did not change the pattern of α-BT-rhodamine labeling, indicating that the functional PDZ domain of PICK1 is needed for affecting α7 nAChR clusters.
Fig. 7.
Viral expression of PICK1 causes a reduction in surface α7 nAChR clusters in cultured hippocampal interneurons. Cultured hippocampal cells were infected with different Sindbis viruses (SV), labeled with α-BT-rhodamine and analyzed by conventional fluorescence microscopy. The panels show examples of EGFP fluorescence (right) and surface α7 nAChR staining by α-BT-rhodamine for a non-infected control neuron, a neuron infected with SV containing EGFP, a neuron infected with SV expressing PICK1-WT (wild-type) and EGFP, and a neuron infected with SV containing PICK1-AA mutant and EGFP. Inserts show magnifications of α-BT-staining of somatic regions indicated by the box (for lower-power images, scale bars are 20 μm). Wild-type PICK1 expression reduces α7 clustering. A quantitative analysis of these effects is shown at the bottom. For each neuron in a group (non-infected, EGFP virus, PICK1-WT-EGFP virus, or PICK1-AA-EGFP virus), four surface areas covering 100 μm2 (comparable to the boxes indicated in the top panels) were randomly chosen per cellular region (s, soma; p, proximal dendrites). The α-BT fluorescence intensity was quantitated and plotted per surface area (***p < 0.0001; **p < 0.0015, by unpaired two-tailed Student’s t-test). α7 surface clustering is reduced by PICK1-WT but not by PICK1-AA in somatic and proximal dendritic areas.
The effects of PICK1 viral expression were quantitatively assessed comparing α-BT cluster levels within groups and per cellular region. On the soma of interneurons, clusters often appeared grouped into larger aggregates, as noted for Fig. 5. Our data revealed a significant reduction in α7 nAChR clusters, measured as the cumulative α-BT signal per surface area, on the somata and proximal dendrites of interneurons expressing wild-type PICK1 (Fig. 7, WT). No effect on α7 clusters was seen in non-infected, EGFP- or PICK1-AA-infected interneurons.
To exclude any side effects of viral infection, the effect of PICK1 on α7 nAChR clustering in interneurons was confirmed by transfection of a EYFP-PICK1 fusion protein (or EYFP alone) into hippocampal primary neurons (11 days in vitro) and by examining α7 cluster distribution using α-BT-rhodamine. The results show, as in the case of virus-infected cells, that interneurons expressing EYFP-PICK1 or EYFP have a healthy morphology indicating that PICK1 expression per se did not harm these cells (Fig. 8). EYFP-PICK1 was observed diffusely and in clusters, as shown previously for myc-tagged PICK1 in hippocampal cultures (Boudin and Craig, 2001). The amount of α7 nAChR clusters on dendrites of transfected interneurons again was measured as the cumulative α-BT signal per surface area. These data showed a significant reduction of the α-BT signal in interneurons expressing EYFP-PICK1 compared to both untransfected interneurons and interneurons expressing EYFP alone (Fig. 8), thus confirming the results from viral expression (Fig. 7). Altogether, these results provide strong evidence that PICK1 reduces clustering of α7 nAChR at the surface of GABAergic interneurons.
Fig. 8.
Expression of PICK1 by magnetofection causes a reduction in surface α7 nAChR clusters in cultured hippocampal interneurons. Cultured hippocampal cells were transfected with EYFP-PICK1 or EYFP constructs using magnetofection, labeled with α-BT-rhodamine and anti-VGAT antibody and analyzed by conventional fluorescence microscopy. The panels show examples of surface α7 nAChR staining by α-BT-rhodamine (left), EYFP fluorescence (middle), and VGAT staining (right) for a non-transfected control interneuron, an interneuron transfected with EYFP, and an interneuron transfected with EYFP-PICK1. PICK1 expression reduces α7 clustering. A quantitative analysis of these effects is shown at the bottom. For each neuron in a group (untransfected, EYFP-PICK1 transfected or EYFP transfected), proximal dendritic surface areas were randomly chosen (the boxes represent examples). The amount of α7 surface clusters on dendrites of transfected neurons was measured as the cumulative α-BT fluorescence area per dendritic surface area. α7 surface clustering is reduced by EYFP-PICK1 but not by EYFP in dendritic areas (*p = 0.0267; unpaired two-tailed Student’s t-test). Scale bar, 20 μm.
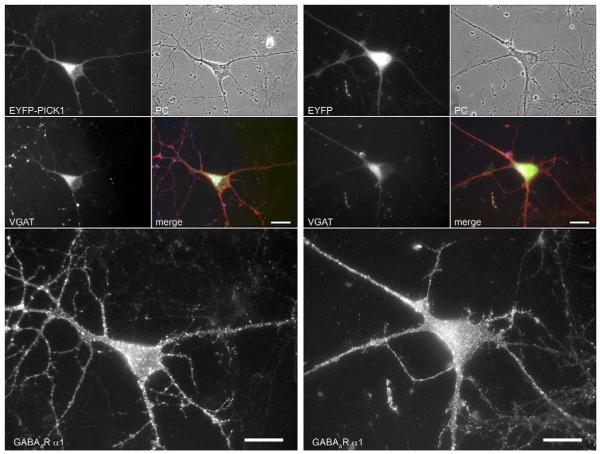
To further ascertain that PICK1 expression does not harm the cells causing non-specific redistribution of other surface receptors, we transfected EYFP-PICK1 or EYFP constructs in cultured neurons using magnetofection and stained the neurons for the GABAA receptor α1 subunit and VGAT as markers for interneurons (Fig. 9). The results show that the GABAA receptor α1 subunit immunofluorescence was unaffected in interneurons after EYFP-PICK1 expression compared to control EYFP expression. Therefore, expression of PICK1 in hippocampal GABAergic interneurons does not have a general effect on surface receptors, but specifically reduces surface clusters of α7 nAChRs.
Fig. 9.
Expression of EYFP-PICK1 does not affect the distribution of GABAA receptors at the surface of interneurons. EYFP-PICK1 or EYFP constructs were expressed in cultured hippocampal neurons using magnetofection. Neurons were stained against GABAA receptor α1 subunit (red; before permeabilization) and with VGAT-antibodies (blue; after permeabilization) as markers for interneurons. The panel shows two representative interneurons, overexpressing EYFP-PICK1 (left) or EYFP (right). The four small images on the top show the EYFP-PICK1 or EYFP signal, phase contrast (PC), VGAT signal and the merged image. The large images below show the GABAA receptor α1 subunit signal. Note the abundant GABAA receptor surface clusters in both interneurons and the healthy morphology of the cells. The GABAA α1 receptor signal is not affected in interneurons after EYFP-PICK1 expression compared to control EYFP expression. Scale bars, 20 μm.
Discussion
This study identifies the first synaptic scaffold protein, PICK1, that interacts with nAChRs in the CNS, exemplified by α7 nAChRs. We show that PICK1 binds to α7 in yeast, heterologous mammalian cells and hippocampal tissue. PICK1 and α7 clusters are detectable, sometimes adjacent and partially overlapping, at the surface of hippocampal GABAergic interneurons, and PICK1 negatively regulates α7 nAChR clustering in these cells.
PICK1 interacts with α7 nAChRs through its PDZ domain and an internal segment of the α7 loop
Very little is known about protein interactions of α7 nAChRs. SFKs bind to the cytoplasmic loop of α7, phosphorylate the receptor and decrease its activity (Charpantier et al., 2005). Ric-3 has been identified as an effector of functional expression and maturation of various nAChRs, including α7 receptors, in vertebrates and invertebrates. Ric-3 protein associates with α7 subunits in a complex, although it remains unknown whether it directly binds to the α7 nAChR and where such a binding region would map within the α7 protein (Ben-Ami et al., 2005; Halevi et al., 2002; Lansdell et al., 2005; Williams et al., 2005).
We identify PICK1 as a binding partner for the α7 cytoplasmic loop, and our experiments strongly suggest that this represents a direct and specific interaction of the two proteins. Thus, we observe PICK1–α7 interaction in yeast, using the cytoplasmic loop of α7asa bait. In recombinant pulldown experiments α7 loop fusion protein (GST) interacts with PICK1 protein that is either expressed in COS cells or in bacteria, and the interaction is also seen in the reverse case, using GST-PICK1 to pull down α7. Furthermore, interaction between PICK1 and α7 receptors is observed in the case of native proteins, because α-BT- or α7-antibody-precipitations bring down, in a specific fashion, PICK1 in lysates from brain and dissected hippocampus. In controls, the loops of other nAChR subunits do not interact with PICK, and α7 nAChRs do not associate with PSD95-proteins or GluR2 receptors. Finally, α7 nAChRs and PICK1 partially co-localize in heterologous cells and can be found adjacent in clusters at the surface of GABAergic interneurons. The combination of these data strongly implies a direct and specific interaction between PICK1 and the α7 loop. An intermediate protein would have to exist in yeast, bacteria, COS cells and neurons; it would have to survive the GST protein purification on glutathione-sepharose, and this is very unlikely.
We mapped the involved binding regions in both α7 and PICK1. Whereas in α7, a C-terminal peptide of the intracellular loop was necessary and sufficient, binding in PICK1 was mediated by its PDZ domain. Although the α7 peptide contains motifs similar to class I and II consensus binding motifs for PDZ-domains, these α7 sequences were not necessary to bind to the PDZ domain of PICK1. Thus the PDZ domain of PICK1 binds to an internal region in the α7 loop independent of consensus motifs. This is similar to Arf1 and Arf3, where the C-terminus also binds to PICK1 independent of consensus sequences (Takeya et al., 2000) — but the binding regions of Arf proteins and α7 do not show particular homology (Fig. 1).
PDZ domains of synaptic scaffolding proteins often bind to short motifs (class I, II or III) at the intracellular C-terminus of transmembrane receptors (Nourry et al., 2003). In this manner, PICK1 interacts with AMPA receptor subunits, mGluR7a, kainate receptors and others, through class I or class II PDZ-binding motifs (Boudin and Craig, 2001; Boudin et al., 2000; Hirbec et al., 2003; Madsen et al., 2005; Torres et al., 2001; Torres et al., 1998; Xia et al., 1999). We expand this range by introducing an interaction of PICK1’s PDZ domain with an internal protein segment in the cytoplasmic loop of α7. Although novel for PICK1, other PDZ domains are well known to bind to internal protein portions (Nourry et al., 2003). Internal recognition can be analogous to C-terminal interactions, i.e. according to the class I, II or III consensus features (Gee et al., 1998), suggesting that many PDZ domains might recognize internal motifs if these are provided in the correct structural context (Harris and Lim, 2001). Nonetheless, internal peptides lacking any consensus features can also be ligands for PDZ domains. One example is the interaction of dishevelled with the receptor Frizzled (Wong et al., 2003), and the PICK1–α7 interaction reported here expands this category.
PICK1 reduces clustering of α7 nAChRs at the surface of hippocampal GABAergic interneurons
To address the role of PICK1 in clustering of surface receptors, two standard tools are most often applied: (i) expression of receptor and PICK1 in heterologous cells to assess whether PICK1 can induce receptor clustering and (ii) overexpression of PICK1 in neurons that endogenously express the receptor to determine whether PICK1 affects native receptor clusters (Torres et al., 1998; Xia et al., 1999; Boudin and Craig, 2001; Boudin et al., 2000; Torres et al., 2001). These studies showed that in heterologous cells, PICK1 induces clustering of GluR2-containing AMPA-Rs, mGluR7a and others. The situation in neurons is more complex, as PICK1 can increase or decrease synaptic clustering of neurotransmitter receptors, depending on receptor subunits and neural cell type (Perez et al., 2001; Terashima et al., 2004; Torres et al., 2001). In our case, unlike any of the receptors described previously, PICK1 expression did not induce or affect clusters of α7 nAChRs in heterologous cells including SH-SY5Y, even though PICK1 itself was clustered, particularly in HEK 293T cells and SH-SY5Y cells. Yet, expression of PICK1 reduced clustering of α7at the surface of GABAergic hippocampal interneurons. This reduction was a specific and most likely direct process as supported by the following findings. First, the reduction was seen by using two entirely different techniques to express PICK1, viral expression or magnetofection. Second, the reduction, in the same way as binding to α7 did, required an intact PICK1 PDZ domain, since the AA mutation or expression of GFP alone had no effect. Third, the reduction did not involve intercellular interactions, because only PICK1-transfected interneurons were affected rather than adjacent non-transfected interneurons. Fourth, the reduction was not simply a follow-up effect of downregulation of GluR2, since we and others (Jonas and Burnashev, 1995; Leranth et al., 1996; Kawai et al., 2002) detected no overlap in the distribution of α7 and GluR2 (data not shown). Fifth, PICK1 expression did not affect surface clustering of GABAA receptors in hippocampal interneurons demonstrating that PICK1 does not have a general effect on surface receptors, but rather specifically reduces α7 surface clusters.
Thus our experiments point toward a specific PICK1–α7 mechanism, mediated by binding between these proteins, that controls α7 clustering at the surface of hippocampal interneurons. PICK1 does not induce α7 nAChR clustering, but interacts with the receptor and negatively regulates or limits α7 clustering. This mechanism may depend on one or several proteins expressed in interneurons that bind(s) to the α7–PICK1 complex and affect its targeting and/or transport processes. In such a manner, α7–PICK1 complexes may have a defined molecular composition in these cells, determining their intracellular targeting and clustering. Consistent with this, α7 clusters in populations of spinal cord neurons differently colocalize with cytoskeletal and lipid rafts components indicating that α7-containing protein complexes can be different between neuron populations (Roth and Berg, 2003).
Our data introduce PICK1 as first intracellular protein that controls clustering of nAChRs in the CNS, exemplified by α7 nAChRs. Since PICK1 does not interact with nAChR subunits α4 and β2 in our tests, PICK1′effects may be specific for the α7 receptors within the family of all nAChRs. Very little is known about clustering mechanisms for other nAChRs in the peripheral and central nervous system, while many players are known that regulate synaptic aggregation of muscle AChRs at the neuromuscular junction, as reviewed recently (Wiesner and Fuhrer, 2006). In chick ciliary ganglion, clustering of heteromeric nAChRs (α3, α5, β2 and β4 subunits) depends on signals within the cytoplasmic loop of α3 and requires postsynaptic functioning of APC protein (Temburni et al., 2004; Williams et al., 1998). PSD-93 and PSD-95 associate with these nAChRs and form a scaffold for nicotinic signaling (Conroy et al., 2003). In rodent superior cervical ganglion, formation and stabilization of cholinergic interneural synapses (containing clustered heteromeric nAChRs) require agrin and PSD-93, respectively (Gingras et al., 2002; Parker et al., 2004). At the neuromuscular junction, agrin/MuSK signaling and many intermediate proteins direct synaptic formation and AChR clustering (reviewed by Strochlic et al., 2005), APC being one requirement for AChR clustering (Wang et al., 2003), and rapsyn acting as an anchor (Gautam et al., 1995).
Possible mechanisms and relevance of PICK1 controlling α7 clustering
The pronounced reduction in surface α7 clustering by PICK1 in interneurons implies that not only receptors at GABAergic synapses are affected but also clusters that most likely represent extrasynaptic receptor aggregates. Regulation by PICK1 thus appears as a common property of all α7 receptor clusters in these cell cultures. There are many possibilities by which PICK1 could reduce α7 clustering. PICK1 could disperse surface receptor aggregates leading to diffuse receptors undetectable by our staining. In addition, PICK1 may reduce delivery of newly synthesized α7 nAChRs to the plasma membrane, or promote receptor internalization. The actions of PICK1 on other neurotransmitter receptors, together with the known protein interactions of PICK1 (Jin et al., 2006; Perez et al., 2001; Takeya et al., 2000), are compatible with any or even a combination of these possibilities. In a static microscopical picture, some clusters of PICK1 and α7 are adjacent and overlap partially, although precise colocalization is low (Fig. 6). Nonetheless, α7 and PICK1 can interact with each other (Figs. 1–4). It is likely, thus, that their interaction is under dynamic regulation and occurs transiently, for example in a “kiss and run” manner. Such transient protein interactions are prominent in membrane fusion events that underlie trafficking and secretion, for example synaptic vesicle exocytosis, recycling of caveolae or fusion between phagosomes and endosomes (Pelkmans and Zerial, 2005; Wightman and Haynes, 2004; Duclos et al., 2000). It is therefore possible that PICK1 acts in trafficking of α7 receptors towards or away from clusters rather than being a static anchor protein for clustered receptors, but more experimental approaches will be necessary to address these issues in detail. Functional expression of α-BT-binding α7 nAChRs is also regulated by palmitoylation of α7 receptors during their assembly in the ER (Drisdel et al., 2004), and tyrosine dephosphorylation can increase levels of α7 receptor at the surface (Cho et al., 2005), although clustering is not affected (Charpantier et al., 2005).
PICK1 was originally isolated as a binding protein of protein kinase C (PKCα) (Staudinger et al., 1995), and previous studies have shown the importance of the PICK1-PKC interaction for targeting and clustering mechanisms of other neurotransmitter receptors. For example, PICK1 targets PKCα to phosphorylate kainate receptors, causing their stabilization at the synapse by GRIP-interaction (Hirbec et al., 2003). Presynaptically, PICK1 binds to the C-terminus of mGluR7a and causes receptor clustering and phosphorylation by PKC (Boudin et al., 2000; Dev et al., 2000). Further work is necessary to elucidate the potential effect of putative PICK1-PKCα interactions on α7 nAChRs.
The neuronal network in the CNS is vulnerable to calcium-induced excitotoxicity, raising the need for control of calcium influx into individual neurons. Due to the fact that the α7 nAChR is highly permeable to calcium ions and involved in neuronal survival (Dajas-Bailador and Wonnacott, 2004; Seguela et al., 1993), the activity, distribution and clustering of this receptor should be precisely controlled. Our results have strong implications for PICK1 to play a role in these processes. Furthermore, controlling α7 clustering on hippocampal GABAergic interneurons could allow PICK1 to control the disinhibition of pyramidal cells in LTP, providing a potential mechanism for the role of α7 in learning, as the activity of postsynaptic α7 receptors on GABAergic interneurons influences hippocampal inhibition (Alkondon et al., 1997; Jones and Yakel, 1997), and as activation of these receptors blocks concurrent STP and LTP induction in pyramidal cells innervated by these interneurons (Ji et al., 2001). Finally, postsynaptic α7 nAChRs on GABAergic interneurons are also important in hippocampal sensory gating (Martin et al., 2004). Auditory gating is diminished with schizophrenia and used as a model for this disease in rodents (Martin et al., 2004; Ripoll et al., 2004). Interestingly, PICK1 polymorphism is associated with schizophrenia (Hong et al., 2004) and recent data implicate PICK1 as a susceptibility gene for schizophrenia (Fujii et al., 2006), while on the other hand many genetic and other studies have linked α7to this disease (Ripoll et al., 2004).
In summary, modulation of α7 nAChR activity and clustering may form one aspect of the various emerging roles of α7 nAChRs ranging from synaptic to systems level, including neuronal survival, nicotine addiction, synaptic plasticity in learning, and neurological disease. While recent progress has identified phosphorylation mechanisms as regulators of α7 nAChR activity (Charpantier et al., 2005; Cho et al., 2005), the present report implies PICK1 to control α7 nAChR clustering in the brain. The precise intracellular mechanisms and the relevance of this control for α7-mediated physiological and pathological processes remain to be investigated.
Experimental methods
Identification and cloning of PICK1 and yeast two-hybrid assay
Yeast two-hybrid (YTH) screening (Fields and Song, 1989) was performed using the Matchmaker System 3 (Clontech, Palo Alto, California) according to the manufacturer’s protocol, in order to identify α7 nAChR-binding proteins. The cytoplasmic loop of rat α7 nAChR cDNA (amino acids 332–467; α7 cDNA was a gift from Dr. Jim Boulter, UCLA, California) was inserted in-frame into the pGBKT7 bait vector. A rat brain cDNA library in vector pACT2 (Clontech) was used. Yeast cells (AH109) were sequentially cotransformed with α7 bait and library prey vectors, and then plated on selection medium lacking Ade, Trp, Leu and His. Two independent full-length PICK1 clones expressing His3, Ade and β-galactosidase activity were isolated. Positive clones were cotransformed with the bait vector or control plasmids into the AH109 yeast strain to confirm the interaction (i) on selection plates, (ii) with the Gal lift filter assay and (iii) using X-α-Gal indicator plates according to the manufacturer’s instructions (Clontech). All bait and prey plasmids used were from PCR products subcloned in frame into pGBKT7, pACT2 or pGADT7 vectors and were confirmed by DNA sequencing. The α7 nAChR bait sequences were PCR amplified and inserted into the bait vector using EcoRI and BamHI restriction sites. For example, the following primer pairs were used for α7 nAChR bait plasmid construction: bait 1 (aa 332–467) 5′-GCGCGAATTCAGAATCATTCTCCTGAAC + 5′-GCGCGGATCCTCACACCACGCAGGCTGC; bait 9 (aa 429–467) 5′-GCGCGAATTCGGGGACCCCGACCTGGCC + 5′-GCGCGGATCCT CACACCACGCAGGCTGC; bait 10 (aa 371–467) 5′-GCGCGAATTCCTGAGTGCA GGTGCTGGG + 5′-GCGCGGATCCTCACACCACGCAGGCTGC. The PICK1 prey sequences were PCR amplified and inserted into the prey vector using EcoRI and BamHI restriction sites. Other information for cDNA constructs is indicated in the Figures. To verify protein expression of bait constructs, yeast protein extracts were prepared of transformed yeast cells using the Urea/SDS method (Clontech) and analysed by Western blotting with the anti-GAL4 DNA-BD monoclonal antibody (Clontech) (data not shown). The point mutations in the cytoplasmic loop of rat α7 nAChR were introduced using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The primer pair to change the motif EVRY to EARA (aa 439–442) was 5′-TCCTGGAGGAGGCCCGCGCCATCGCCAACCGC + 5′-GCGGTTGGCGATGGCGCGGGCCTCCTCCAGGA. The primer pair to change the motif ESEV to EAEA (aa 452–455) was 5′-CTGCCAGGACGAGGCTGAGGCGATCTGCAGTGAATGG + 5′-CCATTCACTGCAGATCGCCTCAGCCTCGTCCTGGCAG. Constructs were verified by sequencing. The NCBI accession numbers are AF327562 for rat PICK1 and L31619 for rat α7nAChR.
Other DNA constructs
C-terminally Flag-tagged mouse α7 nAChR in pCS2+ expression vector was a gift from Dr. Ines Ibanez-Tallon (MDC Berlin-Buch). N-terminally EYFP-tagged rat PICK1 in pRK5 expression vector (referred to as EYFP-PICK1) and the empty control EYFP vector were a gift from Prof. Ann Marie Craig and Dr. Fernanda Laezza (Washington University) (Xia et al., 1999). All other constructs, including EGFP fused to the C-terminus of PICK1 in the vector pEGFP-N1 (Clontech) (PICK1-EGFP) were generated according to standard molecular biology techniques.
Fusion proteins, bacteria, COS-7 transfection and in-vitro binding
Full-length rat PICK1 or the cytoplasmic loop of rat α7 nAChR (aa 319–467) or of α4 nAChR were subcloned in frame into the GST-fusion vector pGEX-2T (Pharmacia, Piscataway, New Jersey). PICK1 was also subcloned into pET-28a(+) vector (carrying a His-tag; Novagen, EMD Biosciences, Darmstadt, Germany); and PICK1 was myc-tagged and cloned into pcDNA3 expression vector (Invitrogen). All constructs were confirmed by sequencing. The Escherichia coli strain DH5α was used to express GST fusion proteins and the strain BL21 to express His-PICK1, in both cases using IPTG as an inducer. GST fusions were purified using glutathione-sepharose beads as described previously (Fuhrer and Hall, 1996). Bacteria were transfected using a standard heat shock procedure. COS-7 cells were transfected with α7 or myc-PICK1 constructs using Fugene 6 Transfection Reagent (Roche Applied Science, Roche Diagnostics Corporation, Indianapolis, IN). Transfected bacteria and COS cells were lysed in bacterial (Smith and Johnson, 1988) and eucaryotic (Fuhrer et al., 1997) cell lysis buffer, respectively, and incubated with purified fusion proteins, i.e. GST-PICK1, GST-α7loop, or GST-α4loop immobilized to beads. Beads were pelleted, washed with lysis buffer, and analyzed by α7-, myc- or His-immunoblotting. Blots were reprobed for GST. For detection, mouse monoclonal anti-myc antibodies (Sigma), goat polyclonal anti-α7 antibodies (Drisdel and Green, 2000) (Santa Cruz Biotechnology), mAb306 (against α7) (Rangwala et al., 1997; Schoepfer et al., 1990), mouse monoclonal anti-GST antibodies (Santa Cruz Biotechnology), and rabbit polyclonal anti-T7 tag antibodies (His-tag; Novagen) were used.
Rat brain preparation and α7 precipitation
Synaptosomes were prepared from dissected adult rat hippocampus as described previously (Carlin et al., 1980). Briefly, tissue was homogenized in buffer A (0.32 M Sucrose, 1 mM NaHCO3, 1 mM MgCl2, 0.5 mM CaCl2) on ice and centrifuged at 1400×g for 10 min at 4 °C, and the supernatant was saved (S1). The pellet (P1) was resuspended in buffer A, centrifuged at 720×g for 10 min at 4 °C and the pellet (P2) discarded. S2 and S1 were combined, centrifuged at 720×g for 10 min at 4 °C and pellets (P3) were discarded. The supernatants S3 were centrifuged at 13,800×g for 10 min at 4 °C and the supernatant (S4) discarded. The pellet (P4) was resuspended in buffer B (50 mM Tris, 150 mM NaCl, 5 mM EDTA; pH 7.4; containing protease inhibitors (Complete Mini protease inhibitor tablets; Roche, Switzerland), and 0.5% Triton X-100 was added. Samples were rotated for 15 min at 4 °C and split into two identical portions. To one sample, free α-BT was added (10 μM final concentration) and both samples were rotated for 60 min at 4 °C.
Alternatively, lysates from adult rat brain membranes were prepared as described previously (Chen and Patrick, 1997). Briefly, brain tissue was homogenized in buffer A1 (50 mM sodium phosphate, 50 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1 mM phenylmethylsulfonyl fluoride) on ice, centrifuged 2 times at 100,000×g for 1 h at 4 °C, and the supernatants were discarded. The pellet was resuspended in ice-cold buffer A2 (buffer A1 plus 2% Triton X-100 and protease inhibitors), rotated for 2 h at 4 °C, and centrifuged at 100,000×g for 1 h at 4 °C. The supernatant was split into two identical samples to which 10 mM nicotine and vehicle, respectively, were added. Samples were rotated for 1 h at 4 °C.
For precipitation with α-BT, 50 μlof α-BT coupled to sepharose beads (Fuhrer and Hall, 1996) were added to both samples (prepared either from hippocampal synaptosomes or from whole brain membranes, see above) for 2 h. Alternatively, for the α7 immunoprecipitation, 1 μl of anti-α7 nAChR antibody mAb319 (Sigma) (Rangwala et al., 1997; Schoepfer et al., 1990) was added for 1 h, followed by Protein G-Sepharose (Amersham Biosciences AB, Uppsala, Sweden). In controls, mAb319 was replaced by an identical amount of rat non-immune IgG. Beads were pelleted, washed with buffer B, and proteins eluted with Lammli buffer at 80 °C and subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes for Western blotting. The anti-PICK1 antibody (Upstate Biotech, New York) was used at 1:500, the anti-α7 antibody (mAb306, mAb319; or ab10096 from Abcam Ltd., Cambridge UK; Charpantier et al., 2005) at 1:1000, the anti-PSD95 family antibody (Upstate) at 1:1000, and the anti-GluR2 antibody (Chemicon, MAB397) at 1:1000. Rat brain microsomal preparations (Upstate) were used as positive controls for the PICK1 signal according to the manufacturer’s instructions (data not shown). HRP-conjugated secondary antibodies (Zymed) were used and detected with enhanced chemiluminescence (SuperSignal West Dura Extended Duration Substrate kit (Pierce)).
Cell culture and immunocytochemistry of primary hippocampal neurons
Rat embryos were obtained from time-pregnant Wistar rats (RCC Laboratory Animal Services, Füllinsdorf, Switzerland). All experiments were approved by the cantonal veterinary office of Zürich. Primary cultures of embryonic day (E)18–19 hippocampal neurons were prepared as described previously (Brunig et al., 2002a,b). The cells were grown in neurobasal medium (Gibco) supplemented with B27 supplement (Gibco), 0.5 mM L-glutamine, and 1.25 mg/ml gentamicin (Gibco) in the presence of a glial feeder cell layer. The cells were plated at 1.5 × 104 per 18 mm glass coverslip previously coated with poly-L-lysine (Sigma) and used for immunocytochemistry after 2–3 weeks.
For the magnetofection method, hippocampal neurons were cultivated as described (Chudotvorova et al., 2005) at a density of 5 × 105 cells per coverslip for 11 days in 5% CO2 and 37 °C in the absence of a glial feeder cell layer in MEM medium (Invitrogen) containing 15% NU serum (BD), 2% B27 supplement, 0.015 M HEPES pH 7.1, 0.45% glucose, 1 mM sodium pyruvate (Invitrogen), and 2 mM L-Glutamine (Gibco).
In all cases, immunocytochemistry was performed according to Brunig et al. (2002a,b). In brief, the living cultures were incubated for 30–60 min at room temperature with 100 nM α-BT coupled to rhodamine, Alexa 488 or Alexa 647 (Molecular Probes) in medium or Ringer’s solution (in mM: CaCl2 2, MgCl2 2, glycine 0.001, with or without TTX 0.0005, glucose 30, HEPES 25, KCl 5, NaCl 119, pH 7.4) (Archibald et al., 1998). They were subsequently washed with Ringer’s solution and fixed with 4% PFA in 0.15 M phosphate buffer for 15 min at RT, followed by washing with PBS and permeabilization for 5 min at RT using 0.2% Triton-X 100 in PBS containing 10% normal goat serum (NGS). Fixed cultures were rinsed extensively with PBS and incubated for 90 min at RT with the following antibodies diluted in PBS containing 10% NGS: rabbit immunoaffinity purified anti-PICK1 (Upstate Biotech., diluted 1:50), rabbit polyclonal or mouse monoclonal anti-VGAT (Synaptic Systems, diluted 1:1000 or 1:500, respectively), rabbit polyclonal anti-GAD65/67 (Affinity, diluted 1:2000), mouse monoclonal anti-bassoon (Stressgen Bioreagents, Ann Arbor, Michigan, 48108 USA, diluted 1:500), mouse monoclonal anti-gephyrin (mAb7a, Connex, Martinsried, Germany, diluted 1:800), or rabbit polyclonal anti-PSD-95 (Cho et al., 1992) (diluted 1:1000). The mouse monoclonal anti-GluR2 antibody against the large N-terminal extracellular domain of GluR2 (Chemicon, diluted 1:200) was incubated on living cultures as described above. Cultures were subsequently washed with PBS and incubated with secondary antibody coupled to Alexa 488, Alexa 350, or rhodamine (Molecular Probes, Jackson Laboratories; diluted 1:200) for 30 min at RT in PBS plus 10% NGS. After washing in PBS, cells were mounted in Mowiol and stored at 4 °C.
COS-7, HEK 293T and SH-SY5Y cells, neuron transfection and staining
Cells were plated onto glass coverslips. COS-7 and HEK 293T cells were used 48 h after transfection or electroporation, and neurons and SH-SY5Y cells were analysed 24 h after magnetofection. COS-7 cells were transfected with HA-tagged PICK1 and nAChR α7 expression constructs using the Fugene transfection reagent (Roche, Indianapolis, IN) according to the manufacturer’s instructions. HEK 293T cells and SH-SY5Y cells stably overexpressing nAChR α7 subunit (Charpantier et al., 2005) were electroporated with different constructs using the nucleofection method according to manufacturer’s instructions (Amaxa Biosystems, Cologne, Germany). Hippocampal neurons were transfected using the magnetofection method at 11 days in vitro with CombiMag (OZ Biosciences) as described earlier (Chudotvorova et al., 2005). The following plasmids were used: EYFP-PICK1, EYFP, PICK1-EGFP, and/or Flag-tagged α7 constructs. Living SH-SY5Y cells and neurons were incubated for 30–60 min with α-BT coupled to rhodamine (Molecular Probes, 100 nM) and/or with a mouse monoclonal anti-Flag antibody (Sigma, diluted 1:1000), and/or with a rabbit polyclonal anti-GABAA receptor α1 subunit antibody (Fritschy and Mohler, 1995)(diluted 1:5000), and subsequently washed with PBS. All cells were fixed with 4% PFA, permeabilized and stained as described above for neuronal cells using the following primary antibodies: mouse (Roche, diluted 1:1000) or rat monoclonal anti-HA (Roche, diluted 1:200), mAb306 (diluted 1:200), or rabbit polyclonal anti-α7 (ab10096 from Abcam Ltd., diluted 1:200).
Viral infection
Neurons were used after 14 days in vitro, transferred to a dish containing conditioned medium without the glia feeder cell layer and were incubated with or without Sindbis virus. We used the same Sindbis constructs and conditions as previously described (Terashima et al., 2004). Incubation was done for 17–22 h (Perez et al., 2001; Terashima et al., 2004), after which cells were incubated with α-BT-rhodamine, washed, fixed and analyzed by epifluorescence or confocal microscopy.
Data analysis
Experiments were analyzed by epifluorescence microscopy (ApoTome, Carl Zeiss AG, Germany) and using a high-resolution digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) or by confocal laser scanning microscopy (TCS 4D; Leica, Deerfield, IL). Images were acquired with a 100× lens (numerical aperture 1.4) at a magnification of 0.11 μm/pixel. Controls in which one or more primary antibodies were omitted indicated no significant cross-contamination among fluorescence channels. Imaging conditions were kept constant for each channel. Contrast-optimized images using the Photoshop software were analyzed with the ImageJ imaging software (NIH) keeping constant threshold levels. Clusters were defined by their intensity (more than twice the intensity of the surrounding membrane) and size (at least 9 adjacent pixels). For display, only minimal contrast adjustments were made.
Quantitative analyses after virus infection (Fig. 7) were performed on randomly selected samples in a total of 121 cells originating from three independent cultures (9 GFP virus-infected cells; 43 non-infected cells; 29 WT virus-infected cells; 40 AA virus-infected cells). Along three membrane regions of each cell (soma, proximal and distal dendrites), four areas, each covering 100 μm2, were randomly chosen per region. The boxes in Fig. 7 show examples of somatic areas. Definitions were: proximal dendrites, dendritic areas from the soma to a distance of about 140 μm; distal dendrites, dendritic areas on smaller branching dendrites further away than 140 μm from the soma. In Fig. 8, segments of 75 μm2 were selected on proximal dendrites from 86 cells from 2 independent cultures (30 nontransfected cells, 31 EYFP transfected cells, 25 EYFP-PICK1 transfected cells). Within each area, the surface covered by α-BT-fluorescence was measured with the ImageJ imaging software (NIH), and values were expressed as mean ± S.E.M.
Acknowledgments
We are grateful to Martin Schwab for helpful discussions, Patric Matter for assistance in experiments with bacterial fusion proteins, Corinne Sidler and Barbara Studler for help with neuronal primary cultures, Christophe Pellegrino from the INMED/INSERM Marseille, France for help with magnetofection, Anne Greet Bittermann from the Laboratory of Electron Microscopy at the University of Zürich for her excellent technical assistance with the confocal microscope, Alain Camilleri for statistical analysis, Jose Maria Mateos for insights into image analysis, and Eva Hochreutener and Roland Schöb for help with illustrations. This work was supported by the Dr. Eric Slack-Gyr Foundation, the Swiss National Science Foundation and the Swiss Foundation for Research on Muscle Diseases.
Footnotes
Available online on ScienceDirect (www.sciencedirect.com).
References
- Alkondon M, Pereira EF, Barbosa CT, Albuquerque EX. Neuronal nicotinic acetylcholine receptor activation modulates gamma-aminobutyric acid release from CA1 neurons of rat hippocampal slices. J. Pharmacol. Exp. Ther. 1997;283:1396–1411. [PubMed] [Google Scholar]
- Alkondon M, Pereira EF, Albuquerque EX. Alpha-bungarotoxin- and methyllycaconitine-sensitive nicotinic receptors mediate fast synaptic transmission in interneurons of rat hippocampal slices. Brain Res. 1998;810:257–263. doi: 10.1016/s0006-8993(98)00880-4. [DOI] [PubMed] [Google Scholar]
- Archibald K, Perry MJ, Molnar E, Henley JM. Surface expression and metabolic half-life of AMPA receptors in cultured rat cerebellar granule cells. Neuropharmacology. 1998;37:1345–1353. doi: 10.1016/s0028-3908(98)00135-x. [DOI] [PubMed] [Google Scholar]
- Ben-Ami HC, Yassin L, Farah H, Michaeli A, Eshel M, Treinin M. RIC-3 affects properties and quantity of nicotinic acetylcholine receptors via a mechanism that does not require the coiled-coil domains. J. Biol. Chem. 2005;280:28053–28060. doi: 10.1074/jbc.M504369200. [DOI] [PubMed] [Google Scholar]
- Berg DK, Conroy WG. Nicotinic alpha 7 receptors: synaptic options and downstream signaling in neurons. J. Neurobiol. 2002;53:512–523. doi: 10.1002/neu.10116. [DOI] [PubMed] [Google Scholar]
- Boudin H, Craig AM. Molecular determinants for PICK1 synaptic aggregation and mGluR7a receptor coclustering: role of the PDZ, coiled-coil, and acidic domains. J. Biol. Chem. 2001;276:30270–30276. doi: 10.1074/jbc.M102991200. [DOI] [PubMed] [Google Scholar]
- Boudin H, Doan A, Xia J, Shigemoto R, Huganir RL, Worley P, Craig AM. Presynaptic clustering of mGluR7a requires the PICK1 PDZ domain binding site. Neuron. 2000;28:485–497. doi: 10.1016/s0896-6273(00)00127-6. [DOI] [PubMed] [Google Scholar]
- Brunig I, Scotti E, Sidler C, Fritschy JM. Intact sorting, targeting, and clustering of gamma-aminobutyric acid A receptor subtypes in hippocampal neurons in vitro. J. Comp. Neurol. 2002a;443:43–55. doi: 10.1002/cne.10102. [DOI] [PubMed] [Google Scholar]
- Brunig I, Suter A, Knuesel I, Luscher B, Fritschy JM. GABAergic terminals are required for postsynaptic clustering of dystrophin but not of GABA(A) receptors and gephyrin. J. Neurosci. 2002b;22:4805–4813. doi: 10.1523/JNEUROSCI.22-12-04805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruses JL, Chauvet N, Rutishauser U. Membrane lipid rafts are necessary for the maintenance of the (alpha)7 nicotinic acetylcholine receptor in somatic spines of ciliary neurons. J. Neurosci. 2001;21:504–512. doi: 10.1523/JNEUROSCI.21-02-00504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpantier E, Wiesner A, Huh KH, Ogier R, Hoda JC, Allaman G, Raggenbass M, Feuerbach D, Bertrand D, Fuhrer C. Alpha7 neuronal nicotinic acetylcholine receptors are negatively regulated by tyrosine phosphorylation and Src-family kinases. J. Neurosci. 2005;25:9836–9849. doi: 10.1523/JNEUROSCI.3497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Patrick JW. The alpha-bungarotoxin-binding nicotinic acetylcholine receptor from rat brain contains only the alpha7 subunit. J. Biol. Chem. 1997;272:24024–24029. doi: 10.1074/jbc.272.38.24024. [DOI] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- Cho CH, Song W, Leitzell K, Teo E, Meleth AD, Quick MW, Lester RA. Rapid upregulation of alpha7 nicotinic acetylcholine receptors by tyrosine dephosphorylation. J. Neurosci. 2005;25:3712–3723. doi: 10.1523/JNEUROSCI.5389-03.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudotvorova I, Ivanov A, Rama S, Hubner CA, Pellegrino C, Ben-Ari Y, Medina I. Early expression of KCC2 in rat hippocampal cultures augments expression of functional GABA synapses. J. Physiol. 2005;566:671–679. doi: 10.1113/jphysiol.2005.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy WG, Liu Z, Nai Q, Coggan JS, Berg DK. PDZ-containing proteins provide a functional postsynaptic scaffold for nicotinic receptors in neurons. Neuron. 2003;38:759–771. doi: 10.1016/s0896-6273(03)00324-6. [DOI] [PubMed] [Google Scholar]
- Cooper ST, Millar NS. Host cell-specific folding and assembly of the neuronal nicotinic acetylcholine receptor alpha7 subunit. J. Neurochem. 1997;68:2140–2151. doi: 10.1046/j.1471-4159.1997.68052140.x. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Yokoyama N, Bianchi LM, Henkemeyer M, Fritzsch B. EphB2 guides axons at the midline and is necessary for normal vestibular function. Neuron. 2000;26:417–430. doi: 10.1016/s0896-6273(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F, Wonnacott S. Nicotinic acetylcholine receptors and the regulation of neuronal signalling. Trends Pharmacol. Sci. 2004;25:317–324. doi: 10.1016/j.tips.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S. The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nakajima Y, Kitano J, Braithwaite SP, Henley JM, Nakanishi S. PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J. Neurosci. 2000;20:7252–7257. doi: 10.1523/JNEUROSCI.20-19-07252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel RC, Green WN. Neuronal alpha-bungarotoxin receptors are alpha7 subunit homomers. J. Neurosci. 2000;20:133–139. doi: 10.1523/JNEUROSCI.20-01-00133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel RC, Manzana E, Green WN. The role of palmitoylation in functional expression of nicotinic alpha7 receptors. J. Neurosci. 2004;24:10502–10510. doi: 10.1523/JNEUROSCI.3315-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S, Diez R, Garin J, Papadopoulou B, Descoteaux A, Stenmark H, Desjardins M. Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J. Cell Sci. 2000;113(Pt. 19):3531–3541. doi: 10.1242/jcs.113.19.3531. [DOI] [PubMed] [Google Scholar]
- Fabian-Fine R, Skehel P, Errington ML, Davies HA, Sher E, Stewart MG, Fine A. Ultrastructural distribution of the alpha7 nicotinic acetylcholine receptor subunit in rat hippocampus. J. Neurosci. 2001;21:7993–8003. doi: 10.1523/JNEUROSCI.21-20-07993.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Frazier CJ, Buhler AV, Weiner JL, Dunwiddie TV. Synaptic potentials mediated via alpha-bungarotoxin-sensitive nicotinic acetylcholine receptors in rat hippocampal interneurons. J. Neurosci. 1998;18:8228–8235. doi: 10.1523/JNEUROSCI.18-20-08228.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Mohler H. GABAA-receptor heterogeneity in the adult rat brain: differential regional and cellular distribution of seven major subunits. J. Comp. Neurol. 1995;359:154–194. doi: 10.1002/cne.903590111. [DOI] [PubMed] [Google Scholar]
- Fuhrer C, Hall ZW. Functional interaction of Src family kinases with the acetylcholine receptor in C2 myotubes. J. Biol. Chem. 1996;271:32474–32481. doi: 10.1074/jbc.271.50.32474. [DOI] [PubMed] [Google Scholar]
- Fuhrer C, Sugiyama JE, Taylor RG, Hall ZW. Association of muscle-specific kinase MuSK with the acetylcholine receptor in mammalian muscle. EMBO J. 1997;16:4951–4960. doi: 10.1093/emboj/16.16.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Maeda K, Hikida T, Mustafa AK, Balkissoon R, Xia J, Yamada T, Ozeki Y, Kawahara R, Okawa M, Huganir RL, Ujike H, Snyder SH, Sawa A. Serine racemase binds to PICK1: potential relevance to schizophrenia. Mol. Psychiatry. 2006;11:150–157. doi: 10.1038/sj.mp.4001776. [DOI] [PubMed] [Google Scholar]
- Gautam M, Noakes PG, Mudd J, Nichol M, Chu GC, Sanes JR, Merlie JP. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature. 1995;377:232–236. doi: 10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- Gee SH, Sekely SA, Lombardo C, Kurakin A, Froehner SC, Kay BK. Cyclic peptides as non-carboxyl-terminal ligands of syntrophin PDZ domains. J. Biol. Chem. 1998;273:21980–21987. doi: 10.1074/jbc.273.34.21980. [DOI] [PubMed] [Google Scholar]
- Gingras J, Rassadi S, Cooper E, Ferns M. Agrin plays an organizing role in the formation of sympathetic synapses. J. Cell Biol. 2002;158:1109–1118. doi: 10.1083/jcb.200203012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevi S, McKay J, Palfreyman M, Yassin L, Eshel M, Jorgensen E, Treinin M. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Hirbec H, Francis JC, Lauri SE, Braithwaite SP, Coussen F, Mulle C, Dev KK, Coutinho V, Meyer G, Isaac JT, Collingridge GL, Henley JM. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron. 2003;37:625–638. doi: 10.1016/s0896-6273(02)01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CJ, Liao DL, Shih HL, Tsai SJ. Association study of PICK1 rs3952 polymorphism and schizophrenia. NeuroReport. 2004;15:1965–1967. doi: 10.1097/00001756-200408260-00026. [DOI] [PubMed] [Google Scholar]
- Huh KH, Fuhrer C. Clustering of nicotinic acetylcholine receptors: from the neuromuscular junction to interneuronal synapses. Mol. Neurobiol. 2002;25:79–112. doi: 10.1385/MN:25:1:079. [DOI] [PubMed] [Google Scholar]
- Ji D, Lape R, Dani JA. Timing and location of nicotinic activity enhances or depresses hippocampal synaptic plasticity. Neuron. 2001;31:131–141. doi: 10.1016/s0896-6273(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Jin W, Ge WP, Xu J, Cao M, Peng L, Yung W, Liao D, Duan S, Zhang M, Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J. Neurosci. 2006;26:2380–3290. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Burnashev N. Molecular mechanisms controlling calcium entry through AMPA-type glutamate receptor channels. Neuron. 1995;15:987–990. doi: 10.1016/0896-6273(95)90087-x. [DOI] [PubMed] [Google Scholar]
- Jones S, Yakel JL. Functional nicotinic ACh receptors on interneurones in the rat hippocampus. J. Physiol. 1997;504(Pt. 3):603–610. doi: 10.1111/j.1469-7793.1997.603bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Sudweeks S, Yakel JL. Nicotinic receptors in the brain: correlating physiology with function. Trends Neurosci. 1999;22:555–561. doi: 10.1016/s0166-2236(99)01471-x. [DOI] [PubMed] [Google Scholar]
- Kawai H, Zago W, Berg DK. Nicotinic alpha 7 receptor clusters on hippocampal GABAergic neurons: regulation by synaptic activity and neurotrophins. J. Neurosci. 2002;22:7903–7912. doi: 10.1523/JNEUROSCI.22-18-07903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdell SJ, Gee VJ, Harkness PC, Doward AI, Baker ER, Gibb AJ, Millar NS. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol. Pharmacol. 2005;68:1431–1438. doi: 10.1124/mol.105.017459. [DOI] [PubMed] [Google Scholar]
- Leranth C, Szeidemann Z, Hsu M, Buzsaki G. AMPA receptors in the rat and primate hippocampus: a possible absence of GluR2/3 subunits in most interneurons. Neuroscience. 1996;70:631–652. doi: 10.1016/s0306-4522(96)83003-x. [DOI] [PubMed] [Google Scholar]
- Levy RB, Aoki C. Alpha7 nicotinic acetylcholine receptors occur at postsynaptic densities of AMPA receptor-positive and -negative excitatory synapses in rat sensory cortex. J. Neurosci. 2002;22:5001–5015. doi: 10.1523/JNEUROSCI.22-12-05001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom J, Anand R, Peng X, Gerzanich V, Wang F, Li Y. Neuronal nicotinic receptor subtypes. Ann. N. Y. Acad. Sci. 1995;757:100–116. doi: 10.1111/j.1749-6632.1995.tb17467.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ford B, Mann MA, Fischbach GD. Neuregulins increase alpha7 nicotinic acetylcholine receptors and enhance excitatory synaptic transmission in GABAergic interneurons of the hippocampus. J. Neurosci. 2001;21:5660–6569. doi: 10.1523/JNEUROSCI.21-15-05660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen KL, Beuming T, Niv MY, Chang CW, Dev KK, Weinstein H, Gether U. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J. Biol. Chem. 2005;280:20539–20548. doi: 10.1074/jbc.M500577200. [DOI] [PubMed] [Google Scholar]
- Martin LF, Kem WR, Freedman R. Alpha-7 nicotinic receptor agonists: potential new candidates for the treatment of schizophrenia. Psychopharmacology (Berl.) 2004;174:54–64. doi: 10.1007/s00213-003-1750-1. [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci. STKE RE7. 2003 doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, Murray TK, Lakics V, Visanji NP, Duty S. The role of neuronal nicotinic acetylcholine receptors in acute and chronic neurodegeneration. Curr. Drug Targets CNS Neurol. Disord. 2002;1:399–411. doi: 10.2174/1568007023339166. [DOI] [PubMed] [Google Scholar]
- Parker MJ, Zhao S, Bredt DS, Sanes JR, Feng G. PSD93 regulates synaptic stability at neuronal cholinergic synapses. J. Neurosci. 2004;24:378–388. doi: 10.1523/JNEUROSCI.3865-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436:128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- Peng X, Katz M, Gerzanich V, Anand R, Lindstrom J. Human alpha 7 acetylcholine receptor: cloning of the alpha 7 subunit from the SH-SY5Y cell line and determination of pharmacological properties of native receptors and functional alpha 7 homomers expressed in Xenopus oocytes. Mol. Pharmacol. 1994;45:546–554. [PubMed] [Google Scholar]
- Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala F, Drisdel RC, Rakhilin S, Ko E, Atluri P, Harkins AB, Fox AP, Salman SS, Green WN. Neuronal alphabungarotoxin receptors differ structurally from other nicotinic acetylcholine receptors. J. Neurosci. 1997;17:8201–8212. doi: 10.1523/JNEUROSCI.17-21-08201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll N, Bronnec M, Bourin M. Nicotinic receptors and schizophrenia. Curr. Med. Res. Opin. 2004;20:1057–1074. doi: 10.1185/030079904125004060. [DOI] [PubMed] [Google Scholar]
- Roth AL, Berg DK. Large clusters of alpha7-containing nicotinic acetylcholine receptors on chick spinal cord neurons. J. Comp. Neurol. 2003;465:195–204. doi: 10.1002/cne.10856. [DOI] [PubMed] [Google Scholar]
- Sadasivam G, Willmann R, Lin S, Erb-Vogtli S, Kong XC, Ruegg MA, Fuhrer C. Src-family kinases stabilize the neuromuscular synapse in vivo via protein interactions, phosphorylation, and cytoskeletal linkage of acetylcholine receptors. J. Neurosci. 2005;25:10479–10493. doi: 10.1523/JNEUROSCI.2103-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepfer R, Conroy WG, Whiting P, Gore M, Lindstrom J. Brain alpha-bungarotoxin binding protein cDNAs and MAbs reveal subtypes of this branch of the ligand-gated ion channel gene superfamily. Neuron. 1990;5:35–48. doi: 10.1016/0896-6273(90)90031-a. [DOI] [PubMed] [Google Scholar]
- Seguela P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J. Neurosci. 1993;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoop RD, Martone ME, Yamada N, Ellisman MH, Berg DK. Neuronal acetylcholine receptors with alpha7 subunits are concentrated on somatic spines for synaptic signaling in embryonic chick ciliary ganglia. J. Neurosci. 1999;19:692–704. doi: 10.1523/JNEUROSCI.19-02-00692.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Lu J, Olson EN. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J. Biol. Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- Steinberg JP, Takamiya K, Shen Y, Xia J, Rubio ME, Yu S, Jin W, Thomas GM, Linden DJ, Huganir RL. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron. 2006;49:845–860. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Strochlic L, Cartaud A, Cartaud J. The synaptic muscle-specific kinase (MuSK) complex: New partners, new functions. BioEssays. 2005;27:1129–1135. doi: 10.1002/bies.20305. [DOI] [PubMed] [Google Scholar]
- Takeya R, Takeshige K, Sumimoto H. Interaction of the PDZ domain of human PICK1 with class I ADP-ribosylation factors. Biochem. Biophys. Res. Commun. 2000;267:149–155. doi: 10.1006/bbrc.1999.1932. [DOI] [PubMed] [Google Scholar]
- Temburni MK, Rosenberg MM, Pathak N, McConnell R, Jacob MH. Neuronal nicotinic synapse assembly requires the adenomatous polyposis coli tumor suppressor protein. J. Neurosci. 2004;24:6776–6784. doi: 10.1523/JNEUROSCI.1826-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima A, Cotton L, Dev KK, Meyer G, Zaman S, Duprat F, Henley JM, Collingridge GL, Isaac JT. Regulation of synaptic strength and AMPA receptor subunit composition by PICK1. J. Neurosci. 2004;24:5381–5390. doi: 10.1523/JNEUROSCI.4378-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- Torres GE, Yao WD, Mohn AR, Quan H, Kim KM, Levey AI, Staudinger J, Caron MG. Functional interaction between monoamine plasma membrane transporters and the synaptic PDZ domain-containing protein PICK1. Neuron. 2001;30:121–134. doi: 10.1016/s0896-6273(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Wang J, Jing Z, Zhang L, Zhou G, Braun J, Yao Y, Wang ZZ. Regulation of acetylcholine receptor clustering by the tumor suppressor APC. Nat. Neurosci. 2003;6:1017–1018. doi: 10.1038/nn1128. [DOI] [PubMed] [Google Scholar]
- Wiesner A, Fuhrer C. Regulation of nicotinic acetylcholine receptors by tyrosine kinases in the peripheral and central nervous system: same players, different roles. Cell. Mol Life Sci. 2006;63:2818–2828. doi: 10.1007/s00018-006-6081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman RM, Haynes CL. Synaptic vesicles really do kiss and run. Nat. Neurosci. 2004;7:321–322. doi: 10.1038/nn0404-321. [DOI] [PubMed] [Google Scholar]
- Williams BM, Temburni MK, Levey MS, Bertrand S, Bertrand D, Jacob MH. The long internal loop of the alpha 3 subunit targets nAChRs to subdomains within individual synapses on neurons in vivo. Nat. Neurosci. 1998;1:557–562. doi: 10.1038/2792. [DOI] [PubMed] [Google Scholar]
- Williams ME, Burton B, Urrutia A, Shcherbatko A, Chavez-Noriega LE, Cohen CJ, Aiyar J. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor alpha7 subunit in mammalian cells. J. Biol. Chem. 2005;280:1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- Willmann R, Pun S, Stallmach L, Sadasivam G, Santos AF, Caroni P, Fuhrer C. Cholesterol and lipid microdomains stabilize the postsynapse at the neuromuscular junction. EMBO J. 2006;25:4050–4060. doi: 10.1038/sj.emboj.7601288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, Mlodzik M, Shi DL, Zheng J. Direct binding of the PDZ domain of dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell. 2003;12:1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Zarei MM, Radcliffe KA, Chen D, Patrick JW, Dani JA. Distributions of nicotinic acetylcholine receptor alpha7 and beta2 subunits on cultured hippocampal neurons. Neuroscience. 1999;88:755–764. doi: 10.1016/s0306-4522(98)00246-2. [DOI] [PubMed] [Google Scholar]