Abstract
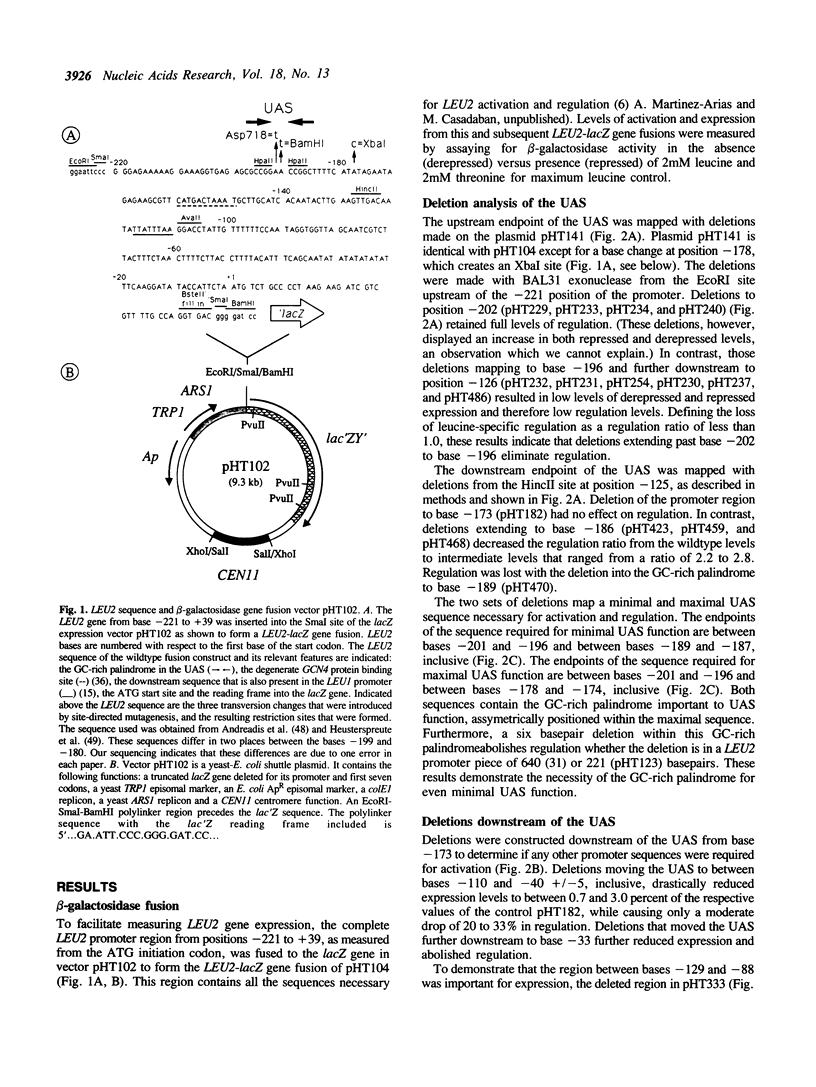
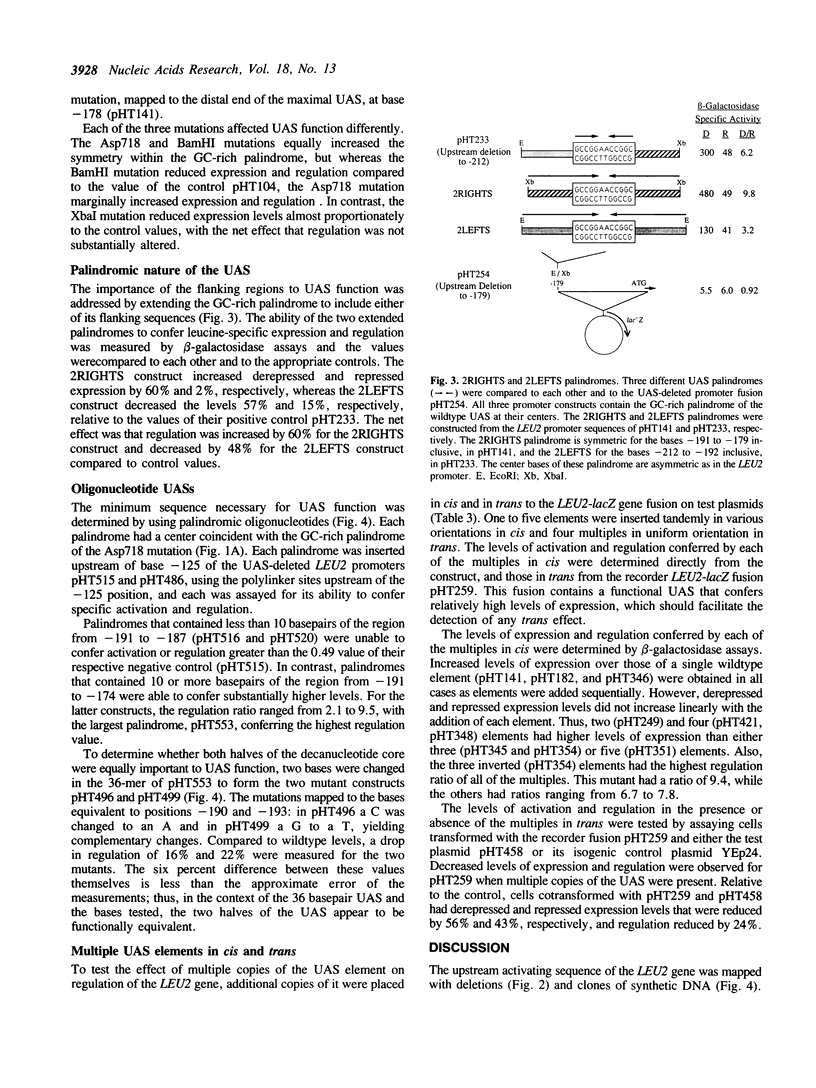
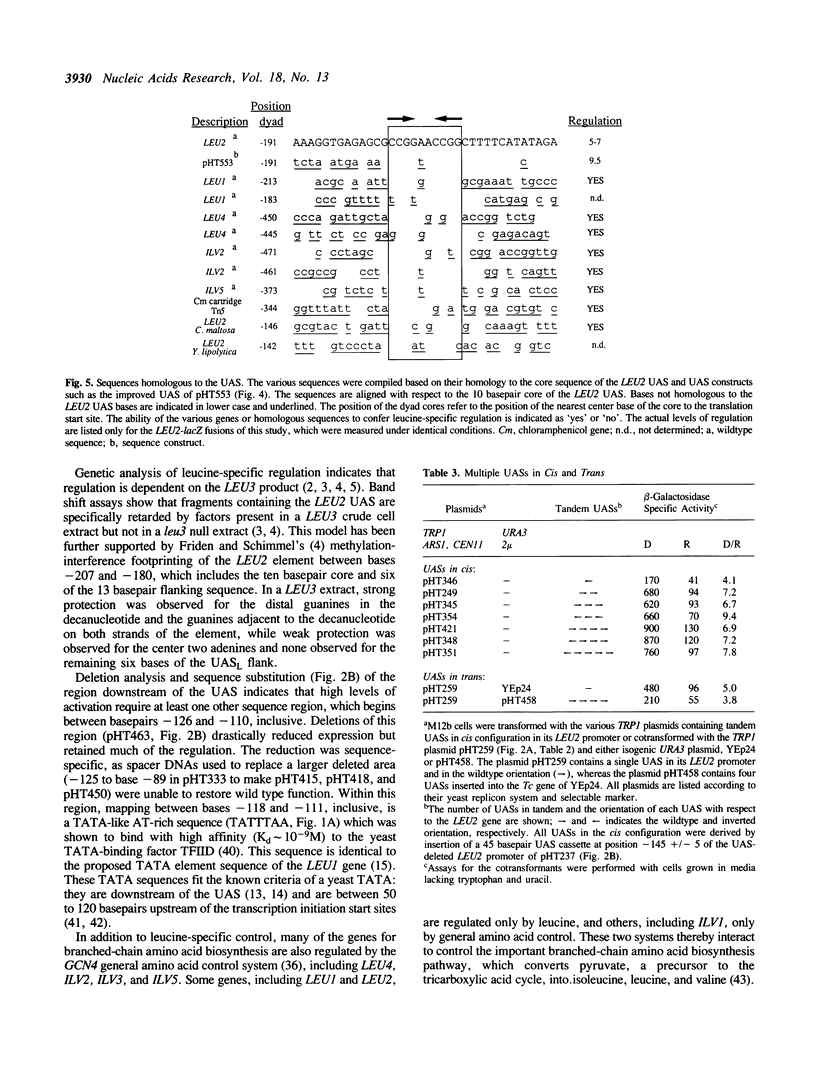
The upstream activating sequence (UAS) conferring leucine-specific regulation of transcription in Saccharomyces cerevisiae was identified by analysis of the LEU2 promoter and by comparison to other genes regulated by leucine. The UAS was localized with deletions and cloned synthetic DNA. Point mutations and sequence rearrangements were used to identify important basepairs and to construct an improved UAS with increased regulation and expression. The improved UAS contains a core ten basepair, GC-rich, palindromic sequence, which is sufficient to confer minimal levels of activation and regulation, within a 36 basepair palindromic sequence which confers maximal activation and regulation. Deletions downstream of the UAS indicated that the UAS must act in conjunction with at least one other site, perhaps a TATAA region, in order to confer high levels of activation. Tandem copies of the UAS in front of LEU2 increased expression and regulation. Tandem UAS elements in trans on a multi-copy 2 mu-based plasmid decreased expression and regulation. These results are consistent with a model that the UAS serves as the DNA-binding site for diffusible activation factor(s), possibly the LEU3 gene product.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Beltzer J. P., Chang L. F., Hinkkanen A. E., Kohlhaw G. B. Structure of yeast LEU4. The 5' flanking region contains features that predict two modes of control and two productive translation starts. J Biol Chem. 1986 Apr 15;261(11):5160–5167. [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisco P. R., Cunningham T. S., Kohlhaw G. B. Cloning, disruption and chromosomal mapping of yeast LEU3, a putative regulatory gene. Genetics. 1987 Jan;115(1):91–99. doi: 10.1093/genetics/115.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. D., Satyanarayana T., Umbarger H. E. Biosynthesis of branched-chain amino acids in yeast: effect of carbon source on leucine biosynthetic enzymes. J Bacteriol. 1975 Mar;121(3):959–969. doi: 10.1128/jb.121.3.959-969.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidow L. S., Kaczmarek F. S., DeZeeuw J. R., Conlon S. W., Lauth M. R., Pereira D. A., Franke A. E. The Yarrowia lipolytica LEU2 gene. Curr Genet. 1987;11(5):377–383. doi: 10.1007/BF00378180. [DOI] [PubMed] [Google Scholar]
- Drain P., Schimmel P. Multiple new genes that determine activity for the first step of leucine biosynthesis in Saccharomyces cerevisiae. Genetics. 1988 May;119(1):13–20. doi: 10.1093/genetics/119.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Dumas K. S., Livak K. J. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 1985 Jun 11;13(11):4011–4027. doi: 10.1093/nar/13.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988 Jul;8(7):2690–2697. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987 Aug;7(8):2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusterspreute M., Ha Thi V., Davison J. Expression of galactokinase as a fusion protein in Escherichia coli and Saccharomyces cerevisiae. DNA. 1984 Oct;3(5):377–386. doi: 10.1089/dna.1984.3.377. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg S., Petersen J. G. Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1988 Mar;13(3):207–217. doi: 10.1007/BF00387766. [DOI] [PubMed] [Google Scholar]
- Hsu Y. P., Schimmel P. Yeast LEU1. Repression of mRNA levels by leucine and relationship of 5'-noncoding region to that of LEU2. J Biol Chem. 1984 Mar 25;259(6):3714–3719. [PubMed] [Google Scholar]
- Kohlhaw G. B., Hsu Y. P., Lemmon R. D., Petes T. D. Transposed LEU2 gene of Saccharomyces cerevisiae is regulated normally. J Bacteriol. 1980 Nov;144(2):852–855. doi: 10.1128/jb.144.2.852-855.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde B., Arcangioli B., Guarente L. A single Saccharomyces cerevisiae upstream activation site (UAS1) has two distinct regions essential for its activity. Mol Cell Biol. 1986 Dec;6(12):4690–4696. doi: 10.1128/mcb.6.12.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Morrison D. A. Transformation and preservation of competent bacterial cells by freezing. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Petersen J. G., Holmberg S. The ILV5 gene of Saccharomyces cerevisiae is highly expressed. Nucleic Acids Res. 1986 Dec 22;14(24):9631–9651. doi: 10.1093/nar/14.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Satyanarayana T., Umbarger H. E., Lindegren G. Biosynthesis of branched-chain amino acids in yeast: correlation of biochemical blocks and genetic lesions in leucine auxotrophs. J Bacteriol. 1968 Dec;96(6):2012–2017. doi: 10.1128/jb.96.6.2012-2017.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srienc F., Bailey J. E., Campbell J. L. Effect of ARS1 mutations on chromosome stability in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Jul;5(7):1676–1684. doi: 10.1128/mcb.5.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Takagi M., Kobayashi N., Sugimoto M., Fujii T., Watari J., Yano K. Nucleotide sequencing analysis of a LEU gene of Candida maltosa which complements leuB mutation of Escherichia coli and leu2 mutation of Saccharomyces cerevisiae. Curr Genet. 1987;11(6-7):451–457. doi: 10.1007/BF00384606. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]



