Summary
Nucleosides are required for DNA and RNA synthesis, and the nucleoside adenosine plays a role in a variety of signaling processes 1,2. Transporting nucleosides across cell membranes provides the major source of nucleosides in many cell types and is also responsible for the termination of adenosine signaling. Due to their hydrophilic nature, nucleosides require a specialized class of integral membrane proteins, known as nucleoside transporters (NTs), for specific transport across cell membranes. In addition to nucleosides, NTs are important determinants for the transport of nucleoside-derived drugs across cell membranes 3–5. A wide range of nucleoside-derived drugs has been shown to depend, at least in part, on NTs for transport across cell membranes including anticancer drugs (e.g., Ara-C and gemcitabine) and antiviral drugs (e.g., AZT and ribavirin) 4,6–13. Concentrative nucleoside transporters (CNTs), members of the solute carrier transporter superfamily SLC28, use an ion gradient to actively transport nucleosides as well as nucleoside-derived drugs against their chemical gradients. The structural basis for selective ion-coupled nucleoside transport by CNTs is unknown. Here we present the crystal structure of a concentrative nucleoside transporter from Vibrio cholerae in complex with uridine at 2.4 Å. Our functional data show that the transporter utilizes a sodium gradient for nucleoside transport like its human orthologs. The structure reveals the overall architecture of this class of transporter, unravels the molecular determinants for nucleoside and sodium binding, and provides a framework for understanding the mechanism of nucleoside and nucleoside drug transport across cell membranes.
Humans have three isoforms of CNT and the substrate specificity and tissue distribution are different among the isoforms 14–16. Knowledge of the mechanism of these transporters would help us not only to understand physiological processes associated with nucleosides but also to provide a framework for future drug design to improve nucleoside drug delivery. The major barrier towards achieving a mechanistic understanding is the lack of atomic structures that reveal the origins of nucleoside specificity and the principles of function of CNTs. For structure determination and functional studies we chose a CNT homolog from Vibrio cholerae (vcCNT) due to its high sequence homology to hCNTs (39% identical to hCNT3, Supplementary Figure 1) and optimal biochemical stability.
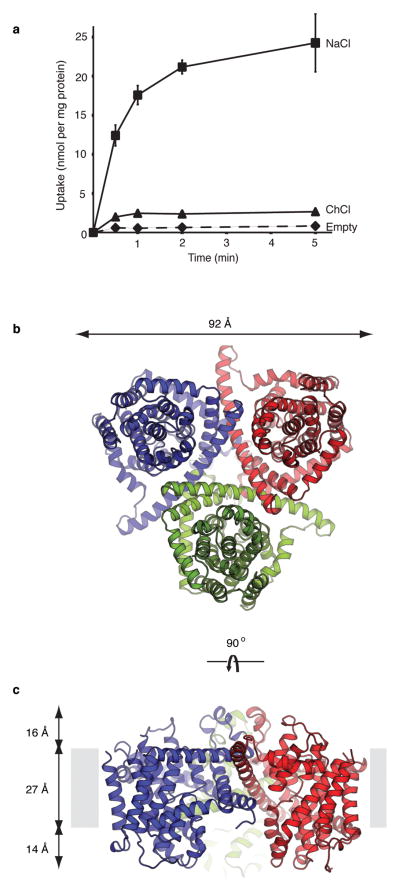
To test whether vcCNT can transport nucleosides, and if so what ion gradient vcCNT utilizes, we carried out a radioactive nucleoside uptake assay using recombinant vcCNT-reconstituted liposomes. Studies have shown that the human CNTs use Na+ and the E. coli CNT uses H+ for nucleoside transport 17–19. Transport activity was measured by monitoring uptake of [5,6-3H]-uridine both in the presence and absence of a Na+ gradient (Figure 1a). We chose uridine because all of the CNTs that have been characterized to date transport uridine 14,17–19. When a Na+ gradient was present, vesicles containing vcCNT accumulated radioactive uridine significantly more than the control empty vesicles. When a Na+ gradient was not present (Na+ is replaced with choline in the solution), uridine uptake was significantly reduced. Further characterization showed that uridine uptake is electrogenic and independent of pH gradients (Supplementary Figure 2). These data clearly demonstrate that vcCNT, like human CNTs, utilizes a Na+ gradient to transport nucleosides.
Figure 1.
vcCNT is a Na+-coupled nucleoside transporter with a trimeric architecture. a, Time course of the uptake of 2.4 μM [5,6-3H]-uridine into vesicles containing vcCNT in the presence of a sodium gradient (squares, NaCl), in the absence of a sodium gradient (triangles, ChCl), and the control empty vesicles in the presence of sodium (diamonds, empty). b, Cartoon representation of the vcCNT trimer viewed from the cytoplasm. Individual protomers are colored blue, red, and green. c, Cartoon representation for the vcCNT trimer viewed parallel to the membrane. The putative membrane bilayer is indicated in c with grey boxes. The dimensions of the putative membrane bilayer, extracellular region, and intracellular region are shown.
We crystallized and solved the structure of vcCNT at 2.4 Å (Supplementary Table 1). Experimental phases to 3.5 Å were obtained by single anomalous dispersion (SAD) from a platinum-soaked crystal (see Methods). The final model contains a single protomer in the asymmetric unit and is of good quality with a free R factor of 22.8% (Supplementary Table 1). vcCNT crystallizes as a homotrimer that is shaped like an inverted triangular basin with its mouth facing the intracellular side and a knob-like structure facing the extracellular side (Figure 1b and 1c). The three-fold axis coincides with the crystallographic 6-fold axis and is perpendicular to the membrane. When viewed from the intracellular side each side of the triangle formed by the trimer is ~92 Å, and when viewed parallel to the membrane the trimer is ~57 Å in height (Figure 1). The membrane-embedded region lies approximately in the middle of the transporter judging from the positions of three amphipathic helices on each protomer. Based on the predicted location of the membrane bilayer, the mouth of the basin penetrates into the membrane plane (Figure 1c and Supplementary Figure 3). The presence of several polar amino acids on the basin surface likely allows bulk aqueous solution to reach deep into the membrane bilayer and access this surface of the transporter.
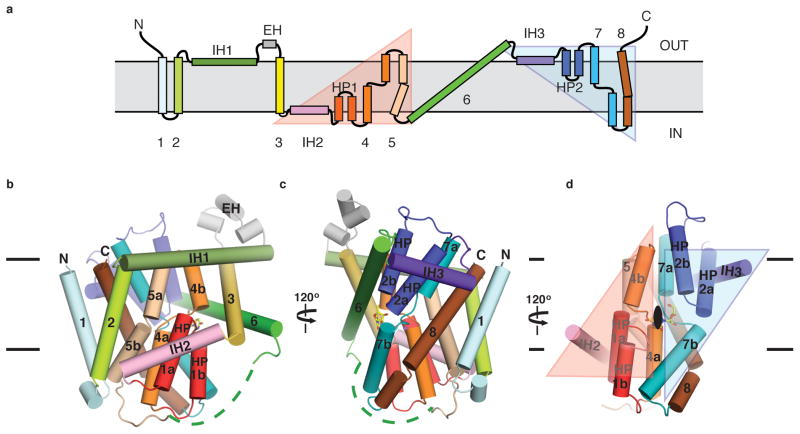
The structure reveals that each protomer contains 8 transmembrane helices (TM1-8), two reentrant helix-turn-helix hairpins (HP1 and HP2) with opposite orientations in the membrane, and three interfacial helices (IH1-3) that run parallel to the membrane (Figure 2a). The orientation of the structure relative to the membrane is consistent with previous accessibility studies 20–22 and conforms to the positive-inside rule (Supplementary Figure 4). hCNTs are predicted to contain three additional N-terminal TMs compared to prokaryotic CNTs, suggesting an 11-TM topology for hCNTs 20.
Figure 2.
Topology and fold of the vcCNT protomer. a, Schematic representation of vcCNT topology. The group of helices under the pink triangular background is related to the group of helices under the cyan triangular background by two-fold pseudo-symmetry with the symmetry axis parallel to the membrane. b, Cartoon representation of the vcCNT protomer fold. Only helices comprising the scaffold domain and the group of helices under the pink triangular background are labeled. c, Same with b but rotated 120°. Only TM1, TM6, and helices comprising the group of helices under the cyan triangular background are labeled. d, Same with c but rotated 120°. The scaffold domain is removed to show the pseudo-two-fold relationship between the two groups of helices colored as pink and cyan triangular backgrounds. The two-fold symmetry axis runs through the nucleoside at the center of the transporter.
Each protomer can be grouped into two subdomains based on their locations (at the outer and inner regions) relative to the center of the protomer. TM1, TM2, IH1, EH (extracellular helices), TM3, and TM6 are located at the outer part of the transporter and they appear to be important for maintaining the overall architecture of the transporter. These helices in the outer region form a scaffold for the transporter (henceforth termed the scaffold domain) (Figure 2). Trimerization contacts are mediated by part of the scaffold domain - IH1, EH, TM3, and TM6 (Figure 2b). IH1 is a 40 Å-long amphipathic helix that is most likely situated at the water-membrane interface, and TM6 is ~60 Å long and tilted almost 60° with respect to the membrane normal. EH protrudes into the extracellular solution and is ~16 Å in length. Since IH1 is involved in trimerization, is constrained to be at the membrane-water interface, and interacts with many TMs (TM2-TM5), it most likely plays an important role in building and maintaining the overall structure of vcCNT. Surrounded by the scaffold domain, many conserved amino acids implicated in nucleoside transport are localized at the inner domain (henceforth termed the transport domain). The transport domain is composed of two structural groups that are related by an internal two-fold pseudo-symmetry. The first group is composed of IH2, HP1, TM4a/b, and TM5 (pink background in Figure 2a), and the second group is composed of IH3, HP2, TM7a/b, and TM8 (cyan background in Figure 2a). These two groups, separated by TM6 in sequence, can be superimposed with an r.m.s.d. of 3.3 Å with the two-fold symmetry operator running though the middle of TM6 and parallel to the membrane (Figure 2d). There is no significant amino acid sequence homology between these two groups (~10% sequence identity). This two-fold relationship positions the tips of HP1, HP2, and unwound regions of TM4 and TM7 at the center of the transport domain, which is located slightly below the middle of the membrane plane (Figure 2d). Sequence alignment of hCNTs and vcCNT reveals high sequence conservation around HP1, HP2, and the two unwound helices (TM4 and TM7), indicating the functional importance of this region (Supplementary Figure 1).
To the best of our knowledge, the overall fold of vcCNT is novel although local structural elements such as helical hairpins and unwound helices have been observed previously 23,24.
The crystal structure suggests that vcCNT adopts a trimeric configuration. To test whether the stoichiometry of vcCNT is trimeric and further validate the physiological relevancy of our crystal structure, we performed structure-guided disulfide bridge cross-linking experiments (Supplementary Figure 5). Our cysteine mutants readily form disulfide cross-linked trimers in both detergent micelles and cell membranes under oxidizing conditions, therefore our crystal structure reflects a physiologically relevant oligomerization state. Given the sequence conservation of most of the amino acids involved in trimerization, we propose that the stoichiometry of both eukaryotic and prokaryotic CNT family members is trimeric (Supplementary Figure 1).
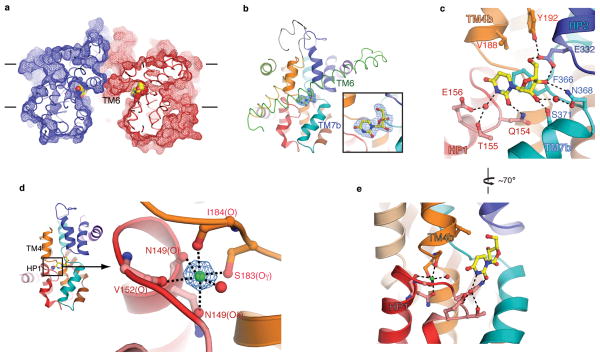
The structure contains three deep clefts (one per subunit) at the intracellular side facing the center of the trimer (Figure 3a). A simulated-annealing OMIT map clearly shows that the electron density in the cleft is that of uridine (Figure 3b). The bound uridine in the cleft faces the intracellular basin of the transporter; however, it is not free to be released into the intracellular solution because TM6 and TM7b partly cover the binding site (Figure 3a and 3b and Supplementary Figure 6).
Figure 3.
Nucleoside binding site and sodium binding site. a, Cut away surface representation of vcCNT viewed parallel to the membrane. The putative membrane bilayer is shown with lines. Uridine is shown as spheres. b, The vcCNT protomer viewed from the center of the trimer. The scaffold domain is shown in ribbon representation and the transport domain is shown in cartoon representation. The blue mesh, covered by TM6 (green), is an Fo-Fc simulated annealing OMIT map, contoured at 4 σ, showing density for uridine. The inset is a zoomed-in image of the simulated annealing OMIT electron density. c, The nucleoside-binding site showing HP1 (red), TM4b (orange), HP2 (blue), and TM7b (cyan). Hydrogen bonds are shown as dashed lines. d, The sodium-binding site is located between HP1 (red) and the unwound region of TM4 (orange). The blue mesh is an Fo-Fc simulated annealing OMIT map, contoured at 6 σ, showing density for sodium. Coordination of the sodium ion is shown as dashed lines. e, The sodium-binding site is near the nucleoside-binding site.
The nucleoside-binding site is located at the center of the internal two-fold symmetry and lined by the tips of HP1 and HP2 and the unwound regions of TM4 and TM7 (Figure 3b and 3c). Inspection of the interactions between uridine and the binding site shows that many polar or charged amino acids from HP1, HP2, TM4, and TM7 interact with the uracil base and ribose. HP1 and TM4b are responsible for interacting with the uracil base. The side chains of Gln 154, Thr 155, and Glu 156 from HP1 interact with the uracil base either directly (Gln 154) or indirectly through a water molecule (Thr 155 and Glu 156) (Figure 3c). Val 188 from TM4b interacts with the uracil base via van der Waals interactions. The involvement of these amino acid residues in the interactions with the nucleoside is consistent with mutational studies by Young and colleagues: the residue corresponding to Gln 154, together with that corresponding to Val 188, is important for nucleoside specificity of hCNTs; Glu 156 is critical for Na+-nucleoside coupled transport 25–27.
HP2 and TM7 are responsible for the interactions with ribose. The side chains of Glu 332 (HP2), Asn 368 (TM7), and Ser 371 (TM7) interact with the ribose either directly (Glu 332 and Ser 371) or indirectly through a water molecule (Asn 368). Mutation of the residue corresponding to Glu 332 in hCNTs was shown to have significant functional effects on both nucleoside binding and the rate of transport 21,26.
Since vcCNT is a sodium-coupled transporter, it must contain at least one sodium-binding site. Initial hints regarding the location of the sodium-binding site came from an Fo-Fc map that shows a strong peak (~8σ) between the tip of HP1 and the unwound region of TM4. Further inspection reveals that this site is octahedrally coordinated by three backbone carbonyls, two side chain hydroxyl groups, and a water molecule (Figure 3d) with the typical distances (~2.4 Å) for Na+ coordination by protein 28. Since it is difficult to distinguish Na+ from H2O based on electron density alone, we performed the valence test that was used to identify the Na+ sites in the crystal structure of LeuT 28,29. The result (ν = 1.02) is consistent with the bound ion in this site being Na+. Therefore we suggest that vcCNT has at least one Na+-binding site at this position between HP1 and the unwound region of TM4.
The sodium-binding site is near the nucleoside-binding site, but the Na+ does not directly interact with the nucleoside (Figure 3e). What is the functional role of Na+ binding at this site? Since key amino acid residues for the interactions with the nucleoside base are localized on HP1 and TM4b, we hypothesize that the role of Na+-binding is to bring HP1 close to TM4 to complete the formation of the nucleoside-binding site.
In the field of sodium-coupled secondary transporters, the current paradigm revolves around the alternating access mechanism 30. Assuming the alternating access model is utilized, our crystal structure probably represents an inward-facing occluded conformation (Figure 3a and Supplementary Figure 6). On the bases of our crystal structure, previous accessibility studies of hCNTs, and studies of the sodium-coupled aspartate transporter GltPh that shares certain structural features with vcCNT (Supplementary Figure 7), we propose a hypothetical model to explain sodium-coupled nucleoside transport by vcCNT (Figure 4). A rigid-body movement of the transport domain across TM6 while the scaffold domain is held in place could allow for the transition from an inward-facing to an outward-facing conformation. Our reasoning in proposing this mechanism is provided in the Supplementary Information.
Figure 4.
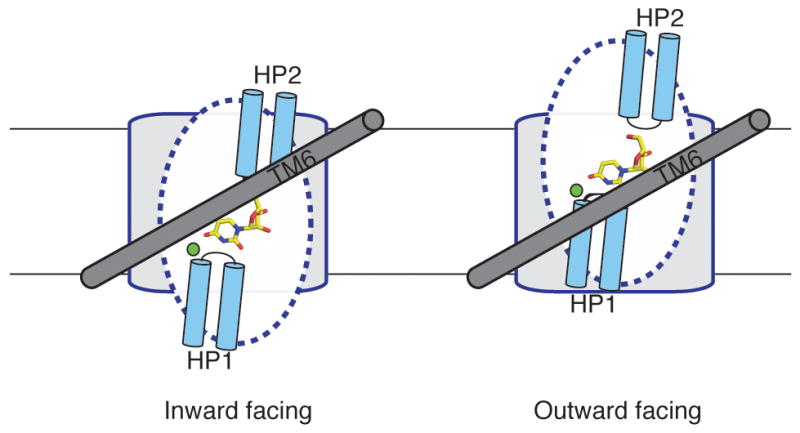
Hypothetical mechanism of nucleoside transport. Only a single protomer is shown for simplicity, viewed from the center of the trimer (same orientation as 3b). On the left, the transporter adopts an inward-facing conformation. Uridine is bound between the tips of HP1 and HP2, and TM6 (grey), serving as a hydrophobic barrier, partially covers uridine. The blue dotted lines demarcate the mobile part of the transport domain and the grey region denotes the scaffold domain. A rigid-body motion of the nucleoside-binding site across TM6 can expose the binding site to the extracellular side.
The crystal structure provides an important step toward understanding the mechanism of nucleoside transport by the CNT family. However, many important issues remain to be elucidated including the structural bases for nucleoside and nucleoside-drug specificity and the conformational changes associated with each step along the transport cycle.
Methods Summary
vcCNT was expressed in E. coli C41 cells. Cells were lysed and protein was solubilized with dodecyl maltoside. Protein was purified by Co2+ affinity chromatography followed by gel filtration chromatography in the presence of decyl maltoside and uridine.
vcCNT was crystallized in the presence of 100 mM CaCl2, 37–42% PEG400, and 100 mM buffer (pH 5.6–9.5). Platinum (Pt) derivatives were prepared by soaking in K2Pt(CNS)6. A partial poly ALA model was built with phases to 5.0 Å from single isomorphous replacement with anomalous scattering (SIRAS). A complete model was built by combining phases from single anomalous dispersion using a new platinum derivative with molecular replacement phases from the partial model (MR-SAD) to 3.5 Å. Structure refinement was then performed against the 2.4 Å native data. The final model is of good quality with good Ramachandran statistics (98% favored and 2% allowed). The model contains residues 2 to 416 (residues 230 to 240 missing), a uridine, 5 decylmaltosides (1 full and 4 partial), a sodium ion and 43 bound waters. Disordered residues Lys 226 and Glu 345 were modeled as Ala.
For the flux assay, protein was reconstituted into lipid vesicles and then vesicles were diluted into buffer containing either choline chloride or NaCl. Flux was initiated by the addition of 2.4 μM [5,6-3H]-uridine and 1 μM valinomycin.
Full methods
Expression and Purification
The vcCNT gene was cloned from Vibrio cholerae genomic DNA into a modified pET26 vector that contains a pelB leader sequence and a PreScission Protease cleavable His10-MBP fusion. The original vector was a gift from Raimund Dutzler (University of Zurich) and was further modified. The vector containing vcCNT was expressed in E. coli C41 (DE3) cells. Cells were lysed by homogenizer (AVESTIN) and protein was solubilized with 30 mM dodecyl maltoside (DDM) for two hours at 4 °C. Solubilized lysates were spun down to remove the insoluble fraction and supernatants were applied to Talon Co2+ affinity resin. After binding, protein was eluted with imidazole and digested overnight with PreScission Protease. The digestion mixture was concentrated and applied to a Superdex 200 size exclusion column in the presence of 5 mM decyl maltoside (DM), 1 mM uridine, 150 mM NaCl, 20 mM Tris pH 8.0, and 2 mM dithiothreitol. Peak fractions corresponding to vcCNT were collected for further experiments.
Crystallization
Initial crystallization conditions were obtained at the high-throughput crystallization-screening laboratory at the Hauptman-Woodward Institute 31. Protein was concentrated to ~10 mg/mL and mixed 1:1 with crystallization solution containing 100 mM CaCl2, 37–42% PEG400, and 100 mM buffer. Crystals grew over a wide pH range (5.6–9.5), but data for structure solution was collected on crystals grown at pH 9.0 (100 mM Tris) and pH 9.5 (100 mM glycine). Crystals were grown using the microbatch-under-oil method. After 10–14 days, crystals were harvested, transferred to cryo solution containing 32.5% PEG400, and flash frozen in liquid nitrogen. Platinum derivatives were prepared by soaking in 2.5 mM K2Pt(CNS)6 for 2–4 h then transferring to cryo solution and flash freezing.
Structure Determination
The data were collected on beam lines 22ID, 22BM, and 24ID-C at the Advanced Photon Source. The data were processed using HKL200032. Crystals of vcCNT diffract to 2.4 Å Bragg spacings and belong to the space group P63. Extensive screening of crystals was performed because approximately 70% of crystals are merohedrally twinned with the twinning operator perpendicular to the crystallographic 6-fold axis leading to the apparent space group of P6322. We found that SeMet-substituted crystals are almost always twinned with significant twinning fractions (20–45%), which necessitated heavy atom-soaked crystals as the choice for a de novo phasing method for the vcCNT structure. Initial phases to 5.0 Å were obtained by single isomorphous replacement with anomalous scattering (SIRAS) from a platinum-soaked crystal. Pt sites were found by SHELXD 33, phasing was calculated to 5.0 Å Bragg spacings using SOLVE 34 with the figure of merit 0.34 and the density was modified by solvent flattening using RESOLVE 35. A partial model was built by manually placing idealized poly-Ala helices into the solvent-flattened electron density map using COOT 36. After further extensive screening of platinum derivatives, we found a derivative that diffracted to 3.1 Å with significant anomalous signal but was non-isomorphous with any of our native data. Platinum sites were found from an anomalous difference Fourier map using the partial model phases. Combined phases of single anomalous dispersion from the platinum derivative and molecular replacement from the partial model (MR-SAD) were calculated at 4.0 Å with the figure of merit 0.35 and extended to 3.5 Å by solvent flattening with PHASER 37 and RESOLVE 35 using the PHENIX interface 38. After iterative cycles of manual adjustment of poly-Ala helices and calculation of combined MR-SAD phases, the electron density map was of excellent quality and allowed us to place side chains into the partial model. We also collected native data to 2.4 Å at a long wavelength (1.6 Å) and identified sulfur sites using an anomalous difference Fourier map, which helped guide the model building. After about 70% of manual model building was complete, molecular replacement was done using the partial model against the 2.4 Å native data for further model building and refinement. Structure refinement was done using PHENIX 38. An anomalous difference Fourier map with the native data to 2.4 Å collected at a long wavelength (1.6 Å) identified six sulfur sites and helped guide the model building. The final model is of good quality with Rwork/Rfree of 19.6/22.8 % and contains residues 2 to 416 (residues 230 to 240 missing), a uridine, 5 DMs (1 full and 4 partial), a sodium ion, and 43 bound waters.
Vesicle Reconstitution and Flux Assay
Protein was reconstituted into lipid vesicles containing 10 mg/mL of 3:1 POPE:POPG at a mass ratio of 1:500 protein:lipid as previously described 39. Vesicles were reconstituted in the presence of 200 mM KCl, 20 mM HEPES pH 7.4, and 100 mM choline chloride. Reconstituted vesicles were then flash frozen and thawed 3 times then extruded through a 1.0 μm filter using the Avanti Mini-Extruder.
For the flux assay, vesicles were diluted 20-fold into buffer containing 200 mM KCl, 20 mM HEPES pH 7.4, and either 100 mM choline chloride or 100 mM NaCl. Flux was initiated by the addition of 2.4 μM [5,6-3H]-uridine and 1 μM valinomycin. All experiments were performed in triplicate at 30 °C. Vesicles were harvested on GF/B glass microfiber filters (Whatman) and counted by scintillation the following day.
Supplementary Material
Acknowledgments
Data for this study were collected at beamlines SER-CAT BM22/ID22 and NE-CAT ID 24-C at the Advanced Photon Source. We thank R. MacKinnon and J. Butterwick for critical reading; R. Lefkowitz and A. Shukla for providing access and technical support for the radioactive flux assay; S. Lockless for advice on experiments; and C. Pemble for help with remote data collection. This work was supported by start-up funds from the Duke University Medical Center, the McKnight Endowment Fund for Neuroscience, the Alfred P. Sloan Foundation, the Klingenstein Fund, the Mallinckrodt foundation, the Basil O’Connor Starter Scholar Research Award 5-FY10-473 from the March of Dimes foundation, and the N.I.H. Director’s New Innovator Award 1 DP2 OD008380-01 (all to S.-Y.L).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Z.J. expressed, purified, and crystallized vcCNT. Z.J. performed radioactive flux and cross-linking experiments. C.-G.C. participated in part of the vcCNT crystallization and generated mutants for crystallization and functional studies. Z.J. and S.-Y. L. collected and processed the data, solved the structure, and wrote the paper. S.-Y.L designed the study. All authors discussed the results and commented on the manuscript.
Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank under accession codes 3TIJ.
References
- 1.King AE, Ackley MA, Cass CE, Young JD, Baldwin SA. Nucleoside transporters: from scavengers to novel therapeutic targets. Trends Pharmacol Sci. 2006;27:416–425. doi: 10.1016/j.tips.2006.06.004. S0165-6147(06)00152-0. [DOI] [PubMed] [Google Scholar]
- 2.Rose JB, Coe IR. Physiology of nucleoside transporters: back to the future. Physiology (Bethesda) 2008;23:41–48. doi: 10.1152/physiol.00036.2007. 23/1/41. [DOI] [PubMed] [Google Scholar]
- 3.Cano-Soldado P, et al. Compensatory effects of the human nucleoside transporters on the response to nucleoside-derived drugs in breast cancer MCF7 cells. Biochem Pharmacol. 2008;75:639–648. doi: 10.1016/j.bcp.2007.10.005. S0006-2952(07)00679-X. [DOI] [PubMed] [Google Scholar]
- 4.Damaraju VL, et al. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene. 2003;22:7524–7536. doi: 10.1038/sj.onc.1206952. [DOI] [PubMed] [Google Scholar]
- 5.Jordheim LP, Dumontet C. Review of recent studies on resistance to cytotoxic deoxynucleoside analogues. Biochim Biophys Acta. 2007;1776:138–159. doi: 10.1016/j.bbcan.2007.07.004. S0304-419X(07)00024-8. [DOI] [PubMed] [Google Scholar]
- 6.Errasti-Murugarren E, Pastor-Anglada M. Drug transporter pharmacogenetics in nucleoside-based therapies. Pharmacogenomics. 2010;11:809–841. doi: 10.2217/pgs.10.70. [DOI] [PubMed] [Google Scholar]
- 7.Mackey JR, Baldwin SA, Young JD, Cass CE. Nucleoside transport and its significance for anticancer drug resistance. Drug Resist Updat. 1998;1:310–324. doi: 10.1016/s1368-7646(98)80047-2. S1368-7646(98)80047-2. [DOI] [PubMed] [Google Scholar]
- 8.Mackey JR, et al. Immunohistochemical variation of human equilibrative nucleoside transporter 1 protein in primary breast cancers. Clin Cancer Res. 2002;8:110–116. [PubMed] [Google Scholar]
- 9.Mackey JR, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58:4349–4357. [PubMed] [Google Scholar]
- 10.Nagai K, Nagasawa K, Fujimoto S. Uptake of the anthracycline pirarubicin into mouse M5076 ovarian sarcoma cells via a sodium-dependent nucleoside transport system. Cancer Chemother Pharmacol. 2005;55:222–230. doi: 10.1007/s00280-004-0861-7. [DOI] [PubMed] [Google Scholar]
- 11.Pastor-Anglada M, Felipe A, Casado FJ. Transport and mode of action of nucleoside derivatives used in chemical and antiviral therapies. Trends Pharmacol Sci. 1998;19:424–430. doi: 10.1016/s0165-6147(98)01253-x. S016561479801253X. [DOI] [PubMed] [Google Scholar]
- 12.Pastor-Anglada M, et al. Nucleoside transporters in chronic lymphocytic leukaemia. Leukemia. 2004;18:385–393. doi: 10.1038/sj.leu.2403271. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, et al. The role of nucleoside transporters in cancer chemotherapy with nucleoside drugs. Cancer Metastasis Rev. 2007;26:85–110. doi: 10.1007/s10555-007-9044-4. [DOI] [PubMed] [Google Scholar]
- 14.Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 2004;447:728–734. doi: 10.1007/s00424-003-1107-y. [DOI] [PubMed] [Google Scholar]
- 15.Molina-Arcas M, Casado FJ, Pastor-Anglada M. Nucleoside transporter proteins. Curr Vasc Pharmacol. 2009;7:426–434. doi: 10.2174/157016109789043892. CVP-Abs-054. [DOI] [PubMed] [Google Scholar]
- 16.Ritzel MW, et al. Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside cotransporter proteins (hCNT3 and mCNT3) broadly selective for purine and pyrimidine nucleosides (system cib) J Biol Chem. 2001;276:2914–2927. doi: 10.1074/jbc.M007746200. [DOI] [PubMed] [Google Scholar]
- 17.Loewen SK, et al. Functional characterization of a H+/nucleoside co-transporter (CaCNT) from Candida albicans, a fungal member of the concentrative nucleoside transporter (CNT) family of membrane proteins. Yeast. 2003;20:661–675. doi: 10.1002/yea.1000. [DOI] [PubMed] [Google Scholar]
- 18.Loewen SK, et al. Transport of physiological nucleosides and anti-viral and anti-neoplastic nucleoside drugs by recombinant Escherichia coli nucleoside-H(+) cotransporter (NupC) produced in Xenopus laevis oocytes. Mol Membr Biol. 2004;21:1–10. doi: 10.1080/0968768031000140836. [DOI] [PubMed] [Google Scholar]
- 19.Xiao G, Wang J, Tangen T, Giacomini KM. A novel proton-dependent nucleoside transporter, CeCNT3, from Caenorhabditis elegans. Mol Pharmacol. 2001;59:339–348. doi: 10.1124/mol.59.2.339. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton SR, et al. Subcellular distribution and membrane topology of the mammalian concentrative Na+-nucleoside cotransporter rCNT1. J Biol Chem. 2001;276:27981–27988. doi: 10.1074/jbc.M100518200. [DOI] [PubMed] [Google Scholar]
- 21.Slugoski MD, et al. Substituted cysteine accessibility method analysisof human concentrative nucleoside transporter hCNT3 reveals a novel discontinuous region of functional importance within the CNT family motif (G/A)XKX3NEFVA(Y/M/F) J Biol Chem. 2009;284:17281–17292. doi: 10.1074/jbc.M109.009704. M109.009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, et al. Cysteine-accessibility analysis of transmembrane domains 11-13 of human concentrative nucleoside transporter 3. Biochem J. 2006;394:389–398. doi: 10.1042/BJ20051476. BJ20051476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y, et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011;473:50–54. doi: 10.1038/nature09939. nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. nature03018. [DOI] [PubMed] [Google Scholar]
- 25.Loewen SK, et al. Identification of amino acid residues responsible for the pyrimidine and purine nucleoside specificities of human concentrative Na(+) nucleoside cotransporters hCNT1 and hCNT2. J Biol Chem. 1999;274:24475–24484. doi: 10.1074/jbc.274.35.24475. [DOI] [PubMed] [Google Scholar]
- 26.Slugoski MD, et al. Conserved glutamate residues Glu-343 and Glu-519 provide mechanistic insights into cation/nucleoside cotransport by human concentrative nucleoside transporter hCNT3. J Biol Chem. 2009;284:17266–17280. doi: 10.1074/jbc.M109.009613. M109.009613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao SY, et al. Conserved glutamate residues are critically involved in Na+/nucleoside cotransport by human concentrative nucleoside transporter 1 (hCNT1) J Biol Chem. 2007;282:30607–30617. doi: 10.1074/jbc.M703285200. M703285200. [DOI] [PubMed] [Google Scholar]
- 28.Nayal M, Di Cera E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J Mol Biol. 1996;256:228–234. doi: 10.1006/jmbi.1996.0081. S0022283696900819. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. nature03978. [DOI] [PubMed] [Google Scholar]
- 30.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koszelak-Rosenblum M, et al. Determination and application of empirically derived detergent phase boundaries to effectively crystallize membrane proteins. Protein Sci. 2009;18:1828–1839. doi: 10.1002/pro.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 33.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 34.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. GR0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SY, Letts JA, MacKinnon R. Functional reconstitution of purified human Hv1 H+ channels. J Mol Biol. 2009;387:1055–1060. doi: 10.1016/j.jmb.2009.02.034. S0022-2836(09)00204-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.