Abstract
We have previously reported the development of small molecule phosphatidylinositol-3,4,5-trisphosphate (PIP3) antagonists (PITs) that block pleckstrin homology (PH) domain interaction, including activation of Akt, and show anti-tumor potential. Here we show that the same molecules inhibit growth factor-induced actin remodeling, lamellipodia formation and, ultimately, cell migration and invasion, consistent with an important role of PIP3 in these processes. In vivo, a PIT-1 analog displays significant inhibition on tumor angiogenesis and metastasis. ADP ribosylation factor 6 (ARF6) was recently identified as an important mediator of cytoskeleton and cell motility, which is regulated by PIP3-dependent membrane translocation of the guanine nucleotide exchange factors (GEFs) such as ADP-ribosylation factor nucleotide binding-site opener (ARNO) and general receptor for 3-phosphoinositides (GRP1). We demonstrate that PITs inhibit PIP3/ARNO or GRP1 PH domain binding and membrane localization, resulting in the inhibition of ARF6 activation. Importantly, we show that expression of the constitutively active mutant of Arf6 attenuates inhibition of lamellipodia formation and cell migration by PITs, confirming that inhibition of Arf6 contributes to inhibition of these processes by PITs. Overall, our studies demonstrate the feasibility of developing specific small molecule targeting PIP3 binding by PH domains as potential anti-cancer agents that can simultaneously interfere with cancer development at multiple points.
Introduction
Phosphatidylinositol-3,4,5-trisphosphate (PIP3) , a lipid product of Phosphatidylinositol-3-kinase (PI3K), controls a complex cellular signaling network regulating cell survival and motility (Park et al., 2008). This makes PIP3 one of the most important second messengers downstream from growth factor and oncogene signals. PIP3 exerts its effect through binding to the plekstrin homology (PH) domains of multiple downstream effector proteins (Park et al., 2008). Dysregulation of PI3K signaling, which is very frequently observed in human tumors (Cantley, 2002; Vivanco and Sawyers, 2002), has made this pathway a very important target for drug discovery. In many cases, regulation of PIP3 targets, such as PDK1 and Akt, and their dowstream signaling pathway has been a primary focus in drug discovery, because of the role of these pathways in the regulation of cancer cell survival and metabolism. Regulation of cell motility is another very important function of PIP3. This effect is exerted through multiple mechanisms, including regulation of Akt and PDK1, as well as a large number of PH domain-containing proteins, such as ADP-ribosylation factor nucleotide binding-site opener (ARNO) and general receptor for 3-phosphoinositides (GRP1), controlling activation of Rho and ADP ribosylation factor (ARF) families of GTPases. Thus, dysregulation of PI3K signaling plays a profound, yet complex, role in the control of the cancer cell invasiveness (Cantley, 2002; Vivanco and Sawyers, 2002). We have recently described two new classes of antagonists of PIP3/PH domain binding, termed PITENINs (PITs) (Miao et al., 2010). In the present work, we have investigated the effects of these molecules on the regulation of actin cytoskeleton and cell motility.
In particular, our initial results suggested that PITs target PH domains of GRP1 and ARNO, in addition to those of Akt and PDK1. GRP1 and ARNO are Guanine Exchange Factors (GEFs) of ARF GTPases (Caumont et al., 2000; Frank et al., 1998a; Frank et al., 1998b; Langille et al., 1999; Santy and Casanova, 2001). Therefore, we have explored the regulation of this family by PITs. ARF is a member of the Ras superfamily of small GTPases. Among the six known ARF isoforms, ARF6 has received significant attention because of its unique role in the regulation of actin remodeling at the plasma membrane, formation of membrane ruffles, and ultimately, its contribution to the regulation of cell motility and endosome recycling (Balana et al., 2005; Donaldson, 2003). ARF6 has also been shown to play important roles in tumor cell invasion in vitro and tumor metastasis in vivo (Balana et al., 2005; Muralidharan-Chari et al., 2009; Tague et al., 2004). Screening of various breast tumor cell lines has revealed a direct correlation between ARF6 protein expression and tumor invasiveness (Hashimoto et al., 2004). In contrast, downregulation of ARF6 by siRNA or expression of dominant-negative ARF6 mutants blocks tumor cell migration and invasion, resulting in reduced metastasis in vivo (Hashimoto et al., 2004; Tague et al., 2004).
ARF6 cycles between an inactive GDP-bound form localizing to the cytosol, and an active GTP-bound form, translocating to the plasma membrane. Activation of ARF6 is stimulated by GEFs, which promote release of GDP and binding of GTP, whereas inactivation is stimulated by GTPase-activating proteins (GAPs) (Gillingham and Munro, 2007; Nie et al., 2003). Cytohesin family proteins ARNO and GRP1 are known phosphoinositide-dependent GEFs with preference for ARF6 in cells and in vivo (Caumont et al., 2000; Frank et al., 1998a; Frank et al., 1998b; Langille et al., 1999; Santy and Casanova, 2001). ARNO and GRP1 possess similar domain architecture, consisting of an N-terminal coiled-coil domain, a catalytic Sec7 domain (ARF-GEF activity), a phosphoinositide-binding PH domain, and a short C-terminal basic amino acid region (Chardin et al., 1996; Jackson and Casanova, 2000).
ARF6 translocates to plasma membrane in response to growth factor stimulation, where it is activated by GEFs and modulates actin remodeling and lamellipodia formation. Membrane translocation of both ARF6 and its GEFs is necessary for their efficient interaction and ARF6 activation (Cavenagh et al., 1996; Macia et al., 2004; Santy and Casanova, 2001; Yang et al., 1998). The recruitment of ARNO and GRP1 to plasma membrane has been shown to be mediated by the binding of their PH domains to phosphoinositides (Chardin et al., 1996; Jackson and Casanova, 2000). Overexpression of ARNO causes cells to develop broad lamellipodia, separate from neighboring cells, and exhibit a dramatic increase in a migratory behavior. In contrast, an ARNO mutant lacking the PH domain was found to no longer undergo translocation to the plasma membrane and was unable to mediate actin reorganization (Frank et al., 1998b). The PH domains of ARNO and GRP1 exhibit a 50- to 100-fold higher affinity for PI(3,4,5)P3 than either PI(4,5)P2 or PI(3,4)P2 (Kavran et al., 1998; Klarlund et al., 1997; Klarlund et al., 1998; Venkateswarlu et al., 1998). There is also report that ARNO binds to PIP2 with an affinity comparable to that for PIP3.
In the current work, we report that PITs inhibit cancer cell migration, invasion and angiogenesis in vitro and cancer angiogenesis and metastasis in vivo. We further show that PITs are effective at inhibiting PIP3/ARNO or GRP1 PH domain binding and their membrane translocation, thereby suppressing ARF6 activation, which in turn contributes to the inhibition of cancer cell migration. Several ARF GEF inhibitors have been previously described. The activity of all known inhibitors is based on the direct or indirect inhibition of protein-protein interaction by Sec7 domains of GEFs (Hafner et al., 2006; Renault et al., 2003; Viaud et al., 2007). Overall, our work presents a new strategy based on the inhibition of PH domain/phosphoinositide binding and suppression of the plasma membrane translocation of GEFs. Conceptually, protein-lipid interactions may be more readily targetable by chemical inhibitors compared to that of protein-protein interactions, which frequently involve interactions of extended flat protein surfaces that are difficult to disrupt by small molecules.
Results
PIT-1 inhibits PIP3 binding to PH domains of GRP1/ARNO, and suppresses ARF6 activation
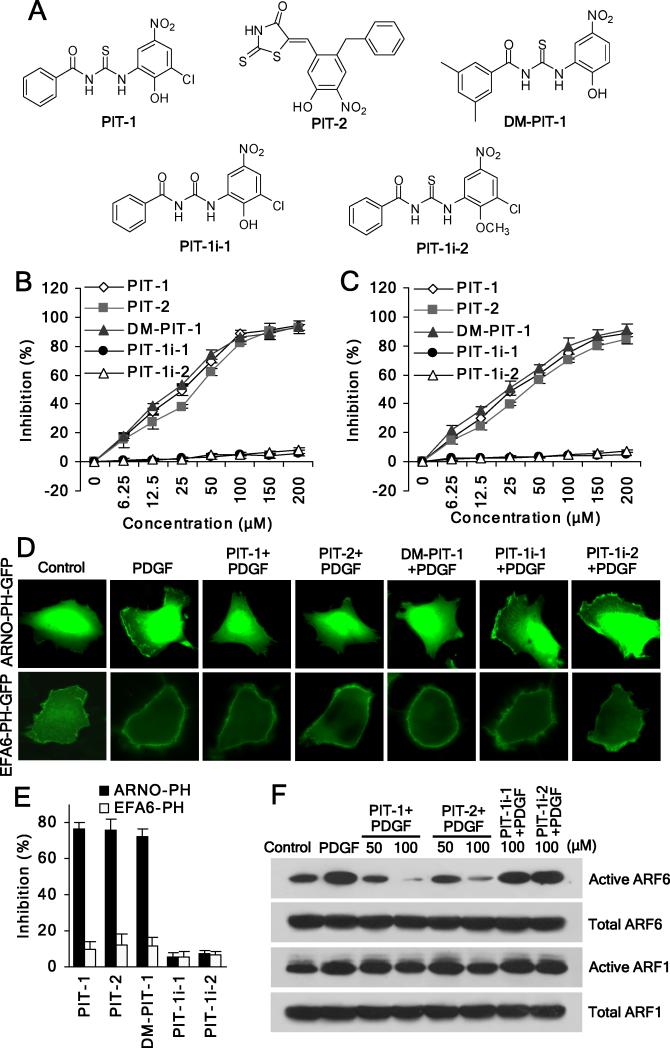
We have previously reported two distinct inhibitors of PIP3/PH domain binding through a screen of ~50,000 diverse small molecules, which were termed PIT-1 and PIT-2 (Fig. 1A) (Miao et al., 2010). They have been characterized as specific PIP3 antagonists with selectivity towards a distinct sub-set of PIP3 specific PH domains (Miao et al., 2010). The finding that PITs are able to inhibit PIP3 binding to PH domains of GRP1 and ARNO attracted our interest because both GRP1 and ARNO are GEFs of ARF6, an GTPase playing important roles in the regulation of lamellipodia formation and cell migration. Importantly, membrane translocation of these GEFs mediated by their PH domains’ binding to phosphoinositides (Chardin et al., 1996; Jackson and Casanova, 2000) is necessary for the efficient ARF6 activation (Cavenagh et al., 1996; Macia et al., 2004; Santy and Casanova, 2001; Yang et al., 1998). Consistent with our prior report (Miao et al., 2010), PIT-1 and PIT-2 effectively inhibited PIP3 binding to PH domains of GRP1 and ARNO in fluorescence polarization (FP) assay using recombinant GRP1 or ARNO PH domains and a fluorescent TMR-labeled PIP3 molecule, while PIT-1i-1 and PIT-1i-2, two inactive analogs of PIT-1, failed to inhibit the binding (Fig. 1B,C).
Figure 1. PIT-1 inhibits PIP3 binding to GRP1/ARNO PH domains, and suppresses ARF6 activation.
(A) Structures of the PITs and inactive analogs.
(B,C) PITs inhibit PIP3/GRP1 or ARNO PH domain binding. TMR-conjugated PIP3 (60 nM) was incubated with GRP1 (B) or ARNO (C) PH domain (100 nM) in the presence of PITs for 40 min, followed by FP measurement.
(D,E) PITs inhibit plasma membrane translocation of ARNO, but not EFA6 PH domain. After transfection, cells were serum starved and incubated with 50 μM PITs or inactive analogues for 2 hr, followed by stimulation with 100 ng/ml PDGF for 5 min. The translocation was analyzed using a fluorescent microscope. The representative fluorescent images are shown (D). The number of cells with membrane located GFP was quantitated by counting in five random fields and the inhibition was calculated (E).
(F) PITs significantly inhibit ARF6 activation. SUM159 cells were serum starved and treated with compounds for 2 hr, followed by stimulation with 100 ng/ml PDGF for 15 min. ARF6 and ARF1 activation assays were performed as described in the Methods section.
To confirm inhibition of the PH domain-dependent functions of these GEFs in the cells, we performed a PH domain translocation assay that measures association of the GFP-fused PH domains with the plasma membrane (Varnai et al., 2005). We used human breast carcinoma SUM159 cells for most of the cellular experiments. Both PIT-1 and PIT-2 significantly inhibited the membrane translocation of GRP1 and ARNO PH domains in response to PDGF stimulation. Neither PIT-1i-1 nor PIT-1i-2 inhibited the translocation of GRP1 and ARNO PH domains (Fig. 1D,E) (Miao et al., 2010). In contrast, PITs failed to inhibit membrane localization/translocation of another ARF6 GEF, EFA6, which has a PH domain with specificity towards PI(4,5)P2 (Macia et al., 2008) (Fig. 1D,E), consistent with their preference towards PIP3-binding PH domains demonstrated in our previous study (Miao et al., 2010). We next examined the effect of PITs on ARF6 activation using an active ARF6 pull-down and detection kit. We found that both PIT-1 and PIT-2 significantly suppressed growth factor-stimulated ARF6 activation, consistent with the FP and membrane translocation results. In contrast, PIT-1i-1 and PIT-1i-2 were again inactive, consistent with the lack of inhibition of PIP3/PH domain binding by these molecules (Fig. 1F). Considering that ARNO/GRP1 family proteins also use ARF1 as a substrate, we also examined the effect of PITs on ARF1 activation. We found PITs also inhibited ARF1 activation, but to a much lesser extent than that of ARF6 activation (Fig. 1F), which might due to the unique membrane location-dependent activation for ARF6 versus cytosolic localization of ARF1.
PIT-1 inhibits actin remodeling, ruffling/lamellipodia formation and cell polarization
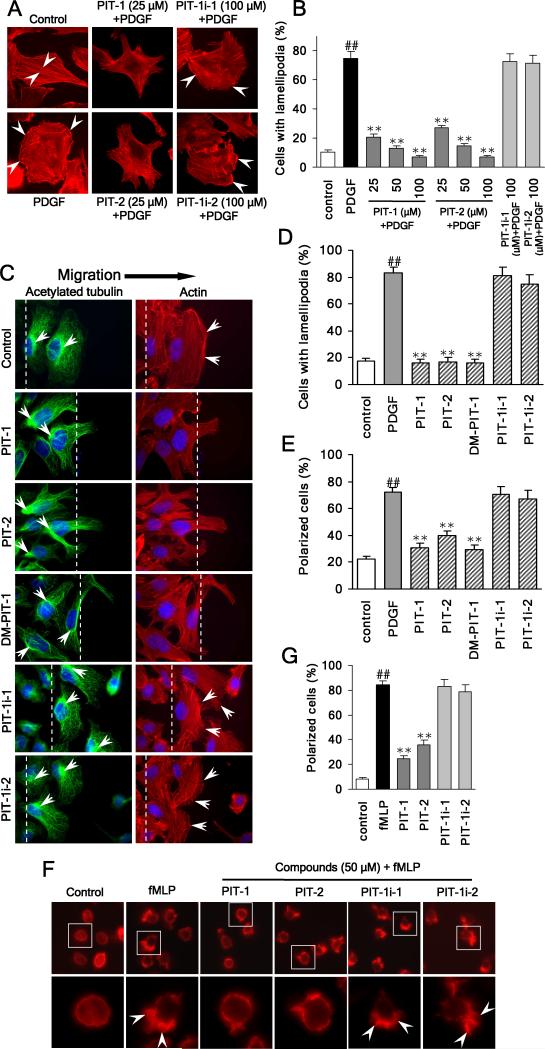
PIP3 is a key modulator in growth factor-stimulated actin remodeling and cell migration (Vivanco and Sawyers, 2002). Thus, we next examined whether PITs influence actin organization and membrane ruffling. We found PDGF-stimulated ruffling was efficiently inhibited by pre-incubation with either PIT-1 or PIT-2 (P<0.01). Confirming the specific mechanism of this activity, no inhibition was observed for inactive PIT-1 analogs, PIT-1i-1 and PIT-1i-2 (Fig. 2A,B). Similar inhibition was also observed using SecinH3, an inhibitor of GRP1/ARNO proteins (Hafner et al., 2006), which suggests that inhibition of ARF6 GEFs/ARF6 pathway may contribute to the inhibition of actin cytoskeleton dynamics by PITs (Fig. S1A,B). Furthermore, treatment with PITs or SecinH3 significantly reduced the amount of F-actin in the cells, including lamellipodia at the leading edge and stress fibers within the cells (Fig. S1A,C), indicative of efficient inhibition of actin polymerization and remodeling.
Figure 2. PIT-1 inhibits lamellipodia formation and cell polarization.
(A) PITs inhibit lamellipodia formation in cancer cells. SUM159 cells were serum starved and treated with compounds for 2 hr, followed by PDGF (100 ng/ml) stimulation for 15 min. Then cells were stained and analyzed using fluorescent microscopy. The representative fluorescent images are shown (A, the stress fibers and lamellipodia are indicated by arrowheads). The number of cells with significant lamellipodia was counted randomly in five fields and the percentage was calculated (B).
(C-E) PITs inhibit cell polarization and lamellipodia formation. Wounds were generated in SUM159 cell monolayers. Then SUM159 cells were serum starved and treated with compounds for 2 hr, followed by stimulation with 100 ng/ml PDGF for 15 min. Then cells were fixed and stained with TRITC-phalloidin for F-actin (red), acetylated tubulin antibody (green) for microtubule organization center (MTOC), and Hoechst for nuclei (blue). Cell polarization was evaluated by analyzing the relative location of the MTOC and nuclei. The cell was considered polarized if MTOC was observed to the wound side of the nuclei. The representative fluorescent images are shown (C, The wound edge is indicated with white lines and the actin ruffles and the MTOC are marked with white arrowheads). The number of polarized cells (E), as well as cells with significant lamellipodia (D) was counted in five random fields under a fluorescent microscope and the percentage was calculated.
(F,G) PITs inhibit fMLP stimulated HL-60 cell polarization. Differentiated HL-60 cells were treated with compounds at indicated concentrations for 2 hr, followed by stimulation with 100 nM fMLP for 3 min. Actin was stained and analyzed using fluorescent microscopy. The representative fluorescent images are shown (F, the leading edge of the polarized cells with lamellipodia is indicated by arrowheads). The number of polarized cells (there is obvious leading edge with lamellipodia) was counted randomly in five fields and the percentage was calculated (G).
##P<0.01 compared with control group, **P<0.01 compared with PDGF/fMLP group.
PITs also efficiently inhibited cell lamellipodia formation in a wound healing assay (Fig. 2C,D). Furthermore, we found that PITs’ treatment resulted in formation of a multiangular radiative cell morphology instead of lamellipodia formed at cell edge in control. This phenotypic change was induced by PIT-1, PIT-2 and SecinH3, but not by PIT-1i-1 and PIT-1i-2 (Fig. S1C).
Dynamic cell polarization in response to extracellular signal gradients plays an important role in mediating directional cell migration. In this regard, PIP3 is known to accumulate at the leading edge and serve as a docking site for PH domain-containing proteins including ARF6 GEFs-ARNO and GRP1, leading to the rearrangement of actin cytoskeleton and formation of lamellipodia at the front edge (Wang et al., 2002). In wound healing assay, both PIT-1 and PIT-2 were effective at preventing cell polarization induced by PDGF stimulation, while both PIT-1i-1 and PIT-1i-2 failed to prevent cell polarization (Fig. 2C,E). Treatment with PIT-1 also resulted in the efficient inhibition of fMLP-stimulated polarization of differentiated HL-60 neutrophils (Fig. 2F,G).
GRP1/ARNO and ARF6 contribute to attenuation of lamellipodia formation by PIT-1
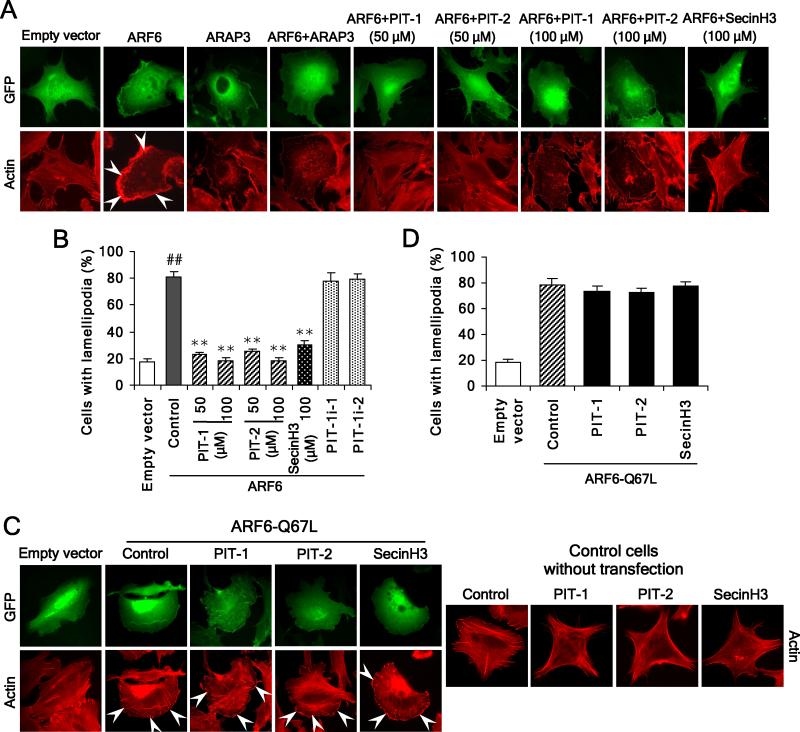
We next characterized the role of GRP1/ARNO-ARF6 to the inhibition of ruffling/lamellipodia formation by PIT-1. We examined the effect of PITs on lamellipodia formation in ARF6-overexpressing cells. Like ARAP3, a GTPase activating protein (GAP) of ARF6, both PIT-1 and PIT-2 significantly inhibited ARF6-induced lamellipodia formation. Similarly, SecinH3 suppressed ARF6-induced lamellipodia formation (Fig. 3A,B). Notably, the inhibition of lamellipodia formation by PITs and SecinH3 was accompanied by acquisition of radial cell morphology (Fig. 3A). By contrast, neither PIT-1 and PIT-2, nor SecinH3, inhibited lamellipodia formation triggered by overexpression of ARF6-Q67L, a constitutively activated ARF6 mutant, which exists mainly in the GTP-bound state (P>0.05) (Fig. 3C,D). Furthermore, the overexpression of ARF6-Q67L significantly abolished morphological alterations induced by these compounds (Fig. 3C).
Figure 3. PIT-1 suppresses ARF6 but not ARF6-Q67L induced lamellipodia formation.
(A,B) PITs inhibit lamellipodia formation induced by ARF6 overexpression. SUM159 cells were transfected with ARF6 or ARAP3, or co-transfected with both. Then cells were serum starved and treated with compounds for 2 hr, followed by stimulation with 100 ng/ml PDGF for 15 min. Cells were stained and analyzed with fluorescent microscopy. The representative fluorescent images are shown (A, the lamellipodia are indicated by arrowheads). The number of cells with significant lamellipodia was counted randomly in five fields and the percentage was calculated (B)
(C,D) PITs fail to inhibit lamellipodia formation induced by ARF6-Q67L overexpression. SUM159 cells were transfected with ARF6-Q67L. Then cells were treated with compounds for 2 hr, followed by staining and fluorescent microscopy analysis. The representative fluorescent images are shown (C) and the number of cells with significant lamellipodia was counted in five random fields and the percentage was calculated (D).
##P<0.01 compared with empty vector group, **P<0.01 compared with ARF6 overexpression control groups.
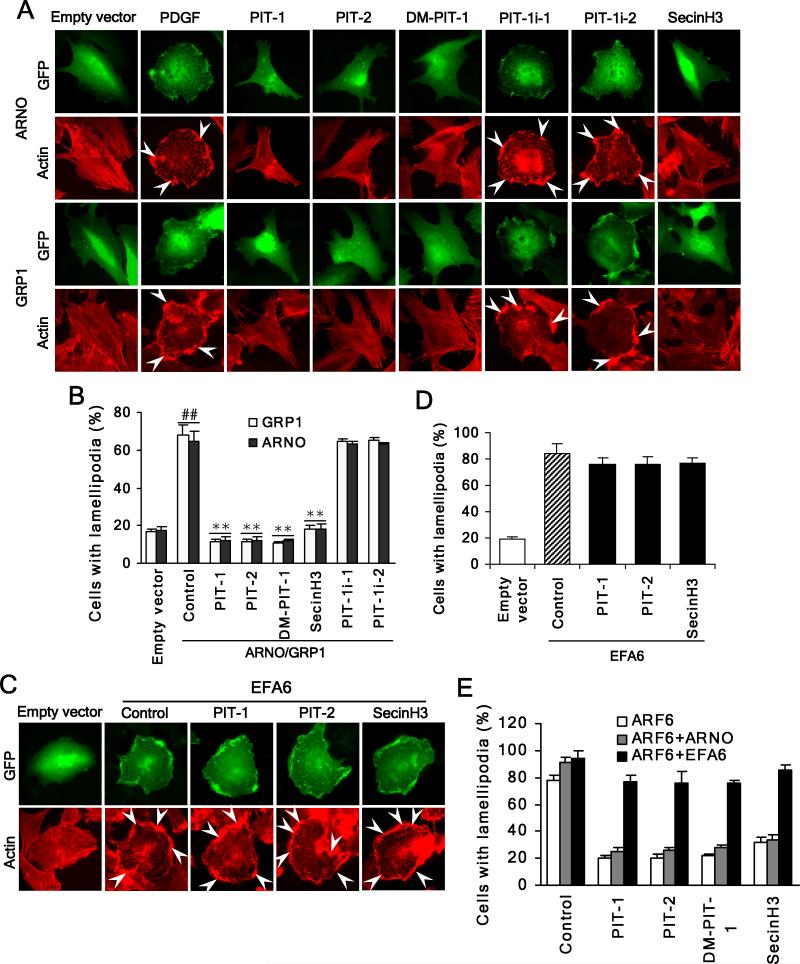
We next explored the contributions of GRP1/ARNO to inhibition of ruffling/lamellipodia formation by PIT-1. GRP1 and ARNO-transfected cells displayed a significantly increased number of peripheral lamellipodia/ruffles with PDGF stimulation. ARNO and GRP1-induced changes were effectively suppressed by PIT-1 and PIT-2, as well as SecinH3, but not by PIT-1i-1 and PIT-1i-2 (Fig. 4A,B), consistent with the inhibition of membrane localization. As an additional control, PIT-1 and PIT-2 had no effect on lamellipodia formation induced by overexpression of PIP2-specific EFA6. SecinH3 was similarly inactive (P>0.05) (Fig. 4C,D), because of its specificity towards cytohesin family versus EFA6 (Hafner et al., 2006). Furthermore, EFA6 efficiently overcame the inhibition of ARF6-induced lamellipodia formation by PITs and SecinH3, while ARNO failed to do that (Fig. 4E). These data are consistent with the inhibition of GRP1/ARNO-ARF6 contributing to the suppression of actin remodeling and ruffling/lamellipodia formation by PITs.
Figure 4. PIT-1 suppresses ARNO/GRP1 but not EFA6 induced lamellipodia formation.
(A,B) PITs inhibit lamellipodia formation induced by ARNO/GRP1 overexpression. SUM159 cells were transfected with full length ARNO or GRP1. Then cells were serum starved and treated with compounds for 2 hr, followed by stimulation with 100 ng/ml PDGF for 15 min. Cells were stained and analyzed with fluorescent microscopy. The representative fluorescent images are shown (A). The number of cells with significant lamellipodia was counted in five random fields and the percentage was calculated (B).
(C,D) PITs fail to inhibit lamellipodia formation induced by EFA6 overexpression. SUM159 cells were transfected with EFA6. Then cells were treated with compounds for 2 hr, followed by staining and fluorescent microscopy analysis. The representative fluorescent images are shown (C) and the number of cells with significant lamellipodia was counted in five random fields and the percentage was calculated (D). (E) EFA6 but not ARNO can overcome inhibition of ARF6 by PITs. SUM159 cells were transfected with ARF6 or co-transfected with ARF6 and EFA6 or ARNO. Then cells were treated with compounds for 2 hr, followed by staining and fluorescent microscopy analysis. The number of cells with significant lamellipodia was counted in five random fields and the percentage was calculated.
##P<0.01 compared with empty vector group, **P<0.01 compared with ARNO/GRP1 overexpression control groups.
PIT-1 inhibits cancer cell migration and invasion
Cell motility is driven by actin rearrangement and lamellipodia formation at the leading edge of cells (Frank et al., 1998b; Vivanco and Sawyers, 2002). PIP3 affects these processes through multiple PH-domain containing proteins, one important group of which are ARF6 GEFs such as GRP1 and ARNO (Caumont et al., 2000; Frank et al., 1998a; Frank et al., 1998b; Langille et al., 1999). The reduction of lamellipodia caused by PITs treatment led us to examine whether PITs can reduce cellular migration and invasion.
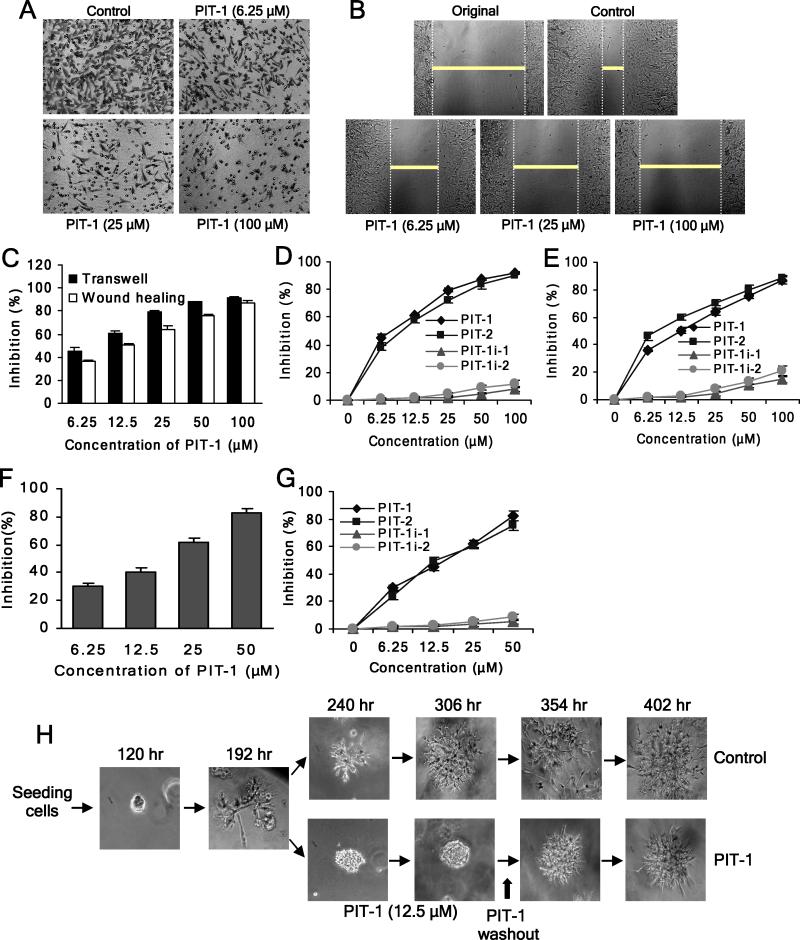
As shown in Fig. 5A,C,D and Fig. S2A, PIT-1 substantially inhibited transwell migration of multiple of human cancer cell lines. While PIT-1 can induce cell death (Miao et al., 2010), inhibition of cell migration was observed at the concentrations of PIT-1 and time points (several hours) insufficient to induce cell death. PIT-1 treatment had no significant cytotoxic effects on the cells in parallel experiments under the same conditions (8 hr) used in migration and invasion experiments (P>0.05) (Fig. S2B). Furthermore, PIT-1 at concentrations of 6.25 and 12.5 μM which are inactive at killing cells even in 48 hr (P>0.05) (Fig. S2C) could significantly suppress cell migration in 8 hr (P<0.01), implying that the effect of PIT-1 on cell motility was not a consequence of cellular toxicity. Similar inhibition of cell migration was observed in a wound healing assay (Fig. 5B,C,E). Consistent with the specific mode of inhibition, PIT-2 also efficiently inhibited cell migration, while neither PIT-1i-1 nor PIT-1i-2 were active (Fig. 5D,E). Moreover, both PIT-3 and PIT-4, two analogs of PIT-1 with increased activity and specificity towards Akt/PDK1, but lacking inhibition of GRP1 and ARNO displayed significantly reduced ability to inhibit cell migration (Table S1, Fig. S2D). Furthermore, both PIT-1 and PIT-2 (but not PIT-1i-1 and PIT-1i-2) also efficiently suppressed cancer cell invasion through matrigel (Fig. 5F,G, Fig. S2E). Consistent with the inhibition of invasion, treatment of SUM159 cells with PIT-1 blocked the acquisition of invasive phenotype by the cells. Notably, this effect was reversible, again indicating that it was not a consequence of cell death (Fig. 5H).
Figure 5. PIT-1 inhibits cancer cell migration and invasion.
(A) PIT-1 inhibits cancer cell migration in a transwell assay. SUM159 cells were treated with PIT-1 for 8 hr. The cells on the lower side of chamber were stained and the representative images are shown, then cells were lysed and colorimetric determination was made at 595 nm.
(B) PIT-1 inhibits cancer cell migration in a wound healing assay. A scratch was introduced into a monolayer of SUM159 cells, followed by treatment with PIT-1 for 8 hr. The width of wounded cell monolayer was measured in five random fields, and the representative images are shown.
(C) Quantitation of the data from the assays in (A) and (B).
(D,E) PIT-1 and PIT-2, but not PIT-1i-1 and PIT-1i-2, inhibit SUM159 cell migration in transwell (D) and wound healing (E) assays.
(F) PIT-1 inhibits cancer cell invasion. SUM159 cells were seeded on a matrigel pre-coated transwell membrane, and the treatment and analysis are similar with transwell assay described above.
(G) PIT-1 and PIT-2, but not PIT-1i-1 and PIT-1i-2, inhibit cancer cell invasion through a matrigel-coated membrane.
(H) PIT-1 reversibly inhibits acquisition of the invasive phenotype of SUM159 cells in a 3-D matrigel matrix. SUM159 cells were seeded in matrigel and incubated for 192 hr, followed by the treatment with PIT-1 (12.5 μM) for 114 hr. Subsequently, PIT-1 was washed out and cells were incubated for additional 96 hr. The representative images are shown.
GRP1/ARNO-ARF6 contribute to suppression of cell migration by PIT-1
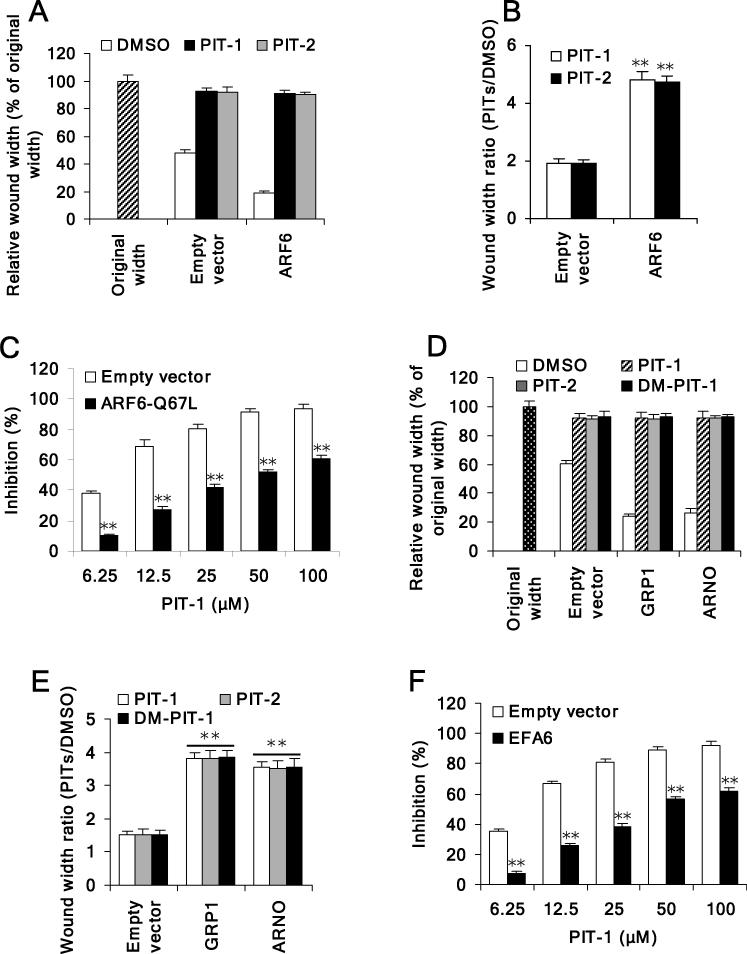
We next explored the contributions of GRP1/ARNO-ARF6 to cell migration inhibition led by PIT-1. We first measured the effect of PITs on cell migration stimulated with ARF6. As shown in Fig. 6A, overexpression of ARF6 led to an increase in cell migration. Both PIT-1 and PIT-2 inhibited migration of ARF6 overexpressing cells more effectively than that of the control cells, consistent with the larger contribution of overexpressed ARF6 to cell migration (Fig. 6B). To further confirm that inhibition of ARF6 contributes to inhibition of cell migration by PIT-1, we measured migration of cells overexpressing constitutively activated ARF6 mutant in the presence of PIT-1. We found that overexpression of ARF6-Q67L indeed significantly attenuated inhibition of cell migration by PIT-1 (Fig. 6C).
Figure 6. PIT-1 suppresses ARF6 or GRP1/ARNO induced cell migration.
(A,B) PITs inhibit ARF6-induced cell migration. SUM159 cells were transfected with ARF6. Scratch wound was generated in cell monolayer, followed by treatment with PITs (50 μM) for 8 hr. The width of wounded cell monolayers was measured in five random fields and expressed as % of original width (A). The wound width ratio (PITs treated groups/DMSO control group) was calculated (B).
(C) Overexpression of ARF6-Q67L attenuates inhibition of cell migration led by PIT-1. SUM159 cells were transfected with ARF6-Q67L. Scratch wound was generated in cell monolayer, followed by treatment with compounds for 8 hr. The width of wounded cell monolayer was measured in five random fields, and the inhibition was calculated.
(D,E) PITs inhibit GRP1/ARNO-induced cell migration. SUM159 cells were transfected with full length GRP1 or ARNO. Scratch wound was generated in cell monolayer, followed by treatment with PITs (50 μM) for 8 hr. The width of wounded cell monolayers was measured in five random fields and expressed as % of original width (D). The wound width ratio (PITs treated groups/DMSO control group) was calculated (E).
(F) Overexpression of EFA6 attenuates inhibition of cell migration by PIT-1. SUM159 cells were transfected with EFA6. Scratch wound was generated in cell monolayer, followed by treatment with compounds for 8 hr. The width of wounded cell monolayer was measured in five random fields, and the inhibition was calculated.
*P<0.05, **P<0.01 compared with empty vector group.
We next examined the effect of PITs on cell migration stimulated with overexpressed GRP1 and ARNO. Overexpression of GRP1 or ARNO led to increased cell migration in wound healing assay (Fig. 6D). Similar to the results in ARF6-overexpressing cells, both PIT-1 and PIT-2 inhibited migration of GRP1 or ARNO overexpressing cells more effectively than that of the control cells (Fig. 6E). In contrast, overexpression of EFA6 could overcome inhibition of cell migration by PIT-1 (Fig. 6F). These data strongly support the notion that inhibition of ARF6 through blocking the activity of its upstream GEFs contributes to actin changes and attenuation of cell migration by PITs.
PIT-1 inhibits endothelial cell migration and angiogenesis in vitro
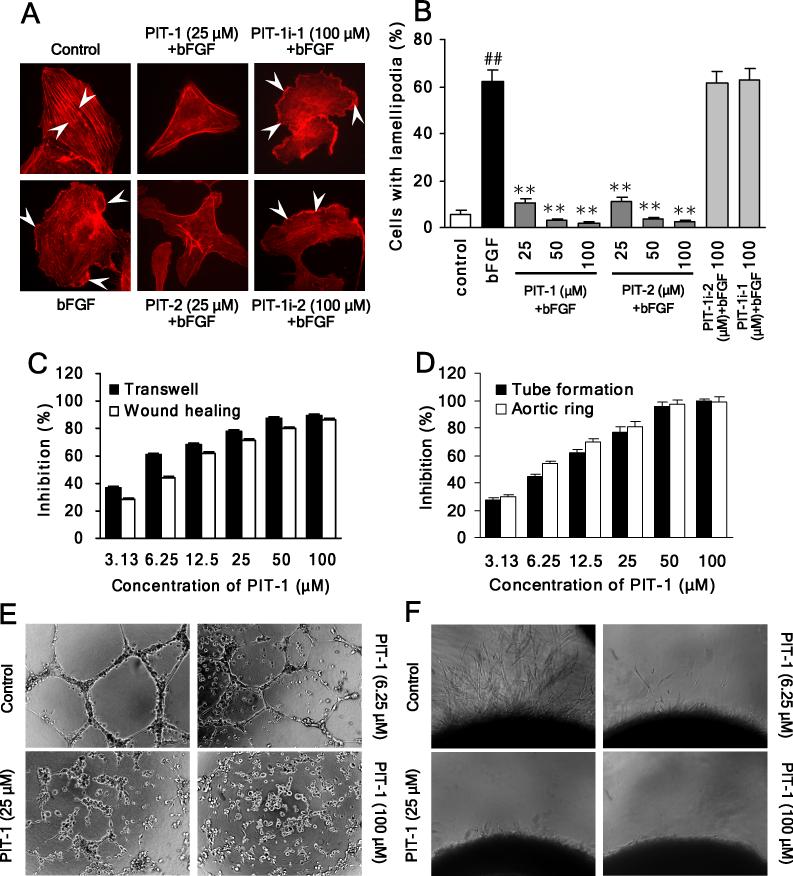
Endothelial cell migration plays a central role in tumor angiogenesis (Avraamides et al., 2008). Considering the efficient inhibition of cancer cell migration by PITs, we also examined whether these molecules may possess an additional anti-tumor modality via suppressing migration of endothelial cells. First, we determined that PIT-1 and PIT-2, but not the inactive analogs, efficiently suppressed actin reorganization and cell ruffling/lamellipodia formation in endothelial cells (Fig. 7A,B), resulting in significant inhibition on migration of HUVEC cells in both wound healing and transwell assays (Fig. 7C). No loss of cell viability was observed under the experimental conditions (P>0.05) (Fig. S2F). Based on these results, we next examined the ability of PIT-1 to attenuate angiogenesis in vitro using endothelial tube formation and aortic ring assays. As shown in Fig. 7D,E, treatment with PIT-1 resulted in efficient inhibition of tube formation. This effect was also observed with PIT-2, but not with PIT-1i-1 (Fig. S3A-C). Microvessel growth from rat aorta sections is the result of a combination of endothelial cell migration and tube formation, and thus provides a close approximation of in vivo angiogenesis. Similar with tube formation assay, PIT-1 treatment remarkably suppressed microvessel outgrowth in aortic ring sprouting experiment (Fig. 7D,F). Overall, these data suggest that PIT-1 is capable of blocking migration of both cancer and endothelial cells, suggesting that this molecule may be capable of blocking both cancer metastasis and angiogenesis.
Figure 7. PIT-1 inhibits angiogenesis in vitro.
(A,B) PITs inhibit lamellipodia formation in HUVEC cells. Cells were serum starved and treated with compounds for 2 hr, followed by bFGF (10 ng/ml) stimulation for 15 min. Then cells were stained and analyzed as described above (A, the stress fibers and lamellipodia are indicated by arrowheads). The number of cells with significant lamellipodia was counted in five random fields and the percentage was calculated (B).
(C) PIT-1 inhibits endothelial cell (HUVEC) migration in transwell and wound healing assays. The cells were treated and analyzed as described above.
(D-F) PIT-1 inhibits tube formation and microvessel outgrowth.
(D) Quantitation of the effects of PIT-1 on endothelial cell tube formation and microvessel outgrowth from (E) and (F).
(E) PIT-1 inhibits tube formation with endothelial cells. HUVEC cells were seeded on matrigel and treated with PIT-1 for 8 hr. The representative images are shown. The tube formation was assessed by counting the number of closed tubes in five random fields from each well and the inhibition was calculated.
(F) PIT-1 suppresses microvessel outgrowth in the aortic ring sprouting experiment. The rat aortic rings were embedded in matrigel and treated with PIT-1 for 6 days, followed by counting the number of microvessel outgrowths under a microscope. The representative images are shown and the inhibition was calculated.
##P<0.01 compared with control group, **P<0.01 compared with bFGF group.
PIT-1 and its analog-inhibition of in vivo cancer angiogenesis and metastasis
Considering the prominent inhibition of in vitro cell migration and angiogenesis by PITs, we next determined if this could contribute to the anti-cancer effects of the previously described dimethyl analog of PIT-1 (DM-PIT-1, Fig. 1A) in vivo (Miao et al., 2010). This analog was used for in vivo experiments due to the improved delivery by loading into long-circulating PEG-PE mixed micelles (DM-PIT-1-M) to facilitate in vivo bioavailability (Miao et al., 2010). DM-PIT-1 inhibited binding of PIP3/GRP1 and ARNO PH domain comparably to PIT-1 and did not display any significant differences from PIT-1 across a range of assays used to characterize the PIT-1 (Fig. 1B-E, Fig. 2C-E, Fig. 4A,B,E, Fig. 6E-G, Fig. S1A,C, Fig. S2G, Table S1).
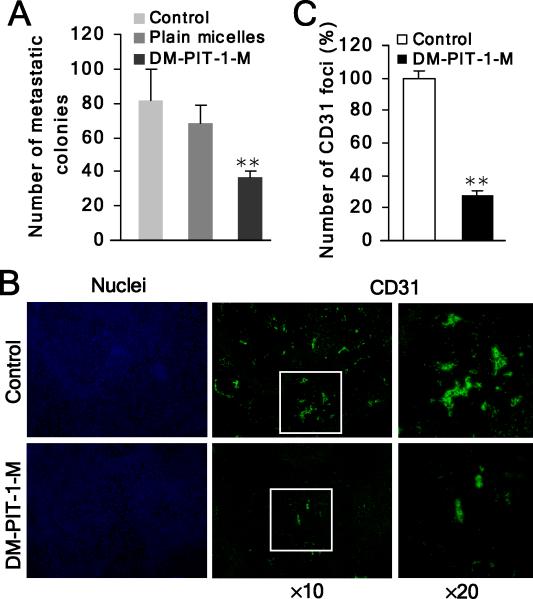
First, we tested the effect of DM-PIT-1-M on experimental pulmonary metastasis formation in vivo. Administration of DM-PIT-1-M (5 mg/kg/day) resulted in a significant suppression on pulmonary metastasis formation of B16-F10 melanoma cells. Drug administration for 5 days resulted in 55.3% reduction in the number of pulmonary metastasis, when analyzed 18 days after melanoma cell injection. The mean number of metastatic colonies in micellar DM-PIT-1 group was 36.6 compared with 81.8 colonies in the control group. The reduction in metastasis was statistically significant (P<0.01) compared to both control and plain micelle groups (no significant difference between control and plain micelles groups, P>0.05) (Fig. 8A). Moreover, no significant loss in body weight or other signs of toxicity after 5 day treatment (P>0.05), suggesting DM-PIT-1-M was well tolerated at the doses used in the experiment (Fig. S3D).
Figure 8. DM-PIT-1 inhibits in vivo cancer angiogenesis and metastasis.
(A) Administration of DM-PIT-1-M (5 mg/kg/day) suppresses pulmonary metastasis formation with B16-F10 melanoma cells. After 5-day drug administration, the mean number of metastatic colonies was counted.
(B,C) DM-PIT-1-M administration suppresses in vivo angiogenesis in tumors evaluated using IH assay with anti-CD31 antibody (green). Nuclei were stained with Hoechst (blue). The representative fluorescent images are shown (B) and the number of CD31 foci was counted in five random fields and expressed as % of control (C).
**P<0.01 compared with control group.
Second, we have previously reported that administration of DM-PIT-1-M inhibited growth, triggered cell death and suppressed Akt signaling in 4T1 syngeneic xenografts (Miao et al., 2010). Considering the significant inhibitory effect of PIT-1 on angiogenesis in vitro, we next examined the extent of angiogenesis in DM-PIT-1-treated tumors using microvasculature marker CD31. The administration of DM-PIT-1-M resulted in a significant attenuation of the number of CD31 loci in pulmonary metastasis tumors of B16-F10 melanoma cells (P<0.01). The DM-PIT-1 treatment induced a 72.1% reduction of CD31 loci number compared with control group, reflecting the marked decrease in cancer angiogenesis in vivo (Fig. 8B,C). Overall, these initial data suggest that DM-PIT-1 shows significant effect in suppressing cancer angiogenesis and metastasis in vivo and may represent a promising starting point for further optimization and characterization in tumor models in vivo.
Discussion
PITs have been developed as small molecule antagonists of PIP3 binding to the Akt PH domain (Miao et al., 2010). However, we also found that PITs display binding to several additional PIP3-binding PH domains, involved in actin skeleton remodeling and cell migration. Considering a well established role of PIP3 signaling in cell migration, this prompted us to further investigate effects of PITs on these processes. In the current work, we show that PITs suppress actin remodeling, ruffling/lamellipodia formation and cell polarization, which leads to the inhibition of migration and invasion of cancer and endothelial cells by PITs at noncytotoxic concentrations. In vitro activities of PIT-1 (DM-PIT-1) translate into pronounced inhibition of in vivo tumor angiogenesis and metastasis. At the same time, DM-PIT-1 is well tolerated upon systemic administration in mice. We also extend our in vitro observations that PITs inhibit PIP3 binding by the PH domains of GRP1 amd ARNO, to show that PITs inhibit membrane translocation of these molecules, leading to the inhibition of ARF6 activity in the cells. Inhibition of ARF6 contributes to the suppression of actin rearrangement and cell migration and invasion by PITs. Overall, we present a new approach to the regulation of PIP3-dependent migration through blocking PIP3 binding by GEF proteins.
ARF6 is the only member of a family of class III ADP ribosylation factors (ARFs) with established role in regulation of cell cytoskeleton and migratory behavior (Balana et al., 2005; Donaldson, 2003; Venkateswarlu et al., 1998). Among the 15 GEFs for ARF proteins, only the ARNO/cytohesin family and EFA6 contain PH domains that regulate their association with the plasma membrane through binding to phosphoinositides (Cox et al., 2004). The higher specificity of these GEFs towards ARF6 is at least partially due to their plasma membrane co-distribution with ARF6 (Cavenagh et al., 1996; Macia et al., 2004; Santy and Casanova, 2001; Yang et al., 1998). Consistently, the mutant of ARNO lacking PH domain failed to promote ARF6-dependent cytoskeletal rearrangements (Frank et al., 1998b). All of these data suggest that it may be possible to develop small molecule inhibitors of GRP1/ARNO PH domain/phopshoinositides binding as ARF6 inhibitors and potential anti-metastatic and anti-angiogenic drug candidates. Our data show that PITs fit into this role and are a promising starting point for future studies.
A number of observations support the specificity of PITs effects on cell migration and actin cytoskeleton. Importantly, our data suggest that activity of PITs in cell migration and invasion experiments parallels that of SecinH3. SecinH3 is a well characterized inhibitor of ARNO/cytohesin family, which, unlike PITs targeting PIP3-dependent membrane localization of GEFs, directly binds to the Sec7 domain of ARNO/cytohesin family GEFs, leading to inhibition of ARFs (Hafner et al., 2006). Notably, we observed an additive effect between PIT-1 and SecinH3 in suppression of cell migration (P<0.05, Fig. S2H). This is consistent with different modes of GEFs’ inhibition by these molecules. In addition, we show that inactive analogs of PIT-1, PIT-1i-1 and PIT-1i-2, lack activity towards ARF6 as well as cell migration. Similarly, several additional PIT-1 analogs, which display increased activity towards Akt and PDK1 and significantly increased cytotoxicity in the cells (Miao et al., 2010), but reduced activity towards ARF6 GEFs, also display reduced inhibition of cell migration (Table S1).
Our data suggest that ARF6 inhibition contributes to the effects of PIT-1 on cell motility. However, the partial attenuation of PIT-1 inhibition of cell migration by ARF6-Q67L likely means that other PIT targets may also play a role in these events. This is not unexpected, because multiple additional PIP3 targets, such as Akt, PDK1 and GEFs for Rho GTPases contribute to the regulation of actin cytoskeleton. Notably, the structure-activity relationship data in Table S1 shows the reduction in the inhibition of migration by the more active and specific Akt/PDK1 inhibitors suggest that, at least in SUM159 cells, Akt is not likely to make major contributions to the inhibition of cell motility by PITs. As shown in Fig. S2J, Akt inhibitor VIII exhibited the strongest inhibition on Akt phosphorylation, but it has much weaker inhibition on cell migration compared with PIT-1 and PI3K inhibitor LY294002 (Fig. S2I). In contrast, PIT-1 efficiently inhibited cell migration at concentrations failed to inhibit Akt phosphorylation (Fig. S2I, J). On the other hand, our initial studies show that PITs could inhibit activation of Rac1, another key molecule in lamellipodia formation (Ridley et al., 1992) (Fig. S3E). The inhibition of Rac1 activation by PITs might partially result from inhibition of ARF6 activation considering that Rac1 is a potential downstream molecule of ARF6 (Esteban et al., 2006; Hu et al., 2009). However, direct inhibition of PIP3 binding of PH domain-containing Rac1 GEFs, such as VAV2 and VAV3 (Hornstein et al., 2004), is another possible mechanism, which will need to be explored in the future work.
Curiously, inhibition of migration occurs at a dose, which is several times lower than the one required to inhibit PIP-PH domain binding of GRP1 or ARNO in vitro. One trivial explanation could be that FP or any other similar assay utilizing bacterially expressed isolated PH domains intrinsically cannot be expected to fully recapitulate complex regulation of cellular full length proteins by PIP3 in its endogenous membrane context. Thus, in vitro affinities calculated based on the FP data (Table S1) should be considered with caution. The more likely possibility is that because PIP3 regulates a significant number of factors involved in actin regulation, including GEFs for ARF6 as well as Rac1 family, it is possible that even incomplete inhibition of each individual step may result in the combined pronounced effect on actin remodeling and cell migration. It is also possible that even though our data show that PITs inhibit GRP1 and ARNO functions in the cells, there may exist additional endogenous PIT-1 targets among PIP3-binding GEFs, which might have higher affinity to PIT-1 than that of either GRP1 or ARNO, further explaining high activity of PITs. This needs to be explored in the future work.
While our data suggest that Akt inhibition does not contribute to the inhibition of cell migration by PITs in vitro, it is also important to consider that angiogenesis is triggered by hypoxia, which, in turn, decreases with reduction in tumor size. The number of observable metastasis will also decline if cells die or fail to proliferate. Thus, in vivo inhibition of angiogenesis and metastasis by DM-PIT-1 may reflect the combined effect of inhibition of Akt, cell proliferation and tumor growth as well as the direct regulation of actin cytoskeleton.
Dysregulation of PI3K signaling contributes to multiple aspects of tumorigenesis, including changes in the regulation of cell survival, metabolism and cell motility. Therefore, it would be attractive to be able to target multiple aspects of PI3K role in tumor development to maximize anti-tumor effects. Our previous study showed that PITs can inhibit Akt PH domain, leading to tumor cell death through apoptosis (Miao et al., 2010). Our current data expands on this approach to show that PITs can also inhibit other PIP3 effectors such as GRP1 and ARNO to achieve additional anti-cancer modalities, i.e. targeting cancer angiogenesis and metastasis. At the same time, targeting PIP3 offers an important benefit of controlling the classes of PI3K effectors targeted for inhibition. Our previous (Miao et al., 2010) and current study show that PITs can be modified to achieve selective inhibition of Akt PH domain. One of the important future directions is to further explore PIT-1/PIT-2 modifications leading to the selectivity towards ARF GEFs, as we have done for Akt inhibitors. Such selective inhibitors will provide useful tools for dissecting the role of PIP3 in the regulation of actin cytoskeleton in vitro and in vivo. Tumor invasiveness and formation of distal metastasis are the primary causes of patient death in many human cancers. One of the important future directions is to further explore PIT-1/PIT-2 modifications leading to the selectivity towards ARF GEFs, as we have done for Akt inhibitors. Such selective inhibitors will provide useful tools for dissecting the role of PIP3 in the regulation of actin cytoskeleton in vitro and in vivo. Tumor invasiveness and formation of distal metastasis are the primary causes of patient death in many human cancers. Therefore, compounds that address these issues are important. Further development of the drugs specifically targeting molecules, such as ARNO and GRP1, that are otherwise non-toxic could be of high clinical value. Such molecules could be combined with current chemotherapies or surgical approaches to achieve maximal therapeutic effect.
Materials and Methods
Materials
PIT-1, DM-PIT-1, PIT-1i-1, and PIT-1i-2 were purchased from Chembridge (San Diego, CA). PIT-2 was purchased from Ryan Scientific (Mt. Pleasant, CA). The purity of all compounds was >90%. Tetramethylrhodamine (TMR)-labeled PIP3 was purchased from Echelon Biosciences. Secondary Alexa 488-conjugated antibodies were purchased from Molecular Probes, and secondary HRP-conjugated antibody for western blot assay was purchased from Southern Biotech. Mouse anti-β-tubulin antibody was purchased from Stressgene. Goat anti-CD31 antibody was purchased from Santa Cruz, bFGF was from R&D Systems, and ARF6 activation kit was from Pierce Biotechnology. PDGF, tetramethyl rhodamine isothiocyanate (TRITC)-labeled phalloidin, mouse anti-acetylated tubulin antibody, and all other reagents and chemicals were purchased from Sigma. Full length GRP1 and ARNO vectors were purchased from ATCC and were fused with GFP by PCR. GRP1 and ARNO PH domain-GFP vectors were generous gifts of Tamas Balla (National Institutes of Health, Bethesda, MD). EFA6 GFP vector was a generous gift of Philippe Chavrier (Institut Curie, Paris, France).
Fluorescent immunocytochemistry
Cells were seeded on coverslips, followed by transfection or treatments with compounds. Then cells were fixed with 4% polyformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 20 min, and blocked with 5% normal serum for 30 min. For detection of tubulin or acetylated tubulin, cells were incubated with primary anti-tubulin or anti-acetylated tubulin antibody (1:200) for 1 hr, and an Alexa Fluor 488-conjugated secondary antibody (1:200) for 30 min in the dark. For detection of actin fibers, cells were incubated with TRITC-labeled phalloidin for 20 min. For detection of cell nuclei, cells were incubated with Hoechst for 10 min. All images were obtained using Nikon TE2000 fluorescent microscope.
Cell migration assays
Transwell model
Migration of cancer and endothelial cells was evaluated using 24-transwell Boyden chamber (Costar, Bedford, MA) with a polystyrene membrane (6.5 mm diameter, 10 μm thickness, and 8 μm pore size). Cells were suspended in serum-free media and seeded in the upper compartment of each well (5×104 cells/well) with or without compounds. The lower compartment contained 600 μl of serum-free media supplemented with fibronectin. After an 8-hr incubation at 37°C, cells were fixed and stained with 0.1% crystal violet. Non-migrating cells on the upper surface of the filter were removed, and the stained cells that migrated to the lower side were photographed using a microscope (Nikon, Japan) in five random fields. Then cells were lysed with 10% acetic acid, and colorimetric determination was made at 595 nm using Wallac Victor 3 plate reader.
Wound healing model
Cancer or endothelial cells were seeded into fibronectin-coated 96-well plates (2×104 cells/well). After reaching confluence, cell monolayers were scratched with a pipette tip to obtain a “wound”. The media and dislodged cells were aspirated, and replaced by fresh serum-free media with or without compounds. After an 8-hr incubation at 37°C, cells were photographed using a microscope in five random fields. The width of wounded cell monolayers in images was measured and the inhibition rates of migration were calculated.
Cell invasion assay
Cell invasion assay was performed using 24-transwell Boyden as described above. The upper chamber was pre-coated with 1 mg/ml of matrigel (BD Biosciences) for 4 hr at 37°C to form a basement membrane. The assays using SUM159 cells were performed and the results were analyzed as previously described for the transwell assay.
Three-dimentional matrigel assay and analysis of the invasive morphology of SUM159 cells was performed according to previous report (Simpson et al., 2004).
Cell polarization assays
SUM159 cells were seeded on fibronectin-coated coverslips. After reaching confluence, cell monolayers were scratched with a pipette tip to obtain a “wound” and were incubated for 2 hr in serum-free media with or without compounds. Then cells were fixed with 4% polyformaldehyde for 30 min at room temperature, permeabilized with 0.1% Triton X-100 for 20 min, and blocked with 5% normal serum for 30 min. For detection of acetylated tubulin, cells were incubated with primary anti-acetylated tubulin antibody (1:200) for 1 hr at room temperature, and then an Alexa Fluor 488-conjugated secondary antibody (1:200) for 30 min in the dark. For detection of actin fibers, cells were incubated with TRITC-labeled phalloidin for 20 min. For detection of cell nuclei, cells were incubated with Hoechst for 10 min.
For HL-60 polarization analysis, HL-60 cells were differentiated with 1.3% DMSO for 6 days, and resuspended in HBSS medium containing 1.8% human serum albumin. The compounds were added at indicated concentrations and incubated for 2 hr, followed by stimulation with 100 nM fMLP for 3 min. Actin staining was performed as described above. All images were obtained using Nikon TE2000 fluorescent microscope.
Tube formation assay
The tube formation assay was performed in 96-well plates. Wells were pre-coated with 70 μl of the Matrigel basement membrane matrix (BD Biosciences) per well for 4 hr at 37°C. HUVEC were suspended in serum-free M199 medium and plated on Matrigel at a density of 2 × 104 cells per well. PIT-1 was added at indicated concentrations (3.125-100 μM). After an 8-hr incubation at 37°C, phase-contrast images of the endothelial tubes were obtained using Nikon TE2000 microscope, and the tube formation was assessed by counting the number of closed tubes in five random fields from each well.
Aortic ring assay
The aortas were isolated from 6-week old Sprague-Dawley rats and immediately transferred to a culture dish with serum-free medium. The fibroadipose tissue around the aortas was carefully removed and the aortas were cut into 1-mm long aortic ring fragments. After three consecutive washes in serum-free medium, the aortic rings were embedded into 70 μl Matrigel in 96-well plate and fed with 100 μl of serum-free M199 medium with or without PIT-1 at different concentrations (3.125-100 μM). The medium was replaced every 24 hr. Phase-contrast images were obtained on day 6, and the numbers of microvessel outgrowths per ring were counted.
Transient transfection
SUM159 cells were seeded on coverslips in 24-well plate at a density of 5×104 cells/well and incubated overnight. Then transient transfection was carried out using TransIT-LT1 Transfection Reagent (Mirrus Bio) according to the manufacturer's recommendations. Twenty-four hr after transfection, cells were subjected to translocation or wound healing or fluorescent immunocytochemistry analysis with or without compound treatment or growth factors stimulation.
Fluorescence polarization (FP) assay
Briefly, Akt, GRP1 and ARNO PH domain (100 nM) were incubated with TMR-labeled PIP3 (60 nM) in the buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5 mM b-ME for 40 min at 15°C in the dark with or without PIT compounds at various concentrations (6.25-200 μM). FP values were determined using Wallac Victor 3 plate reader. The inhibition rates were calculated as: inhibition = [1– (compounds treated group/control group)] ×100%.
ARF6, ARF1 and Rac1 activation assays
SUM159 cells were serum starved overnight, and treated with different compounds at a variety of concentrations, followed by stimulation with 100 ng/ml PDGF for 15 min. Next, cells were rinsed once with PBS, and 1 ml lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 1 mM DTT, 5% glycerol, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 1 mM PMSF) was added per plate. The cells were scraped and transferred to a microcentrifuge tube, vortexed briefly and incubated on ice for 5 min, followed by a centrifugation at 16,000×g at 4°C for 15 min. The supernatant was collected. A sample of the lysate was used to test the total protein level of ARF6, ARF1 or Rac1. Next, 700 μl lysate, 50 μl glutathione resin and 100 μg GST-GGA3-PBD (for ARF6 and ARF1) or GST-Pak1-PBD (for Rac1) were mixed together, and the reaction mixture was incubated at 4°C for 1 hr with gentle rocking. The beads were then washed three times with lysis buffer, and the proteins were eluted by boiling beads in 50 μl of the 2×SDS sample buffer at 95-100°C for 5 min. The samples were subjected to western blot analysis using anti-ARF6, anti-ARF1 or anti-Rac1 monoclonal antibody (1:1,000 dilution) (BD Biosciences).
Suppression of in vivo cancer metastasis
Animals
Male C57BL/6 mice, 6-8 weeks old at time of arrival, were obtained from Charles River Laboratories. Animals were housed in sterilized cages and were provided sterile food and water. Animals were acclimated up to two weeks before tumor cells inoculation. All procedures using mice were approved by the Northeastern University Animal Care and Use Committee according to established guidelines.
Cell culture and animal tumor model
Subculture of primary B16-F10 cells was isolated from mouse metastatic lungs (experimental lung metastasis model), also named B16-F10-L1. Subconfluent proliferating cells at passage #7 were harvested by trypsinization and resuspended in DMEM medium at a concentration of 1.0×106 cells/ml. 100 μl of cell suspension was injected intravenously (i.v.) into the lateral tail vein of C57BL/6 mice for induction of experimental lung metastasis. Randomization of mice was based on their body weights. A total of 21 mice were divided into three groups: control, plain micelles and micellar DM-PIT-1 (DM-PIT-1-M). The control group received PBS injected intraperitoneally (i.p.). Mice in the third group were treated i.p. for 5 consecutive days with a daily injection of 5 mg/kg of micellar DM-PIT-1. The plain PEG-PE micelles were prepared using the same lipid component, and used at the same concentration as DM-PIT-1-loaded micelles to the second group. The drug treatment was started 24 hr after tumor inoculation. The body weights of mice were measured at last.
To monitor the lung colonization, mice were euthanized on day 14 and their lungs were harvested and examined for colonies. Nodules of the metastatic tumor lesions in the lungs were carefully counted by visual inspection and photographed using a Conon PowerShot SD780 IS camera. Metastases, which were visible on the lung surface, were counted and scored according to their size (small < 1 mm and large > 1 mm). Lung samples, from representative animals, were collected for IH evaluation.
Statistics
Student's t test and analysis of variance (ANOVA) were performed using StatView (SAS Institute, Cary, NC). P<0.05 was considered significant and P<0.01 as highly significant. The data shown are representatives of at least three independent experiments with similar results, and the data points represent the mean of at least triplicate measurements with error bars corresponding to standard deviation.
Supplementary Material
Acknowledgements
We thank Dr. Joan Brugge (Harvard Medical School, Boston, MA), Dr. Tamas Balla (National Institutes of Health, Bethesda, MD) and Dr. Philippe Chavrier (Institut Curie, Paris, France) for the gifts of plasmids and cells. We thank Dr. Jeff Peterson (Fox Chase Cancer Center, Philadelphia, PA) for providing valuable comments. This work was supported by Smith Family Award for Excellence in Biomedical Research and National Institute on Aging Mentored Research Scientist Career Development Award (to AD), an Innovator Award from US Army (DAMD17-02-1-0403), National Cancer Institute (RO1 CA34722/PO1-50661) (to BSS) and the National Institute on Aging (R37 AG012859) (to JY).
References
- Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8:604–17. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balana ME, Niedergang F, Subtil A, Alcover A, Chavrier P, Dautry-Varsat A. ARF6 GTPase controls bacterial invasion by actin remodelling. J Cell Sci. 2005;118:2201–10. doi: 10.1242/jcs.02351. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Caumont AS, Vitale N, Gensse M, Galas MC, Casanova JE, Bader MF. Identification of a plasma membrane-associated guanine nucleotide exchange factor for ARF6 in chromaffin cells. Possible role in the regulated exocytotic pathway. J Biol Chem. 2000;275:15637–44. doi: 10.1074/jbc.M908347199. [DOI] [PubMed] [Google Scholar]
- Cavenagh MM, Whitney JA, Carroll K, Zhang C, Boman AL, Rosenwald AG, et al. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–74. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, et al. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–4. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 2004;15:1487–505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–6. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Esteban PF, Yoon HY, Becker J, Dorsey SG, Caprari P, Palko ME, et al. A kinase-deficient TrkC receptor isoform activates Arf6-Rac1 signaling through the scaffold protein tamalin. J Cell Biol. 2006;173:291–9. doi: 10.1083/jcb.200512013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Upender S, Hansen SH, Casanova JE. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem. 1998a;273:23–7. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- Frank SR, Hatfield JC, Casanova JE. Remodeling of the actin cytoskeleton is coordinately regulated by protein kinase C and the ADP-ribosylation factor nucleotide exchange factor ARNO. Mol Biol Cell. 1998b;9:3133–46. doi: 10.1091/mbc.9.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Hafner M, Schmitz A, Grune I, Srivatsan SG, Paul B, Kolanus W, et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–4. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, et al. Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci U S A. 2004;101:6647–52. doi: 10.1073/pnas.0401753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein I, Alcover A, Katzav S. Vav proteins, masters of the world of cytoskeleton organization. Cell Signal. 2004;16:1–11. doi: 10.1016/s0898-6568(03)00110-4. [DOI] [PubMed] [Google Scholar]
- Hu B, Shi B, Jarzynka MJ, Yiin JJ, D'Souza-Schorey C, Cheng SY. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res. 2009;69:794–801. doi: 10.1158/0008-5472.CAN-08-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–7. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, et al. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–30. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Rameh LE, Cantley LC, Buxton JM, Holik JJ, Sakelis C, et al. Regulation of GRP1-catalyzed ADP ribosylation factor guanine nucleotide exchange by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:1859–62. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- Langille SE, Patki V, Klarlund JK, Buxton JM, Holik JJ, Chawla A, et al. ADP-ribosylation factor 6 as a target of guanine nucleotide exchange factor GRP1. J Biol Chem. 1999;274:27099–104. doi: 10.1074/jbc.274.38.27099. [DOI] [PubMed] [Google Scholar]
- Macia E, Luton F, Partisani M, Cherfils J, Chardin P, Franco M. The GDP-bound form of Arf6 is located at the plasma membrane. J Cell Sci. 2004;117:2389–98. doi: 10.1242/jcs.01090. [DOI] [PubMed] [Google Scholar]
- Macia E, Partisani M, Favard C, Mortier E, Zimmermann P, Carlier MF, et al. The pleckstrin homology domain of the Arf6-specific exchange factor EFA6 localizes to the plasma membrane by interacting with phosphatidylinositol 4,5-bisphosphate and F-actin. J Biol Chem. 2008;283:19836–44. doi: 10.1074/jbc.M800781200. [DOI] [PubMed] [Google Scholar]
- Miao B, Skidan I, Yang J, Lugovskoy A, Reibarkh M, Long K, et al. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proc Natl Acad Sci U S A. 2010;107:20126–31. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V, Hoover H, Clancy J, Schweitzer J, Suckow MA, Schroeder V, et al. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res. 2009;69:2201–9. doi: 10.1158/0008-5472.CAN-08-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hirsch DS, Randazzo PA. Arf and its many interactors. Curr Opin Cell Biol. 2003;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Park WS, Heo WD, Whalen JH, O'Rourke NA, Bryan HM, Meyer T, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–92. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–30. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–10. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson KJ, Dugan AS, Mercurio AM. Functional analysis of the contribution of RhoA and RhoC GTPases to invasive breast carcinoma. Cancer Res. 2004;64:8694–701. doi: 10.1158/0008-5472.CAN-04-2247. [DOI] [PubMed] [Google Scholar]
- Tague SE, Muralidharan V, D'Souza-Schorey C. ADP-ribosylation factor 6 regulates tumor cell invasion through the activation of the MEK/ERK signaling pathway. Proc Natl Acad Sci U S A. 2004;101:9671–6. doi: 10.1073/pnas.0403531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Bondeva T, Tamas P, Toth B, Buday L, Hunyady L, et al. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J Cell Sci. 2005;118:4879–88. doi: 10.1242/jcs.02606. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavare JM, Cullen PJ. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998;8:463–6. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- Viaud J, Zeghouf M, Barelli H, Zeeh JC, Padilla A, Guibert B, et al. Structure-based discovery of an inhibitor of Arf activation by Sec7 domains through targeting of protein-protein complexes. Proc Natl Acad Sci U S A. 2007;104:10370–5. doi: 10.1073/pnas.0700773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol. 2002;4:513–8. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- Yang CZ, Heimberg H, D'Souza-Schorey C, Mueckler MM, Stahl PD. Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J Biol Chem. 1998;273:4006–11. doi: 10.1074/jbc.273.7.4006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.