Abstract
The tertiary structure of nucleic acids results from an equilibrium between electrostatic interactions of phosphates, stacking interactions of bases, hydrogen bonds between polar atoms and water molecules. Water interactions with ribonucleic acid play a key role in its structure formation, stabilization and dynamics. We used high hydrostatic pressure and osmotic pressure to analyze changes in RNA hydration. We analyzed the lead catalyzed hydrolysis of tRNAPhe from S. cerevisiae as well as hydrolytic activity of leadzyme. Pb(II) induced hydrolysis of the single phosphodiester bond in tRNAPhe is accompanied by release of 98 water molecules, while other molecule, leadzyme releases 86.
Keywords: Leadzyme, tRNAPhe, High hydrostatic pressure, RNA hydration, RNA structure
Introduction
The three-dimensional structure of nucleic acids results from an equilibrium between electrostatic interactions of negatively charged phosphates, stacking interactions of nucleic acid bases, hydrogen bonds between polar atoms and water surrounding the molecule, as well as the conformational energy of the sugar–phosphate backbone [1]. A key role in the structure of nucleic acids plays water [2]. Dehydration of DNA results in a conformational change from B-form to A-form [3]. Although hydration and its role in macromolecular structure and activity has been extensively studied in the case of RNA still many questions remain unanswered [4]. To shed more light on that problem, we examined hydration changes which accompany hydrolysis of yeast phenylalanine specific transfer RNA (tRNAPhe) with Pb(II). Yeast tRNAPhe can be very useful model for this study because its three-dimensional structure has been determined in many crystallographic studies (Fig. 1a) [5–8]. Moreover it undergoes specific lead(II) induced hydrolysis between D17 and G18 [9]. To solve the problem we analyzed the hydrolysis reaction at high hydrostatic and osmotic pressure (OP).
Fig. 1.
a Superposition of yeast tRNAPhe structure—grey (PDB: 1TRA) and tRNAPhe with Pb2+—black (PDB: 1TN2), in separate box is shown the details of reaction site structure; b HHP effect on Pb2+ induced yeast tRNAPhe hydrolysis. The autoradiogram of the 10% PAGE with 7 M urea of the cleavage reaction of 6.7 μM (300,000 cpm) [3′-32P]tRNAPhe induced with 0.5 mM Pb2+ in 10 mM Tris–HCl pH 6.8, 10 mM MgCl2 and 0.4 M NaCl at 22°C, L alkaline ladder; T1 limited hydrolysis with RNase T1; K control without Pb2+; K ap hydrolysis with Pb2+ at AP for 120 min; at 400, 600, 800 and 200 MPa, 1–4 respectively at 1, 2, 4 and 16 h. Arrows depict location of the cleavage product on the gel, ap ambient pressure (0.1 MPa)
High hydrostatic pressure (HHP) is a convenient tool for study hydration because it allows to induce changes without addition external agents, e.g., salts. Both HHP as well as OP affect the structure of water [10, 11].
The mechanism of that cleavage proposed on the basis of crystal structure of yeast tRNAPhe soaked with Pb(II) starts with abstraction of H+ from a ribose 2′-OH group by Pb(II) bound hydroxyl group. RNA hydrolysis is pH dependent and the optimum for Pb(II) is 7.5 while for Mg(II) is 8.5 [12–14]. The Pb(II) ion bound in the D-loop replaces a Mg(II) from its site located in close vicinity to it in the native structure [15]. The molecule of tRNA is extensively hydrated. In the crystal structure of yeast tRNAPhe 120 water molecules were precisely identified that interact directly with the molecule, about 14% bind adjacent or non-adjacent phosphate atoms, 10% 2′-OH of the ribose and either the N3 of purines or the O2 of pyrimidines [5]. Molecular dynamics analysis showed ca. 820 water molecules in first hydration layer of tRNA, while 269 were estimated for h = 0.2 g—the value determining boundary below which the conformational motion are strongly suppressed (h—hydration in grams of water per g of tRNA) [16]. Most of water molecules in tRNA are clustered around Mg2+ ions and regions of strong negative electrostatic potential near the junctions between D and T loops [16]. X-ray analysis of the 7 bp long tRNASer acceptor-stem structure showed that hydration shell consists of 75 water loci per duplex [17]. Many of the interactions observed between EF-Tu-GTP and aatRNA backbone clearly involve the solvent as integral part [18].
Other RNA that specifically binds Pb(II) and exhibits hydrolytic activity was found by in vitro selection method [19, 20]. Its consensus sequence motif called the “leadzyme” consists of a short duplex with an internal bulge loop. Leadzyme activity depends on Pb2+ only [21–23]. Leadzyme forms an A form duplex with one non-Watson–Crick C–A base pair flanking the 5′-end of three-nucleotide bulge, 5′-GAG-3′ at substrate strand. The scissile phosphodiester bond is located at the junction between helical part and the bulge. The first step of RNA hydrolysis catalyzed with leadzyme proceeds according to mechanism proposed for tRNAPhe, yielding a free 5′-hydroxyl and a 2′,3′-cyclic phosphodiester. That step is analogous to the products of reaction catalyzed by natural small ribozymes such as the hammerhead and hairpin, but in the second step the cyclic phosphate is hydrolyzed to a 3′-phosphate in a manner similar to ribonuclease A. This is not observed in neither for tRNAPhe nor the hammerhead or hairpin ribozyme reaction [24–26].
In the paper we show that HHP inhibits the hydrolysis of tRNAPhe with Pb(II), but does not affects significantly a spontaneous cleavage (not Pb(II) dependent) of double stranded RNA. In contrast to hammerhead and HDV ribozymes, leadzyme was not active without Pb2+ at HHP. Hydrolysis of single phosphodiester–sugar bond of tRNAPhe and leadzyme targeted RNA is accompanied by release of 98 and 86 water molecules as estimated by osmotic stress, respectively.
Materials and methods
RNA synthesis
Reaction was performed on a PCR-Mate EP model 391 DNA synthesizer (Applied Biosystems) with 2-0-triisopropylsilyl-protected phosphoramidite synthons. After deprotection, the oligoribonucleotides were desalted on a Qiagen Tip 500 column as described earlier [27]. This method was used for the preparation of the following oligonucleotides: L1: 5′-GGACGAGCCAG; L2: 5′-CUGGGAAGUCC.
3′-end RNA labeling
6 μL of mixture containing 0.16 mM Cp, 20 μCi [γ-32P]ATP and 10 U T4 polynucleotide kinase and 1 × PNK buffer (Promega) was incubated at 37°C for 45 min and 99°C for 2 min. Than 0.8 nmol of tRNAPhe and 10 U of T4 RNA ligase (Boehringer) in of 20 μL of ligase buffer (50 mM Tris-HCl pH 8.2, 5 mM ATP, 10% DMSO, 20 μg/mL BSA, 30 mM DTT, 10MgCl2,) were added and incubated 16 h at 4°C.
5′-end RNA labeling
5 μg of RNA was mix with [γ-32P]ATP and 6 U T4 polynucleotide kinase (Promega) and kinase buffer (Promega) and incubated 30 min at 37°C. Reaction mixture was then separated in a 15% (tRNA) and 24% (L1) polyacrylamide gel with 7 M urea. The radioactive band was cut out, the RNA was eluted with buffer and precipitated with ethanol [28].
Lead induced hydrolysis of tRNAPhe
A mixture (49.6 μL) composed of 3′-end-labeled tRNAPhe (300,000 cpm), tRNAPhe (30 μg), 27.5 μL NaCl (0.1 M) and 7 μL Tris-HCI pH 6.8 (0.1 M) was incubated for 2 min at 70°C. Then, 7 μL of MgC12 (0.1 M) was added, and samples were cooled slowly (15 min) to 22°C after that 15.4 μL of Pb(CH3OO)2 was added to final concentration of 0.5 mM.
The leadzyme catalyzed hydrolysis of RNA
Reaction was carried out in 15 mM MOPS (3-(N-morpholino)propanesulfonic acid) pH 7.5 at 22°C. The labeled L1 (30000 cpm, 1 mM) and L2 (1 mM) were mixed together and heated up to 60°C for 2 min, cooled slowly (1°C/min) and incubated at 22°C for 30 min. The cleavage reaction was initiated by the addition of Pb2+ (25 mM) and carried out at 22°C. Reaction was stopped by addition of an equal volume of loading buffer (25 mM sodium citrate pH 5, 1 mM EDTA, 7 M urea, 0.1% xylene cyanol, and 0.1% bromophenol blue) and stored on ice.
Analysis of HHP effect on hydrolysis reaction
The effect of HHP on Pb(II) induced hydrolysis of tRNAPhe was investigated by subjecting 15 μL of the reaction mixtures to pressure ranging from 0.1 to 800 MPa. Aliquots were removed after 1, 2, 4, 16 h and quenched with loading buffer and analyzed on 10% PAGE.
For analysis of the influence of HHP on leadzyme catalyzed hydrolysis of L1 15 μL of reaction mixture was subjected to pressure ranging from 0.1 to 800 MPa. Aliquots were removed, and reaction was quenched at various times (0–90 min at 0.1 MPa and 60–180 min at 200–800 MPa).
For technical reasons the time necessary to load the chamber, achieve and release a desired pressure after experiment and stop the reaction takes 10 min. The control reaction was carried out in time necessary to operate the HHP apparatus to subtract the hydrolysis carried out during that time. Every analysis was repeated three times. The radioactivity level present in samples was measured and approximately the same value was loaded on the gels.
Pressuring was carried out in Teflon vessels placed in a high pressure cell (Unipress, Warsaw).
Kinetics of the cleavage reaction at hydrostatic pressure
The activation volume of the reaction (ΔV≠) was calculated from the equation: k = A exp −(PΔV≠/RT). Using the software Kleidagraph, the kinetics toward equilibrium were fitted to the exponential equation: x = xeq(1−e−kobs t), where xeq is the fraction of cleaved RNA at equilibrium. The reaction volume change (ΔV) was calculated using the equation: K = A exp −(PΔV/RT), k and K are the rate and the equilibrium constants of the reaction respectively, R is the universal gas constant (8.314 cm3 MPa °K−1 mol−1), T the temperature (°K) and P the pressure (MPa) [29].
Kinetics of the cleavage reaction under OP
The effect of OP was analyzed by including OP agents, namely polyethylene glycol 400 in the cleavage medium, to final concentrations 0, 2.5, 5, 7.5% (v/v). Aliquots were removed at different time (30–120 min), quenched with loading buffer and analyzed on 24% for L1 and 10% PAGE for tRNAPhe. The number of water molecules released upon RNA cleavage was calculated using the equation: δkTln(kΠ/k0)/δΠosm = ΔVw = ΔNw (30 Å3); kΠ is the observed cleavage rate constant (kobs) at OP Π, k0 is the kobs in the absence of added solute. k is the Boltzmann constant and T the temperature (°K). ΔVw is the change of water volume (30 Å3 the molecular volume of water and ΔNw change in the number of associated water molecules) [29].
Electrophoresis, autoradiography and calculations of hydrolysis yield
tRNAPhe and L1 hydrolysis products were analyzed on 10 and 24% polyacrylamide gels with 7 M urea, pH 8.3, respectively. The limited hydrolysis of tRNAPhe and L1 with T1 RNase under denaturing conditions was done in 20 mM sodium acetate buffer pH 4.5, containing 7 M urea and 1 mM EDTA for 20 min at 55°C [28]. Alkaline hydrolysis was carried out in 50 mM NaOH and 1 mM EDTA for 1 min in case of tRNAPhe and 18 min for L1. For quantitative analysis, the radioactivity of 32P on the gels was counted in a Typhoon 8,600 Imager with ImageQuant software (Molecular Dynamics). The rate constants were calculated by plotting the natural log of the amount of cleaved substrates versus time (5–120 min).
Results and discussion
HHP reversibly modifies hydrophobic and ionic interactions and finally changes the solvation of macromolecules. It can modify the equilibrium constant of a reaction (Keq) if it is accompanied by a significant volume change (ΔV). It can also influence the velocity of reactional processes that involve a significant activation volume (ΔV≠). These parameters (ΔV and ΔV≠) can be directly measured from variation of the reaction equilibrium and rate constants as a function of pressure [30, 31]. ΔV and ΔV≠ values allow to discuss changes in the hydration of the molecule effected by conformational changes caused by single bond breakage [31]. OP can also affect the hydration of molecule which results in conformational changes of the enzyme and thus its activity. Analysis of these changes provides information about changes in hydration [29].
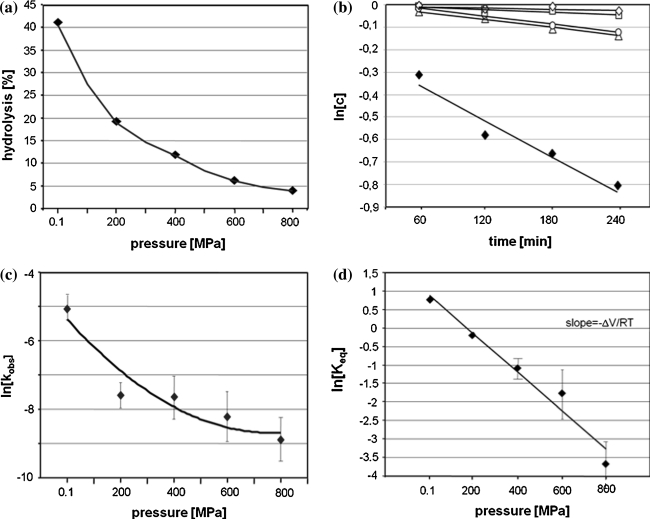
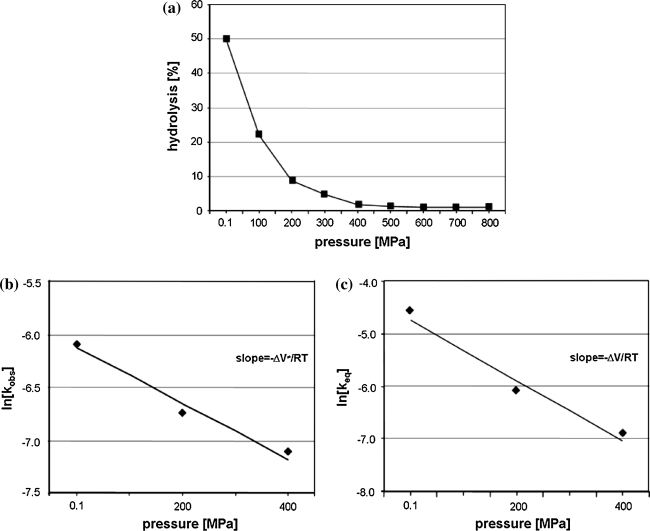
In our study we used yeast tRNAPhe which is specifically cleaved with Pb(II) ions between D17-G18 (major cleavage) however some minor cleavages appear as well at D16, D17, G19 and G20 depending on reaction condition [12–15]. Our experiments were carried out in the way which allows obtaining mainly a single cleavage at HHP, although the cleavage at G19 was still observed at 200 MPa. Lead(II) induced tRNAPhe hydrolysis reaction carried out at ambient pressure (AP) for 120 min proceeds with 43% yield (Figs. 1, 2a) with the observed cleavage rate constant (kobs) of 6.3 × 10−3 min−1 (Table 1; Fig. 2b). To determine the effect of HHP on tRNAPhe cleavage with Pb2+ the reaction mixture was pressured from 0.1 to 800 MPa for 60–240 min (Fig. 2a, b). HHP decreases considerably the kobs (Table 1) of Pb(II) induced hydrolysis which is reflected by positive value of ΔV≠. In the case of the non-linear plot of lnkobs versus pressure (Fig. 2c) kobs can be expressed by the quadratic equation [32]:
where Δβ≠ is the activation isothermal compressibility. To understand the relevance of compressibility. One should remember that the partial molar volume of a macromolecule in solution is the sum of: the constitutive atomic volumes (Va), the volume of cavities in the interior due to imperfect atomic packing (Vc) and the volume change of the solvent due to hydration of solvent-accessible amino-acid groups (ΔVh) [32]. Activation volume, calculated from the plot by curve fitting to above equation, amounts 37.6 cm3/mol while Δβ≠app 0.064 cm3/mol. The extrapolation of kinetic data allowed to estimate the equilibrium constant (Keq) at each pressure (Fig. 2d) and calculate ΔV, which amounts 10.2 cm3/mol.
Fig. 2.
HHP dependence of yeast tRNAPhe cleavage induced with Pb2+. a Hydrolysis of tRNAPhe with Pb(II) dependent on pressure carried out for 2 h. b Determination of kobs by plotting the natural logarithm of product concentration at pressure: 0.1 MPa (filled diamonds). 200 MPa (open triangle), 400 MPa (open circle), 600 MPa (open square), 800 MPa (open diamonds) as a function of time. c Determination of the activation volume changes associated to the hydrolysis reaction. The ΔV≠ was calculated from the plot by curve fitting to equation: lnkobs = lnk0 − ΔV≠P/RT + Δβ≠P2/2RT. d Linear decrease of the theoretical equilibrium constants logarithms with increasing hydrostatic pressures. Keq was calculated as the ratio of cleaved and uncleaved fractions at equilibrium. The percentages of cleaved products were obtained from the extrapolation of each exponential. Change of the reaction volume (ΔV) was calculated from the slope of the curve RTlnKeq/pressure
Table 1.
Kinetic and volumetric parameters for RNA hydrolysis at pressure 0.1–800 MPa
| p (MPa) | kobs × 10−3 (min−1) | ||
|---|---|---|---|
| tRNAPhe | L1 specific hydrol. | L1 spontaneous hydrol. | |
| 0.1 | 6.30 | 10.0 | 0.84 |
| 100 | – | 0.72 | 0.30 |
| 200 | 0.50 | 0.18 | 0.20 |
| 300 | – | 0.12 | 0.16 |
| 400 | 0.48 | 0.08 | 0.13 |
| 600 | 0.26 | – | 0.08 |
| 800 | 0.13 | – | 0.03 |
| ΔV≠/ΔV (cm3/mol) | 39.5/10.2 | 30.0/24.9 | 10.5/2.5 |
| ΔVw (cm3/mol) | −1,760 | −2,100 | −540 |
| Water released | 98 | 86 | 30 |
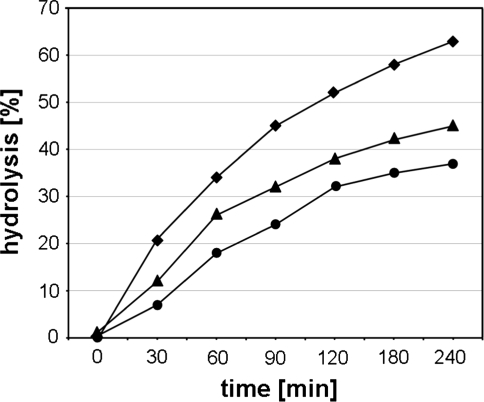
Analysis of the tRNAPhe hydrolysis at 0.1 MPa followed the pressuring at 200 and 800 MPa for 2 h showed that the reaction is almost fully reversible (kobs amounts 6.2, 6.0 and 5.8 × 10−3 min−1 for 0.1, 200 and 800 MPa, respectively) (Fig. 3). It means that observed decrease of hydrolysis at HHP does not result from irreversible changes in tRNA structure.
Fig. 3.
Reversibility of the effects of HHP on yeast tRNAPhe hydrolysis with Pb(II). Kinetics of reactions carried out at AP (diamonds) and after 2 h of reaction under pressure 200 MPa (triangle) and 800 MPa (circles). The reaction mixtures were exposed to AP and allowed to react for 4 h
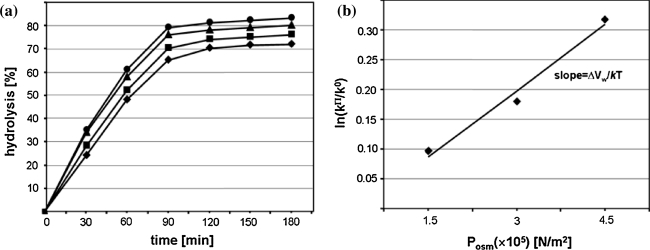
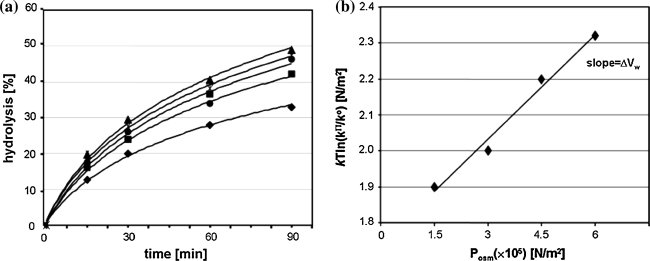
Analysis of reaction kinetics carried out in condition of osmotic stress enables to calculate the number of exchanged water molecules [29]. OP acts opposite to HHP which is reflected by negative value of ΔVw = −1,760 cm3/mol (Fig. 4a, b) corresponding to 98 water molecules released from tRNAPhe molecule as a consequence of Pb(II) induced cleavage. After cleavage, tRNA still migrates as a single band on non-denaturing gel (data not shown) which means that the structure is fully stabilized by hydrogen bonds. Analysis of free tRNAPhe crystals soaked with Pb(II) has shown that ion binding does not induce significant structural changes, however superposition of that molecule with free tRNAPhe shows small differences in atom position (Fig. 1a) but in the crystal the cleavage of the P–O bond was not detected [13]. FTIR spectra of yeast tRNAPhe recorded up to 1,400 MPa showed an increase of molecule hydration and some conformational changes [33]. Similarly analysis of DNA at pressure up to 1,380 MPa showed only subtle changes in its structure and increase of the number of bound water molecules from 57 to 76 [34]. However, change in the number of water molecules associated with tRNAPhe must be accompanied by change in the nucleic acid conformation.
Fig. 4.
OP dependence of tRNAPhe hydrolysis catalyzed with Pb(II). a Hydrolysis of tRNAPhe in the presence of increasing concentrations of PEG 400: 0% (diamonds), 2.5% (v/v) (squares), 5% (triangles), 7.5% (circles); b effect of OP on the observed cleavage rate constant: ln(kΠ/k0) plotted as a function of OP. kΠ and k0, the observed rate constant of the reaction under osmotic stress and in standard conditions, respectively; k the Boltzmann constant, T the absolute temperature (K)
It was shown that the Pb(II) induced cleavage between D17 and G18 occurs in tRNA consisting of two (not ligated) fragments 1–36 and 38–76. The prerequisite for hydrolysis is, however, the presence almost whole molecule having both TΨC and DHU [35]. It means that this reaction depends on proper three-dimensional structure of tRNAPhe.
To evaluate whether the observed amount of water expelled from tRNA molecule was located rather in the vicinity of the reaction site or scattered throughout the whole body of tRNA we used smaller RNA molecule. Because in the case of yeast tRNAPhe specific cleavage with Pb(II) is highly structure dependent and occurs only in the presence of whole molecule, any fragment of tRNA could not be used.
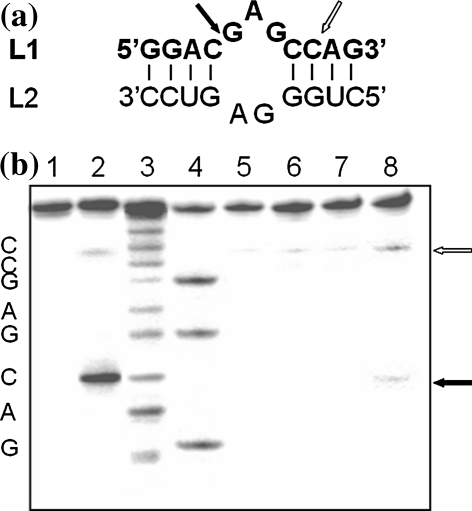
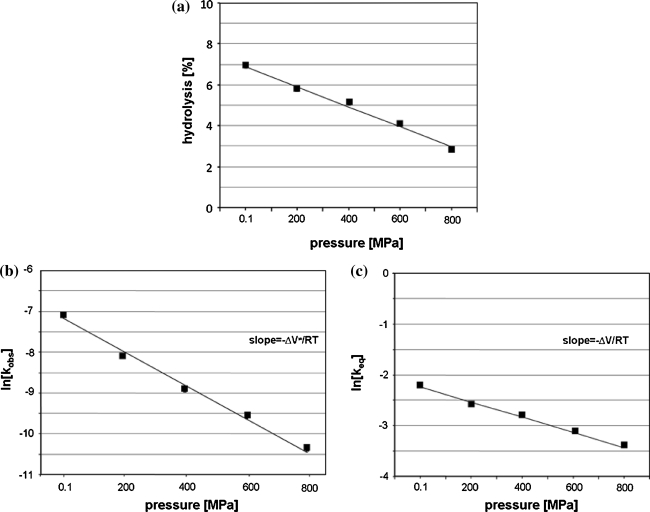
The only known molecule, except of yeast tRNAPhe, which is specifically hydrolyzed with Pb(II) is leadzyme. The one used in our work consists of an asymmetric internal loop of five-purine nucleotides surrounded by two short double RNA helices (Fig. 5a). It catalyzes specific hydrolysis of its target L1 at a C-G phosphodiester bond on the longer fragment of the loop (Fig. 5a, b) [19, 20, 27]. The yield of that cleavage at 0.1 MPa reaches a maximum of 52% after 2 h with kobs of 1 × 10−2 min−1 (Table 1). An increase of hydrostatic pressure up to 800 MPa decreases the hydrolysis efficiency (Fig. 6a; Table 1). In contrast to tRNAPhe leadzyme catalyzed reaction is almost completely inhibited at 400 MPa therefore calculation was carried out in the range 0.1–400 MPa. In that pressure range the plots of lnkobs and lnKeq versus pressure are linear. The ΔV≠ and ΔV amount 30.0 and 24.9 cm3/mol, respectively (Fig. 6b, c; Table 1).
Fig. 5.
Secondary structure of leadzyme construct used in the work. a Black arrow points the position of leadzyme catalyzed hydrolysis, open arrow shows spontaneous cleavage. b 24% PAGE with 7 M urea L1 substrate hydrolysis catalyzed with L2 at HHP. The hydrolysis reaction of 1 mM [32P] labeled L1 with 1 mM L2 was carried in 15 mM NaMOPS pH 7.5 buffer containing 25 mM Pb2+. Lanes: 1 control, 2 reaction at 0.1 MPa for 2 h, 3 alkaline hydrolysis, 4 limited hydrolysis with RNase T1, 5−8 hydrolysis of L1 at 400 MPa for 0.5, 1, 2 and 4 h
Fig. 6.
Hydrostatic pressure dependence of the leadzyme catalyzed cleavage reaction of L1. a Carried out in MOPS containing Pb(II) for 2 h. Determination of the activation volume (ΔV≠) (b) and volume change (ΔV) (c) accompanying the specific hydrolysis reaction, calculated from the slope of the curve
In the presence of increasing concentration of PEG400 (0–7.5%) the rate of leadzyme catalyzed cleavage reaction increases (Fig. 7a). Based on the plot of ln[kπ/ko] versus OP (Fig. 7b) it was calculated that the formation of the transition state involves the release of 116 water molecules per RNA molecule. However in addition to the specific lead-dependent cut at the C–G bond of the target RNA an additional cleavage between C–A in the double stranded fragment was observed. This cleavage is Pb(II) independent and occurs due to spontaneous hydrolysis of RNA (Fig. 5a, b) [27]. The cleavage of bond between C–A in RNA depends on conformational context [36]. To check the hydration changes accompanying that spontaneous hydrolysis we analyzed the effect of HHP on L1 hydrolysis carried out in the absence of Pb(II). The ΔV≠ and ΔV for that cleavage is 10.5 and 2.5 cm3/mol, respectively (Fig. 8; Table 1). Determination of ΔVw allows calculation that non-specific cleavage is accompanied by release of 30 water molecules (Fig. 9a, b; Table 1). It means that Pb(II) induced hydrolysis causes release of 86 water molecules, which is pretty close to the amount obtained for tRNAPhe.
Fig. 7.
OP dependence of leadzyme catalyzed hydrolysis reaction of L1. a Kinetics of cleavage reaction was carried out in increasing concentration of PEG 400: 0% (diamonds), 2.5% (v/v) (squares), 5% (circles), 7.5% (crosses) and 10% (triangles). b Determination of ΔVw for Pb(II) catalyzed L1 hydrolysis
Fig. 8.
Hydrostatic pressure dependence of spontaneous hydrolysis of L1 (a) carried out in MOPS without Pb(II). Determination of ΔV≠ (b) and ΔV (c) from the slope of ln[kobs] and ln[Keq] plotted as a function of pressure
Fig. 9.
OP dependence of spontaneous hydrolysis of L1. a Kinetics of cleavage reaction was carried out in increasing concentration of PEG 400: 0% (diamonds), 2.5% (v/v) (squares), 5% (circles), 7.5% (crosses) and 10% (triangles). b Determination of ΔVw for spontaneous hydrolysis
For the hairpin ribozyme it has been shown that formation of transition state is accompanied by reduction of solvent accessible area (saa) by 1,570 Å2 and release of 78 water molecules (~H2O/20 Å2) [30]. Single base covers an area of approximately 177 Å2 [37] thus saa in the case of hairpin corresponds to 8.8 bases. Simple calculation gives ~11 bases (98H2O × 20 Å2 = 1,960 Å2) for tRNAPhe, ~9.7 bases (86H2O × 20 Å2 = 1,720 Å2) for leadzyme and ~3.4 (30H2O × 20 Å2 = 600 Å2) in the case of non-specific hydrolysis in helical part of leadzyme. It suggests that water release observed as a result of P–O cleavage was located around the reaction site rather than scattered throughout the molecule. In the case of structurally more simple molecule, like leadzyme, that nine bases form loop and two flanking bp. In more complicated structure like tRNAPhe saa comprises 11 bases, but one have to keep in mind that reaction center consists of two interacting loops TΨC binding Pb2+ and DHU (target). However that fragment of tRNAPhe molecule is more hydrated than the remaining part as shown in crystal structure [6].
ΔV≠ value for leadzyme catalyzed reaction is close to that obtained for tRNAPhe, but ΔV = 24.9 cm3/mol for leadzyme is almost two times greater than observed for tRNAPhe. It may explain almost a complete inhibition of leadzyme activity at 400 MPa, while for tRNAPhe ca. 2.5% of hydrolysis was still detected at 800 MPa after 2 h (Figs. 2a, 6a).
The non-linear plot of lnkobs versus pressure for tRNAPhe means that the activation isothermal compressibility factor (Δβ≠) cannot be neglected (Fig. 2c) [32]. Considering the differences between that plot for tRNAPhe and leadzyme (Figs. 2b, 6b, 8b) one has to keep in mind difference in the size of both molecules as well as in mechanism of the reaction. The plot of lnkobs versus pressure depends on: (i) pressure-induced quaternary structure change (dissociation of subunits, changes of the architecture of binding and active sites), (ii) changes in compressibility (including changes in hydration) and (iii) changes in the rate-determining step [32]. The pressure induced unfolding and dissociation of subunits which are generally observed for proteins does not occur in the case of nucleic acids [38, 39]. So positive value of isothermal compressibility changes (Δβ≠app 0.064 cm3/mol) is associated with compression of cavities in the macromolecule interior, changes in intramolecular packing and increase in molecule hydration [32]. An increase of yeast tRNAPhe hydration and changes of its structure were observed with FTIR [33]. However previous considerations does not exclude the influence of pressure on rate-determining step. The first step of Pb(II) induced hydrolysis of RNA is the same in both cases tRNAPhe and leadzyme [15, 20]. Spontaneous hydrolysis observed in helical fragment of RNA proceeds according to the same mechanism [40]. While in the case of small metalloribozyme the reaction proceeds according to two-step mechanism and in the second step the 2′,3′-cyclic phosphodiester is further converted to 3′-monophosphate [20]. So the difference observed between tRNAPhe and leadzyme might results from the different mechanism of reaction.
Our results show that cleavage of RNA with Pb(II) is pressure inhibited while spontaneous cut of RNA occurring in helical region is relatively less HHP dependent. It means that structural changes of RNA during in Pb(II) induced hydrolysis are bigger. Hydrolysis of single phosphodiester bond in the loop of RNA is accompanied by three times higher hydration changes comparing to cleavage in helix. The first step of volume change of tRNAPhe caused by increasing pressure is connected with decrease of water volume while further effects arise from tRNA conformational changes.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Abbreviations
- HHP
High hydrostatic pressure
- OP
Osmotic pressure
- AP
Ambient pressure
References
- 1.Westhof E. Water: an integral part of nucleic acid structure. Annu Rev Biophys Biophys Chem. 1988;17:125–144. doi: 10.1146/annurev.bb.17.060188.001013. [DOI] [PubMed] [Google Scholar]
- 2.Auffinger P, Hashem Y. Nucleic acid solvation: from outside to insight. Curr Opin Struct Biol. 2007;17:325–333. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Dickerson RE, Drew HR, Conner BN, Wing RM, Fratini AV, Kopka ML. The anatomy of A-, B-, and Z-DNA. Science. 1982;216:475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 4.Caliskan G, Briber RM, Thirumalai D, Garcia-Sakai V, Woodson SA. Dynamic transition in tRNA is solvent induced. J Am Chem Soc. 2006;128:32–33. doi: 10.1021/ja056444i. [DOI] [PubMed] [Google Scholar]
- 5.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: Cleavage by Mg2+ in 15-year old crystals. J Mol Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 6.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/S1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M. The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5:59–68. doi: 10.1016/S0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 8.Bilgin N, Ehrenberg M, Ebel C, Zaccai G, Sayers Z, Koch MH, Svergun DI, Barberato C, Volkov V, Nissen P, Nyborg J. Solution structure of the ternary complex between aminoacyl-tRNA, elongation factor Tu, and guanosine triphosphate. Biochemistry. 1998;37:8163–8172. doi: 10.1021/bi9802869. [DOI] [PubMed] [Google Scholar]
- 9.Krzyzosiak WJ, Marciniec T, Wiewiorowski M, Romby P, Ebel JP, Giege R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- 10.Winter R. Advances in high pressure bioscience and biotechnology II. Heidelberg: Springer; 2003. [Google Scholar]
- 11.Mozhaev VV, Heremans K, Frank J, Masson P, Balny C. High pressure effects on protein structure and function. Proteins Struct Funct Genet. 1996;24:81–91. doi: 10.1002/(SICI)1097-0134(199601)24:1<81::AID-PROT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Ciesiołka J, Wrzesiński J, Górnicki P, Podkowiński J, Krzyżosiak WJ. Analysis of magnesium, europium and lead binding sites in methionine initiator and elongator tRNAs by specific metal-ion-induced cleavages. Eur J Biochem. 1989;186:71–77. doi: 10.1111/j.1432-1033.1989.tb15179.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown RS, Dewan JC, Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985;24:4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- 14.Rubin JR, Sundaralingam M. Lead ion binding and RNA chain hydrolysis in phenylalanine tRNA. J Biomol Struct Dyn. 1983;1:639–646. doi: 10.1080/07391102.1983.10507471. [DOI] [PubMed] [Google Scholar]
- 15.Brown RS, Hingerty BE, Dewan JC, Klug A. Pb(II)-catalysed cleavage of the sugar–phosphate backbone of yeast tRNAPhe—implications for lead toxicity and self-splicing RNA. Nature. 1983;303:543–546. doi: 10.1038/303543a0. [DOI] [PubMed] [Google Scholar]
- 16.Roh JH, Briber RM, Damjanovic A, Thirumalai D, Woodson SA, Sokolov AP. Dynamics of tRNA different levels of hydration. Biophys J. 2009;96:2755–2762. doi: 10.1016/j.bpj.2008.12.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichert A, Fürste JP, Schreiber A, Perbandt M, Betzel Ch, Erdmann VA, Förster Ch. The 1.2 Å crystal structure of an E. coli tRNASer acceptor stem microhelix reveals two magnesium binding sites. Biochem Biophys Res Commun. 2009;386:368–373. doi: 10.1016/j.bbrc.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 18.Nissen P, Kjeldgaard M, Thirup S, Clark BFC, Nyborg J. The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie. 1996;78:921–933. doi: 10.1016/S0300-9084(97)86714-4. [DOI] [PubMed] [Google Scholar]
- 19.Pan T, Uhlenbeck OC. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+ Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- 20.Pan T, Uhlenbeck OC. A small metalloribozyme with a two-step mechanism Nature. 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- 21.Pan T, Dichtl B, Uhlenbeck OC. Properties of an in vitro selected Pb2+ cleavage motif. Biochemistry. 1994;33:9561–9565. doi: 10.1021/bi00198a023. [DOI] [PubMed] [Google Scholar]
- 22.Sugimoto N, Ohmichi T. Site-specific cleavage reaction catalyzed by leadzyme is enhanced by combined effect of lead and rare earth ions. FEBS Lett. 1996;393:97–100. doi: 10.1016/0014-5793(96)00860-5. [DOI] [PubMed] [Google Scholar]
- 23.Ohmichi T, Sugimoto N. Role of Nd3+ and Pb2+ on the RNA cleavage reaction by a small ribozyme. Biochemistry. 1997;36:3514–3521. doi: 10.1021/bi962030d. [DOI] [PubMed] [Google Scholar]
- 24.Wedekind JE, McKay DB. Crystal structure of the leadzyme at 1.8 Å resolution: metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry. 2003;42:9554–9563. doi: 10.1021/bi0300783. [DOI] [PubMed] [Google Scholar]
- 25.Koizumi M, Ohtsuka E. Effects of phosphorothioate and 2-amino groups in hammerhead ribozymes on cleavage rates and Mg2+ binding. Biochemistry. 1991;30:5145–5150. doi: 10.1021/bi00235a005. [DOI] [PubMed] [Google Scholar]
- 26.Slim G, Gait MJ. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res. 1991;19:1183–1188. doi: 10.1093/nar/19.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barciszewska MZ, Wyszko E, Bald R, Erdmann VA, Barciszewski J. 5S rRNA is a leadzyme. A molecular basis for lead toxicity. J Biochem. 2003;133:309–315. doi: 10.1093/jb/mvg042. [DOI] [PubMed] [Google Scholar]
- 28.Barciszewska MZ, Erdmann VA, Barciszewski J. The dynamic conformation of plant cytoplasmic 5S rRNAs. Phytochemistry. 1994;37:113–117. doi: 10.1016/0031-9422(94)85008-9. [DOI] [PubMed] [Google Scholar]
- 29.Parsegian VA, Rand RP, Rau DC. Macromolecules and water: probing with osmotic stress. Methods Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 30.Tobe S, Heams T, Vergne J, Herve G, Maurel MC. The catalytic mechanism of hairpin ribozyme studied by hydrostatic pressure. Nucleic Acids Res. 2005;33:2557–2564. doi: 10.1093/nar/gki552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalikian TV, Sarvazyan AP, Breslauer KJ. Hydration and partial compressibility of biological compounds. Biophys Chem. 1994;51:89–107. doi: 10.1016/0301-4622(94)85007-0. [DOI] [PubMed] [Google Scholar]
- 32.Masson P, Balny C. Linear and non-linear pressure dependence of enzyme, catalytic parameters. Biochim Biophys Acta. 2005;1724:440–450. doi: 10.1016/j.bbagen.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Giel-Pietraszuk M, Barciszewski J. A nature of conformational changes of yeast tRNAPhe High hydrostatic pressure effects. Int J Biol Macromol. 2005;37:109–114. doi: 10.1016/j.ijbiomac.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Girard E, Prange T, Dhaussy A-C, Migianu-Griffoni E, Lecouvey M, Chervin JC, Mezouar M, Kahn R, Fourme R. Adaptation of the base-paired double-helix molecular architecture to extreme pressure. Nucleic Acids Res. 2007;35:4800–4808. doi: 10.1093/nar/gkm511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng HY, Termini J. Catalytic RNA reactions of yeast tRNA(Phe) fragments. Biochemistry. 1992;31:10518–10528. doi: 10.1021/bi00158a015. [DOI] [PubMed] [Google Scholar]
- 36.Kierzek R. Nonenzymatic hydrolysis of oligoribonucleotides. Nucleic Acids Res. 1992;20:5079–5084. doi: 10.1093/nar/20.19.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferre-D’Amare AR, Doudna JA. RNA FOLDS: insights from recent crystal structures. Annu Rev Biophys Biomol Struct. 1999;28:57–73. doi: 10.1146/annurev.biophys.28.1.57. [DOI] [PubMed] [Google Scholar]
- 38.Rayan G, Macgregor RB., Jr Pressure-induced helix-coil transition of DNA copolymers is linked to water activity. Biophys Chem. 2009;144:62–66. doi: 10.1016/j.bpc.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Macgregor RB., Jr Effect of hydrostatic pressure on nucleic acids. Biopolymers. 1998;48:253–263. doi: 10.1002/(SICI)1097-0282(1998)48:4<253::AID-BIP5>3.3.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 40.Perrin CL. On the mechanism of buffer-catalyzed hydrolysis of RNA models. J Org Chem. 1995;60:1239–1243. doi: 10.1021/jo00110a030. [DOI] [Google Scholar]