Abstract
Fukutin is the gene responsible for Fukuyama-type congenital muscular dystrophy (FCMD), an autosomal recessive disease associated with central nervous system (CNS) and eye anomalies. Fukutin is involved in basement membrane formation via the glycosylation of α-dystroglycan (α-DG), and hypoglycosylation of α-DG provokes the muscular, CNS and eye lesions of FCMD. Astrocytes play an important role in the pathogenesis of the CNS lesions, but the post-transcriptional regulation of fukutin mRNA has not been elucidated. In this study, we investigated the characteristics of fukutin mRNA using an astrocytoma cell line that expresses fukutin and glycosylated α-DG. The glycosylation of α-DG was considered to be increased by over-expression of fukutin and decreased by knockdown of fukutin. Knockdown of Musashi-1, one of the RNA-binding proteins involved in the regulation of neuronal differentiation, induced a decrease in fukutin mRNA. Immunoprecipitation and ELISA-based RNA-binding assay demonstrated possible binding between fukutin mRNA and Musashi-1 protein. A relationship between fukutin mRNA and vimentin protein was also proposed. In situ hybridization for fukutin mRNA showed a positive cytoplasmic reaction including cytoplasmic processes. From these results, fukutin mRNA is suggested to be a localized mRNA up-regulated by Musashi-1 and to be a component of a mRNA-protein complex which includes Musashi-1 and (presumably) vimentin proteins.
Keywords: astrocytoma, fukutin, mRNA, Musashi-1, vimentin
Fukutin is the gene responsible for Fukuyama-type congenital muscular dystrophy (FCMD) (Kobayashi et al. 1998), an autosomal recessive disease, showing muscular dystrophy, and CNS and eye anomalies (Fukuyama et al. 1960; Osawa et al. 1997). A representative CNS lesion is cobblestone lissencephaly or type II lissencephaly of the cerebrum and cerebellum. Fukutin is considered to be related to the glycosylation of α-dystroglycan (α-DG) (Hayashi et al. 2001; Michele & Campbell 2003; Martin 2005; Schessl et al. 2006; Yamamoto et al. 2010), in association with protein O-linked mannose β1,2-N-acetylglucosaminyltransferase (POMGnT1) (Xiong et al. 2006). α-DG is one of the components of the dystrophin-glycoprotein complex, which is involved in basement membrane formation, linking intracellular and extracellular proteins (Michele & Campbell 2003; Martin 2005; Schessl et al. 2006). The glycosylated α-DG is a receptor of extracellular matrix proteins such as laminin (Michele & Campbell 2003; Martin 2005; Schessl et al. 2006).
In FCMD patients, hypoglycosylation of α-DG was observed in the sarcolemma of striated muscle and in the glia limitans of the CNS where the basement membrane is formed (Hayashi et al. 2001; Yamamoto et al. 2004, 2010). A fragile basement membrane causes muscular dystrophy and disruptions of the glia limitans. During the foetal period immature neurons over-migrate through the disruptions, which is considered to result in cobblestone lissencephaly. Because the glia limitans is formed by astrocytic endfeet and covered with the basement membrane, the functions of fukutin in astrocytes appear to be important for the pathogenesis of the CNS lesions of FCMD (Yamamoto et al. 2004, 2010).
Thus, the pathomechanism of CNS lesions of FCMD and functions of fukutin have been elucidated gradually, but it is still unclear how fukutin is regulated after transcription. A founder mutation of FCMD is a 3-kb insertion in the 3′-untranslated region (3′-UTR) of the fukutin gene (Kobayashi et al. 1998). The insertion is considered to cause a decrease in transcription and/or instability of mRNA (Kobayashi et al. 1998), but the precise mechanism is unknown. Recent studies have revealed diverse and dynamic roles of RNA in the maintenance and regulation of cells. The post-transcriptional RNA regulation includes alternative splicing, polyadenylation, RNA editing and regulation by RNA-binding proteins, which generate RNA and protein diversity in eukaryotes (Bolognani & Perrone-Bizzozero 2008; Tazi et al. 2009; Farajollahi & Maas 2010; Licatalosi & Darnell 2010). A defect or imbalance of these mechanisms can be a cause of some diseases including neurological disorders (Bolognani & Perrone-Bizzozero 2008; Tazi et al. 2009; Farajollahi & Maas 2010; Licatalosi & Darnell 2010).
Because astrocytes play an important role in the pathogenesis of CNS lesions of FCMD, it could be interesting to elucidate the post-transcriptional regulation of fukutin mRNA in astrocytes. In this study, characteristics of fukutin mRNA were investigated using an astrocytoma cell line.
Materials and methods
Cell line
A cell line from a human astrocytoma (1321N1) (Macintyre et al. 1972; Foster & Perkins 1977) was purchased from European Collection of Cell Cultures (Wiltshire, SP4 OJG, UK). Cells were grown in DMEM (Invitrogen, Carlsbad, CA, USA), supplemented with 10% foetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 2 mM glutamine (Invitrogen) and 1% penicillin-streptomycin (Invitrogen). Cells were maintained at 37 °C in a humidified incubator with a CO2 atmosphere.
Transfection of fukutin cDNA in astrocytoma cells
RNA was extracted from normal striated muscle obtained by autopsy at Tokyo Women's Medical University that was performed after the family members granted informed consent in accordance with the Helsinki Declaration. After reverse transcription, 2 ng/μl cDNA was amplified by PCR using pfx50 DNA polymerase (Invitrogen). The sequences of the primers of the fukutin cDNA (GenBank: AB008226) were as follows: 5′-CACCATGAGTAGAATCAATAAGAACGTGGTTTTG-3′ (coding sense) and 5′- ATATAACTGGATAACCTCAT -3′ (anticoding sense). The reaction mixture was amplified for 35 cycles in a Zymoreactor thermo-cycler (ATTO Co., Tokyo, Japan). The amplification profiles consisted of denaturing at 94 °C for 15 s, annealing at 62 °C for 30 s and extension at 68 °C for 60 s. After electrophoresis of the PCR products using 1.2% agarose gels, gels containing the DNA were cut and the DNA was purified using a PureLink Quick Gel Extraction Kit (Invitrogen). To make a stable transformant, a plasmid containing fukutin cDNA was cloned using a pcDNA 3.1 Directional TOPO Expression Kit, according to the manufacturer's instructions (Invitrogen). The plasmid was transfected into astrocytoma cells using lipofectamine2000 (Invitrogen) and Opti-MEM (Invitrogen). The transfection was confirmed by western blotting, reverse transcriptase polymerase chain reaction (RT-PCR) and sequencing performed by Takara (Tokyo, Japan). The cells transfected with fukutin cDNA were maintained in the above culture medium with an additional supplement of geneticin (800 μg/ml; Invitrogen).
RNAi
RNAi was performed on original astrocytoma cells, as previously described (Yamamoto et al. 2008). Stealth siRNA duplexes for fukutin and for Musashi-1 mRNA were designed and synthesized by Invitrogen. The target sense for fukutin was 5′-UUUGGAAGGGAACAAAUUUCCUGUC-3′ (F697) and that for Musashi-1 was 5′-CGGUGGAGGACGUGAAGCAAUAUUU-3′ (M426). Scrambled negative control Stealth™ RNA (SNC; Invitrogen) was used as a negative control. Omission of siRNA was always performed as a negative control in each experiment. Astrocytoma cells were plated one day before transfection at a density of about 200,000 cells in each 35-mm dish. Antibiotics were omitted from the medium. siRNA (40 nM final concentration) was transfected into the cells using lipofectamineMAX (Invitrogen) and Opti-MEM (Invitrogen) according to the manufacturer's instructions. The culture medium was changed to the regular formulation one day after transfection. Cells were harvested 4 days after transfection. Suppression of each gene was confirmed by RT-PCR or western blotting.
RNA extraction and semi-quantitative RT-PCR
RNA was extracted from cultured cells by the acid guanidium thiocyanate-phenol-chloroform method. The reverse transcription was performed using M-MLV reverse transcriptase (Life Technologies, Rockville, MD, USA) at 42 °C for 2 h, or using PrimeScript RT-PCR kit (Takara).
cDNA at 2 ng/μl was amplified by PCR using the AccuPrime Taq DNA polymerase system (Invitrogen). The sequences of the primers for 112–731 bp of fukutin cDNA (GenBank: AB008226) were 5′-ATGAGTAGAATCAATAAGAA-3′ (coding sense) and 5′- AACTGTAACTTTCGGAAGGG -3′ (anticoding sense), those for 395–883 bp of DG (GenBank: XM_018223) were 5′-ATGAGGATGTCTGTGGGCCT-3′ (coding sense) and 5′-ATCACTGTGGTCTTCAGGGT-3′ (anticoding sense), those for β-actin were 5′-TGGCACCCAGCACAATGAA-3′ (coding sense) and 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ (anticoding sense), and those for GAPDH were 5′-TTTGGTCGTATTGGGCGCCT-3′ (coding sense) and 5′-GTGGTCATGAGTCCTTCCAC-3′ (anticoding sense). The reaction mixture was amplified for 35 cycles in a Zymoreactor thermo-cycler (ATTO). The amplification profiles consisted of denaturing at 94 °C for 1.5 min, annealing at 56 °C for 1.5 min and extension at 72 °C for 1 min. PCR products were electrophoresed using 1.2% agarose gels, stained with ethidium bromide and photographed. The density of each band was measured using ATTO CS Analyzer, version 3.00.1007, and the ratio of each product against that of β-actin or GAPDH was calculated.
Immunohistochemistry
For immunohistochemistry of α-DG, a cellblock was made. Original astrocytoma cells were detached using a cell scraper (Iwaki, Tokyo Japan), fixed in 4% paraformaldehyde/phosphate-buffered saline (pH 7.6) for 30 min at 4 °C and then collected by centrifugation. Cells were gathered using 2% agarose, embedded in paraffin in a routine manner and sectioned at 3 μm. Sections were treated with 3% H2O2 in PBS for 10 min to block endogenous peroxidase and were transferred to normal serum for 30 min. The sections were incubated with the anti-α-DG antibody against the core peptide of α-DG (α-DG-p, sheep, polyclonal, 1:1000, kindly provided by Dr. S. Kröger) (Herrmann et al. 2000) overnight at 4 °C or with the anti-α-DG antibody against the laminin-binding site of glycosylated epitope of α-DG (IIH6C4, monoclonal, 1:500; Upstate Biotechnology, Lake Placid, NY, USA) (Ervasti & Campbell 1993) for two nights at 4 °C. For detection, the Vectastain ABC System (Vector Laboratories, Burlingame, CA, USA) was employed for α-DG-p, and the EnVision System (DAKO, Glostrup, Denmark) was for IIH6C4. Colour was developed with chromogen diaminobenzidine tetrahydrochloride, and the slides were counterstained using haematoxylin. For the negative control experiment, the primary antibody was omitted from the solution.
Astrocytoma cells cultured on culture slides (BD Falcon) were fixed in 95% ethanol for 5 min at RT and incubated with PBS containing 2% skim milk and 1% Triton X-100 for 20 min at RT. Cells were double-stained using the anti-fukutin antibody (rabbit, polyclonal, 1:50; Yamamoto et al. 2002) and vimentin (Code No M725, mouse, monoclonal, 1:200; DAKO) antibody. The slides were incubated with primary antibody at 4 °C, overnight, and then with Cy3-conjugated donkey anti-rabbit IgG (1:200; Jackson Immunoresearch Laboratory, West Grove, PA, USA) and FITC-conjugated donkey anti-mouse IgG (1:200; Jackson Immunoresearch) at room temperature (RT) for 1 h. The nucleus was counterstained by DAPI. For the negative control experiment, the primary antibody was omitted from the solution. Specimens were examined using a fluorescence microscope (Nikon ECLIPSE TS100; Nikon, Tokyo, Japan).
Protein extraction and western blotting
Cultured cells were suspended in RIPA buffer (20 mM Tris–Cl, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitor cocktail (Complete, Mini; Roche Diagnostics, Mannheim, Germany). After centrifugation at 12,500 g, 4 °C for 60 min, the supernatant was used. The specimen was electrophoresed in a 9% SDS-polyacrylamide gel and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). After being treated at 4 °C overnight with TBS (100 mM Tris–Cl, pH 7.4, 150 mM NaCl) containing 0.2% Tween-20, 5% skim milk and 2% porcine serum, the membrane was incubated with a primary antibody at RT for 1 h and then with a secondary antibody. Immunoreactive signals were visualized with a chemiluminescense detection system using ECL kits (Amersham, Buckinghamshire, UK).
Primary antibodies used were as follows: anti-fukutin antisera (anti-N1; rabbit, polyclonal, kindly provided by Dr. Saito; Saito et al. 2000), anti-Musashi-1 (Clone: 282613, 1:500; R&D Systems, Minneapolis, MN, USA), vimentin (1:500; DAKO) and anti-β-actin (I-19:sc-1616, 1:300; Santa Cruz, Santa Cruz, CA, USA) antibodies. Blots processed with the omission of the primary antibodies served as negative reaction controls.
Examination of laminin-binding ability
Laminin-binding ability of astrocytoma cells was examined as an indirect method to evaluate the glycosylation of α-DG (Yamamoto et al. 2008) because quantitative analyses using western blotting and immunohistochemistry were difficult owing to the small amount of glycosylated α-DG in the cells used.
After trypsinization, cells were inoculated on 35-mm laminin-coated dishes (BD Bioscience, Bedford, MA, USA) with DMEM (Invitrogen), supplemented with 10% foetal bovine serum (Thermo Fisher Scientific), 2 mM glutamine (Invitrogen) and 1% penicillin-streptomycin (Invitrogen) and cultured at 37 °C. Cells were allowed to attach gradually to the surface of the dish and showed elongated appearances with time, so that elongated cells were regarded as attached cells, and rounded cells as non-attached cells. The ratio between attached and non-attached cells was calculated for more than 300 cells. The examination was performed from two to six h after the inoculation. For cells on which RNAi was performed, cells harvested one day after the transfection of siRNA were used. Cells treated with lipofectamineMAX were used as negative controls.
RNA immunoprecipitation
Twenty-five microlitres of the Pierce protein G agarose (Thermo Fisher Scientific) was incubated with 5 μg of anti-Musashi-1 (Clone: 282613; R&D Systems), anti-Musashi-1 (H-45: sc-98845; Santa Cruz) or anti-vimentin (DAKO) antibody, or rabbit immunoglobulin fraction (negative control, Code No: ×0903; DAKO) in 250 μl of washing buffer (50 mM Tris–Cl, pH7.5, 150 mM NaCl. 0.05% NP-40), at 4 °C, for at least 4 h with constant rotation. Cultured astrocytoma cells (110 mg for Musashi-1 and 30–40 mg for vimentin) were suspended in 1 ml of extraction buffer (10 mM PIPES, pH6.8, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, 0.2 mg/ml PMSF) and incubated on ice for 5 min. After centrifugation at 1000 g for 5 min, the supernatant was diluted with an equal volume of washing buffer with NaCl and bovine serum albumin added to final concentrations of 150 mM and 3% respectively. The solution was precleared with 25 μl of protein G agarose at 4 °C for 1 h. After the preclearing, salmon sperm DNA was added to the supernatant (final concentration: 100 μg/ml), and the solution was immunoprecipitated with antibody-conjugated protein G agarose at 4 °C with rotation (overnight for Musashi-1 and 3 h for vimentin). After the immunoprecipitation, the protein G agarose was washed 10 times with washing buffer and eluted with 50 μl of elution buffer (50 mM NaHCO3, 1% SDS). RNA was extracted from the eluted solution using RNeasy Kit (Qiagen, Valencia, CA, USA) following the instructions and reverse-transcribed using PrimeScript RT-PCR kit (Takara). PCR was performed using the primers of fukutin and POMGnT1. cDNA obtained by Musashi-1 immunoprecipitation was amplified twice by the manner described in the RT-PCR section. For fukutin, the primers for 112–731 bp were used in the first amplification, and those for 112–578 bp [5′-ATGAGTAGAATCAATAAGAA-3′ (coding sense) and 5′-GTTCCAGAGAGTGAGTCTATCC -3′ (anticoding sense)] were used in the second amplification. Primers for POMGnT1 (GenBank: AB057356) were 5′-ATGGACGACTGGAAGCCCAG-3′ (coding sense) and 5′-GTGCAGATGAGCACTCGGCC-3′ (anticoding sense), covering 95–621 bp. cDNA obtained by vimentin-immunoprecipitation was amplified using the primers for 112–731 bp of fukutin: amplification profiles of PCR consisted of denaturing at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 68 °C for 1 min. Some of the immunoprecipitated materials were examined by western blotting using anti-Musashi-1 and anti-vimentin antibodies.
ELISA-based RNA-binding assay
Microtitre plates were incubated with antibodies (1:100 for anti-Musashi-1 and 1:500 for anti-vimentin antibodies) at 37 °C for 1 h. After washing with TBS, the plates were incubated with cell extract (20 mg/ml) using RIPA buffer at 37 °C for 1 h. For the extract, astrocytoma cells transfected with fukutin were used. The plates were washed with TBS, incubated with RNA at 4 °C for 3 h, washed with TBS and then incubated with digoxigenin-labelled probe (100 ng/ml) at 42 °C overnight. The RNA was extracted from astrocytoma cells and the probe was made by RT-PCR of fukutin mRNA covering 112–731 bp. After washing with TBS, plates were treated with TBS with 3% skim milk at 37 °C for 1 h and then with alkaline phosphatase-conjugated anti-digoxigenin antibody (1:1000; Roche) at RT for 1 h. The colour was developed using an alkaline phosphatase substrate kit (Bio-Rad, Richmond, CA, USA). The titre was measured using a Benchmark microplate reader (Bio-Rad).
In situ hybridization
In situ hybridization was performed according to the previously described manner (Yamamoto et al. 2002) on astrocytoma cells, cultured on culture slides (BD Falcon, Bedford, MA, USA) and fixed in 95% ethanol for 5 min at RT. Basic protein was removed by incubation in 0.2 N HCl at RT for 15 min. After rinsing in 2× SSC (0.3 M NaCl and 0.03 M sodium citrate, pH 7.0), the slides were treated with 10 μg/ml proteinase K (Amresco, Solon, OH, USA) in 2 mM CaCl2, 20 mM Tris–HCl, pH 7.4, at 37 °C for 20 min, rinsed in phosphate-buffered saline (PBS) and refixed in 4% paraformaldehyde-PBS for 5 min. After rinsing in PBS, the slides were acetylated in freshly made 0.1 M triethanolamine solution, pH 8.0, containing 0.25% acetic anhydride for 10 min, washed three times in 2× SSC for 5 min, dehydrated and air-dried. Hybridization was carried out in a moist chamber at 42 °C for 15 h. The hybridization mixture contained 2 μg/ml digoxigenin-labelled cDNA probe, 50% formamide, 5× SSC, 5× Denhardt's solution (0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), 0.1% SDS, 100 μg/ml sonicated salmon sperm DNA and 10% dextran sulphate. The probe was made by RT-PCR of fukutin mRNA covering 552–1497 bp (Yamamoto et al. 2002). The slides were washed twice in 2× SSC, RT for 30 min, washed twice with 50% formamide/1× SSC at 45 °C for 10 min and incubated with an alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) at 4 °C overnight. The colour was developed using NBT/BCIP according to the manufacturer's instructions (Roche), and the sections were counterstained with methyl green. Hybridization solution without addition of the probe was used as a negative control.
Results
Expression of fukutin and α-DG in astrocytoma cells
The RT-PCR demonstrated the expression of fukutin and DG mRNA on original astrocytoma cells (Figure 1a). The cytoplasm and cell membrane were positive for α-DG-p, while the cell membrane was faintly positive for IIH6C4. A negative control did not show a positive reaction (Figure 1b).
Figure 1.
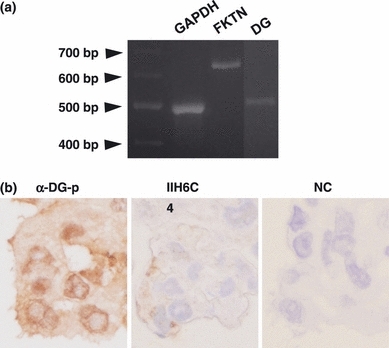
(a) RT-PCR on astrocytoma cells. Both fukutin and DG mRNA were expressed. (b) Immunohistochemistry on a cell block of astrocytoma cells. The cytoplasm and cell membrane are positive for α-DG-p, while the cell membrane is faintly positive for IIH6C4. A negative control does not show a positive reaction. FKTN, fukutin; DG, dystroglycan; NC, negative control.
Alteration of laminin binding in fukutin-over-expressed and -suppressed astrocytoma cells
Astrocytoma cells transfected with fukutin changed their shapes to a more epithelioid appearance (Figure 2a). The transfection of fukutin was confirmed by PCR using the primers of fukutin and vector, and by western blotting using anti-fukutin antibody (Figure 2b). On examination of laminin-binding ability, the ratio between attached and non-attached cells was significantly increased for fukutin-over-expressed cells (Figure 2c).
Figure 2.
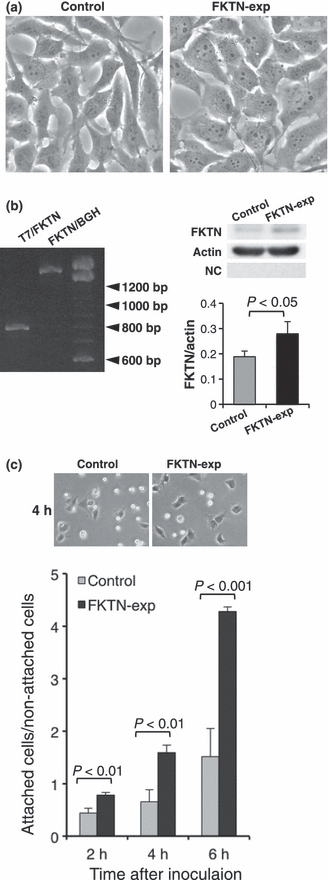
Examination on fukutin-over-expressed astrocytoma cells. (a) Morphology of astrocytoma cells after transfection of fukutin. Cells became epithelioid after transfection of fukutin. (b) Confirmation of transfection by PCR and western blotting. PCR using primers of fukutin and vector shows a band, and western blotting of fukutin revealed a stronger reaction as a band at about 60 kDa. There are no bands in a negative control. (c) On examination of laminin-binding ability, fukutin-over-expressed cells attached more to the laminin-coated dish than in the control. FKTN-exp, fukutin-over-expressed cells; T7 and BGH, elements of vector; NC, negative control.
After knockdown of fukutin, astrocytoma cells became more fusiform with marked elongation of cytoplasmic processes (Figure 3a). RT-PCR product of fukutin was significantly reduced after the knockdown (Figure 3b). The knockdown was also confirmed by real-time PCR (data not shown). The ratio between attached and non-attached cells was significantly decreased in fukutin-suppressed cells (Figure 3c).
Figure 3.
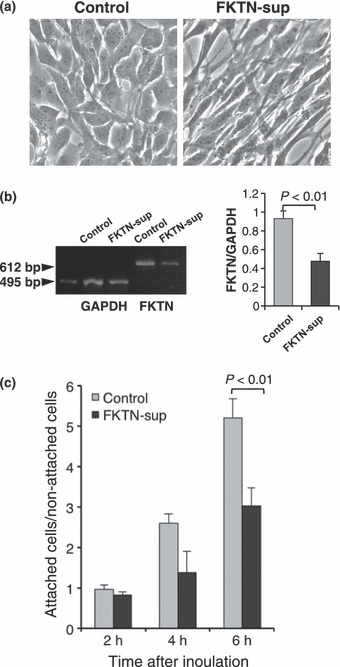
Examination on fukutin-suppressed astrocytoma cells. (a) Morphology of astrocytoma cells after suppression of fukutin by RNAi. Cells become fusiform with elongation of cytoplasmic processes after knockdown of fukutin. (b) Confirmation of knockdown by RT-PCR. Expression of fukutin is reduced after knockdown. (c) On examination of laminin-binding ability, fukutin-suppressed cells attached less to the laminin-coated dish than in the control. FKTN-sup, fukutin-suppressed cells.
RNAi for Musashi-1
Astrocytoma cells tended to exhibit elongated cytoplasmic processes upon knockdown of Musashi-1, although the shape of cells did not change as much as seen in fukutin-suppressed cells (Figure 4a). Western blotting revealed a significant decrease of Musashi-1 expression by the knockdown (Figure 4b). RT-PCR on Musashi-1-suppressed cells demonstrated a significant decrease of fukutin mRNA (Figure 4c).
Figure 4.
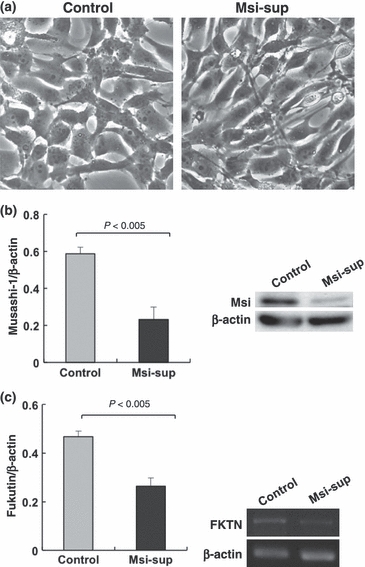
RNAi for Musashi-1 in astrocytoma cells. (a) Morphology of astrocytoma cells. Cells exhibit slightly elongated appearance after knockdown of Musashi-1. (b) Confirmation of knockdown by western blotting. Expression of Musashi-1 is significantly reduced after knockdown. (c) RT-PCR of fukutin in astrocytoma cells. After knockdown of Musashi-1, fukutin expression is significantly decreased. Msi-sup, Musashi-1-suppressed cells.
Relationship between fukutin mRNA and Musashi-1 protein
In the RNA immunoprecipitation, fukutin cDNA was amplified in the material immunoprecipitated using two types of anti-Musashi-1 antibodies. POMGnT1 was not amplified (Figure 5a). Although a weak band was seen in the negative control specimen using rabbit immunoglobulin for immunoprecipitation, a band from specimens using anti-Musashi-1 antibodies appeared to be stronger. In the western blotting, a band at about 39 kDa was observed in the material immunoprecipitated using anti-Musashi-1 antibody, and not in the negative control material using rabbit immunoglobulin. However, a 39 kDa band was also seen in the reaction without primary antibody (Figure 5a). This is probably because the molecular weight of Musashi-1 protein overlapped with that of the antibodies for immunoprecipitation. Moreover, the amount of Musashi-1 protein might have been too small to be detected because two sessions of PCR were needed to reveal a band of fukutin cDNA.
Figure 5.
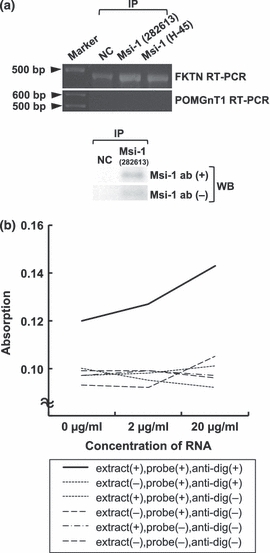
RNA immunoprecipitation and ELISA-based RNA-binding assay for Musashi-1. (a) RT-PCR in materials after RNA immunoprecipitation using two types of anti-Musashi-1 antibodies. In RT-PCR for fukutin, stronger reaction is seen in materials using anti-Musashi-1 antibodies than in the negative control. No bands are seen in RT-PCR for POMGnT1. In western blotting, a band at about 39 kDa is seen in a material immunoprecipitated with Musashi-1, but a weak band is also observed in a reaction without primary antibody. (b) ELISA-based RNA-binding assay showed an increase of absorption only in a solution containing RNA, extract, probe and anti-digoxigenin antibody. Msi-1, Musashi-1; FKTN, fukutin; POMGnT1, protein O-linked mannose β1,2-N-acetylglucosaminyltransferase; NC, negative control; IP, immunoprecipitation; WB, western blotting.
The ELISA-based RNA-binding assay showed an increase of positive reaction only in material treated with anti-Musashi-1 antibody, cellular extract, RNA and anti-digoxigenin antibody (Figure 5b).
Relationship between fukutin mRNA and vimentin protein
In the RNA immunoprecipitation, fukutin cDNA was amplified in the material after the immunoprecipitation using anti-vimentin antibody (Figure 6a). POMGnT1 was not amplified. No apparent amplification of fukutin cDNA was observed in the negative control specimen using rabbit immunoglobulin for immunoprecipitation. The material immunoprecipitated with anti-vimentin antibody exhibited a band at about 57 kDa by the western blotting using anti-vimentin antibody (Figure 6a). A band was not seen in the material for the negative control, or in the reaction without primary antibody.
Figure 6.
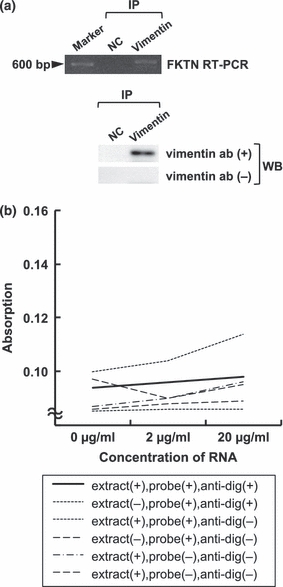
RNA immunoprecipitation and ELISA-based RNA-binding assay for vimentin. (a) RNA immunoprecipitation using anti-vimentin antibody. In RT-PCR for fukutin, a band is observed only in a material using anti-vimentin antibody. Western blotting of vimentin reveals a band at about 57 kDa only in a material using anti-vimentin antibody. (b) ELISA-based RNA-binding assay shows no apparent increase of absorption. FKTN, fukutin; NC, negative control; IP, immunoprecipitation; WB, western blotting.
The ELISA-based RNA-binding assay did not show an increase of positive reaction even in material treated with anti-Musashi-1 antibody, cellular extract, RNA and anti-digoxigenin antibody (Figure 6b).
In in situ hybridization, positive reaction was predominantly observed in the perikarya, but the cytoplasm including cytoplasmic processes was also stained (Figure 7a). Negative control did not show a positive reaction (Figure 7b).
Figure 7.
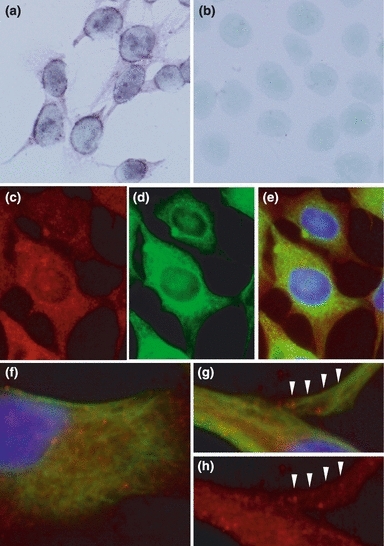
In situ hybridization of fukutin and double-immunohistochemical staining of fukutin and vimentin in astrocytoma cells. In in situ hybridization, positive reaction is predominantly observed in the perikarya, but the cytoplasm including cytoplasmic processes is also stained (a). A negative control does not show positive reaction (b). In immunohistochemistry, fukutin (c) and vimentin (d) are widely distributed in the cytoplasm: (e) is a merged photograph of (c) and (d). Positive reaction of fukutin (red colour) is granular and that of vimentin (green colour) is filamentous (f–h). These two positive deposits appear to be partially co-localized or closely associated even in a peripheral part of cytoplasmic process (arrowheads).
In immunohistochemistry, both fukutin and vimentin were positive in the cytoplasm (Figure 7c–e). Immunoreaction for vimentin was filamentous and that for fukutin was granular. These two immunodeposits looked partially co-localized, even in peripheral areas of cytoplasmic processes (Figure 7f–h).
Discussion
Previous studies have demonstrated that fukutin is associated with the glycosylation of α-DG. The CNS lesions of FCMD mainly result from disruptions of the glia limitans during the foetal period, which is evoked by a fragile basement membrane because of the hypoglycosylation of α-DG. With regard to the pathogenesis of CNS lesions, it is best to study normal human astrocytes, especially those from a foetus. However, we chose an astrocytoma cell line for this study, because astrocytoma cells are considered to retain the basic characteristics of astrocytes and are easily used for various experiments: the cells express fukutin and α-DG containing the laminin-binding site of the glycosylated epitopes. On examination of laminin-binding ability, fukutin-over-expressed astrocytoma cells showed an increase of laminin-binding ability and fukutin-suppressed cells showed a decrease. In a previous study, we showed that the antibody against the laminin-binding site of the glycosylated α-DG (IIH6C4) (Ervasti & Campbell 1993) inhibited adherence of cultured rat astrocytes to a laminin-coated dish (Yamamoto et al. 2006). Fukutin is considered to be related to the glycosylation of α-DG in astrocytoma cells as well, and the cells can be applicable as a substitute of normal astrocytes.
Fukutin mRNA consists of 7349 bp with an open reading frame of 1383 bp beginning at base 112. On founder mutation of FCMD, the retrotransposal 3-kb sequence is inserted into the 3′-UTR of fukutin mRNA (Kobayashi et al. 1998). Among RNA regulation mechanisms, there is an interaction between cis-acting elements in the 3′-UTR and sequence-specific RNA-binding proteins (Bolognani & Perrone-Bizzozero 2008). An interaction between AU-rich elements (ARE) in the 3′-UTR and ARE-binding proteins such as Hu is a good example (Bolognani & Perrone-Bizzozero 2008). RNA-binding proteins related to the CNS include fragile X mental retardation protein (FMRP), cytoplasmic polyadenylation element binding factor and neuro-oncological ventral antigen (Nova) (Ule & Darnell 2006). We chose Musashi-1 for this study because it is one of the markers of stem cells (Okano et al. 2005), and fukutin tends to be expressed more in the foetal brain (Saito et al. 2000; Yamamoto et al. 2002; Hiroi et al. 2011).
Musashi-1 is involved in the regulation of neuronal differentiation and is expressed in an astrocyte lineage as well (Sakakibara & Okano 1997). It is related to the notch signal via n-numb mRNA, keeping neuronal cells immature by suppressing n-numb mRNA (Sakakibara & Okano 1997; Imai et al. 2001; Okano et al. 2005). Because 3′-UTR of fukutin mRNA contains Musashi-1-binding sequences, (G/A)UnAGU (Imai et al. 2001; Okano et al. 2005) and GU3–6(G/AG) (Okabe et al. 2001), binding of Musashi-1 protein to fukutin mRNA may occur.
When Musashi-1 was knocked down by RNAi, fukutin mRNA was reduced in astrocytoma cells, suggesting that Musashi-1 up-regulates fukutin mRNA. In RNA immunoprecipitation, materials immunoprecipitated by two types of anti-Musashi-1 antibodies showed amplification of fukutin cDNA, but POMGnT1 was not amplified. Together with the result of RNAi and ELISA-based RNA-binding assay, it seems appropriate to suggest that Musashi-1 protein binds to fukutin mRNA to regulate fukutin mRNA.
mRNAs are generally translated to proteins in the cell body, but some mRNAs, such as those relating to the maintenance of cell polarity, asymmetrical segregation and synaptic plasticity, have a distinct characteristic (López de Heredia & Jansen 2004; Ule & Darnell 2006). Such mRNAs, called localized mRNAs, are generally characterized by having signals recognized by RNA-binding proteins in the 3′-UTR region, and by being transported to peripheral areas of a cell such as dendrites (López de Heredia & Jansen 2004): a complex consisting of mRNA and several proteins moves along the microtubules using a molecular motor such as kinesin and dynein (López de Heredia & Jansen 2004; Ule & Darnell 2006). Musashi-1 is one of the RNA-binding proteins and plays an essential role in regulating asymmetric cell division (Okabe et al. 2001; Okano et al. 2005). Because this experiment showed binding of fukutin mRNA and Musashi-1 protein, as well as peripheral localization of fukutin mRNA, fukutin mRNA is suggested to be one of the localized mRNAs.
Astrocytes have many important roles in the CNS, being involved in interactions with neurons and in maintaining the blood–brain barrier (BBB). A system of prompt responses against quick changes of environments, like synaptic plasticity, should be necessary for astrocytes as well. The basement membrane is formed at the glia limitans and the BBB in the CNS, with expression of glycosylated α-DG. The glycosylated α-DG is a receptor for several extracellular matrix proteins (Michele & Campbell 2003; Martin 2005; Schessl et al. 2006) and for some microorganisms (Cao et al. 1998; Rambukkana et al. 1998; Tayeh et al. 2010) and is localized at astrocytic endfeet. The glycosylation of α-DG could be a phenomenon to adapt to varying circumstances at the most peripheral part of a cell, which is reasonable as fukutin mRNA is a localized mRNA.
During experiments with RNA immunoprecipitation, we incidentally found a possible relationship between fukutin mRNA and vimentin protein. Because ELISA-based RNA-binding assay showed no positive result, they seem to bind indirectly. Vimentin is supposed to be a component of a fukutin mRNA-protein complex. Interestingly, intermediate filaments are dynamic and motile elements in various cells (Helfand et al. 2004, 2005). These filaments rapidly move along the microtubules, coupling with a molecular motor, such as kinesin or dynein (Helfand et al. 2004; Chang et al. 2006), like localized mRNAs described above. Vimentin is one of the representative intermediate filaments and classified as a type III intermediate filament (Helfand et al. 2004; Chang et al. 2006). It makes a complex with several proteins such as Ndel1, Lis-1 and aCOP and moves along the microtubules (Shim et al. 2008). Fukutin mRNA might form a complex including Musashi-1 and vimentin proteins. On the other hand, fukutin mRNA might be regulated by several RNA-binding proteins, because the long 3′-UTR of fukutin mRNA contains at least Musashi-1 binding sequences and ARE. There is a possibility that vimentin is a component of other fukutin mRNA-protein complex. Further investigation is necessary to clarify this point.
Arc, the immediate early gene product related to synaptic plasticity, is transported along the microtubules forming a complex called messenger ribonucleoprotein particle (mRNP) to a site of synaptic activity and undergoes local translation (Bramham et al. 2010). The mRNP includes Arc mRNA and several proteins such as FMRP and Purα. Kinesin is a motor of this complex. The transcription of Arc is regulated by cyclic AMP response element binding protein (CREB) (Bramham et al. 2010). A CRE-like sequence has been found in the fukutin gene promoter, and the transcription of fukutin may be regulated by CREB (Fang et al. 2005). A regulation system of fukutin appears to share common characteristics with that of Ark.
Acknowledgments
The authors wish to thank Dr Stephan Kröger, Physiologiches Institut, Ludwig-Maximilians-Universität, for kindly providing the anti-α-DG antibody, and to Dr Yoshiaki Saito, Department of Child Neurology, National Center Hospital, National Center of Neurology and Psychiatry, for the anti-fukutin antisera. The authors are also grateful to Mr. Mizuho Karita, Mr. Hideyuki Takeiri, Mr. Fumiaki Muramatsu, Mrs. Noriko Sakayori and Mr. Shuichi Iwasaki for their excellent technical assistance and help in the preparation of the paper.
References
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Alme MN, Bittins M, et al. The Arc of synaptic memory. Exp. Brain Res. 2010;200:125–140. doi: 10.1007/s00221-009-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Henry MD, Borrow P, et al. Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- Chang L, Shav-Tal Y, Trcek T, Singer RH, Goldman RD. Assembling an intermediate filament network by dynamic cotranslation. J. Cell Biol. 2006;172:747–758. doi: 10.1083/jcb.200511033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linkder between laminin and actin. J. Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Sodja C, Chartier J, et al. Identification of a functional CRE in the promoter of Fukuyama congenital muscular dystrophy gene fukutin. Mol. Brain Res. 2005;136:1–11. doi: 10.1016/j.molbrainres.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JS, Perkins JP. Glucocorticoids increase the responsiveness of cells in culture to prostaglandin E1. Proc. Natl Acad. Sci. USA. 1977;74:4816–4820. doi: 10.1073/pnas.74.11.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama Y, Kawazura M, Haruna H. A peculiar form of congenital progressive muscular dystrophy. Paediatr. Univ. Tokyo. 1960;4:5–8. [Google Scholar]
- Hayashi YK, Ogawa M, Tagawa K, et al. Selective deficiency of α-dystroglycan in Fukuyama-type congenital muscular dystrophy. Neurology. 2001;57:115–121. doi: 10.1212/wnl.57.1.115. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Chang L, Goldman RD. Intermediate filaments are dynamic and motile elements of cellular architecture. J. Cell Sci. 2004;117:133–141. doi: 10.1242/jcs.00936. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Chou Y-H, Shumaker DK, Goldman RD. Intermediate filament proteins participate in signal transduction. Trends Cell Biol. 2005;15:568–570. doi: 10.1016/j.tcb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Herrmann R, Straub V, Blank M, et al. Dissociation of the dystroglycan complex in caveolin-3-deficient limb girdle muscular dystrophy. Hum. Mol. Genet. 2000;9:2335–2340. doi: 10.1093/oxfordjournals.hmg.a018926. [DOI] [PubMed] [Google Scholar]
- Hiroi A, Yamamoto T, Shibata N, Osawa M, Kobayashi M. Roles of fukutin, the gene responsible for Fukuyama type congenital muscular dystrophy, in neurons: possible involvement in synaptic function and neuronal migration. Acta Histochem. Cytochem. 2011;44:91–101. doi: 10.1267/ahc.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, et al. The neural RNA-binding protein Musashi-1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Heredia M, Jansen R-P. mRNA localization and the cytoskeleton. Curr. Opin. Cell Biol. 2004;16:80–85. doi: 10.1016/j.ceb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Macintyre EH, Pontén J, Vatter AE. The ultrastructure of human and murine astrocytes and of human fibroblasts in culture. Acta Pathol. Microbiol. Scand. 1972;80:267–283. doi: 10.1111/j.1699-0463.1972.tb02176.x. [DOI] [PubMed] [Google Scholar]
- Martin PT. The dystroglycanopathies: the new disorders of O-linked glycosylation. Semin Pediatr. Neurol. 2005;12:152–158. doi: 10.1016/j.spen.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michele DE, Campbell KP. Dystrophin-glycoprotein complex: post-transcriptional processing and dystroglycan function. J. Biol. Chem. 2003;278:15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- Okano H, Kawahara H, Toriya M, Nakano K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Osawa M, Sumida S, Suzuki N. Fukuyama type congenital muscular dystrophy. In: Fukuyama Y, Osawa M, Saito K, et al., editors. Congenital Muscular Dystrophies. Amsterdam: Elsevier; 1997. pp. 31–68. [Google Scholar]
- Rambukkana A, Yamada H, Zanazzi G, et al. Role of α-Dystroglycan as a Schwann Cell Receptor for Mycobacterium leprae. Science. 1998;282:2076–2079. doi: 10.1126/science.282.5396.2076. [DOI] [PubMed] [Google Scholar]
- Saito Y, Mizuguchi M, Oka A, Takashima S. Fukutin protein is expressed in neurons of the normal developing human brain but is reduced in Fukuyama-type congenital muscular dystrophy brain. Ann. Neurol. 2000;47:756–764. [PubMed] [Google Scholar]
- Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J. Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schessl J, Zou Y, Bönnemann CG. Congenital muscular dystrophies and the extracellular matrix. Semin. Pediatr. Neurol. 2006;13:80–89. doi: 10.1016/j.spen.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Shim SY, Samuels BA, Wang J, et al. Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J. Biol. Chem. 2008;283:12232–12240. doi: 10.1074/jbc.M710200200. [DOI] [PubMed] [Google Scholar]
- Tayeh A, Tatard C, Kako-Ouraga S, Duplantier JM, Dobigny G. Rodent host cell/Lassa virus interactions: evolution and expression of alpha-dystroglycan, LARGE-1 and LARGE-2 genes, with special emphasis on the Mastomys genus. Infect. Genet. Evol. 2010;10:1262–1270. doi: 10.1016/j.meegid.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim. Biophys. Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Xiong H, Kobayashi K, Tachikawa M, et al. Molecular interaction between fukutin and POMGnT1 in the glycosylation pathway of α-dystroglycan. Biochem. Biophys. Res. Commun. 2006;350:935–941. doi: 10.1016/j.bbrc.2006.09.129. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kato Y, Karita M, et al. Fukutin expression in glial cells and neurons: implication in the brain lesions of Fukuyama congenital muscular dystrophy. Acta Neuropathol. 2002;104:217–224. doi: 10.1007/s00401-002-0542-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kato Y, Kawaguchi M, Shibata N, Kobayashi M. Expression and localization of fukutin, POMGnT1 and POMT1 in the central nervous system: consideration for functions of fukutin. Med. Electron Microsc. 2004;37:200–207. doi: 10.1007/s00795-004-0260-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kawaguchi M, Sakayori N, et al. Intracellular binding of fukutin and α-dystroglycan: relation to glycosylation of α-dystroglycan. Neurosci. Res. 2006;56:391–399. doi: 10.1016/j.neures.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kato Y, Shibata N, Sawada T, Osawa M, Kobayashi M. A role of fukutin, a gene responsible for Fukuyama type congenital muscular dystrophy, in cancer cells: a possible role to suppress cell proliferation. Int. J. Exp. Pathol. 2008;89:332–341. doi: 10.1111/j.1365-2613.2008.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Shibata N, Saito Y, Osawa M, Kobayashi M. Functions of fukutin, a gene responsible for Fukuyama type congenital muscular dystrophy, in neuromuscular and other somatic organs. Cent. Nerv. Syst. Agents Med. Chem. 2010;10:169–179. doi: 10.2174/187152410791196369. [DOI] [PubMed] [Google Scholar]


