Abstract
Background
Chemokines and their receptors play a critical role in leukocyte trafficking, and inhibition of select chemokines has been shown to attenuate kidney disease and allograft rejection in animal models. We therefore evaluated chemokine and chemokine receptor transcripts in human renal allograft biopsies, correlating transcript levels with clinical course and immunohistochemical analysis to relate chemokine expression to relevant clinical human disease phenotypes.
Methods
Renal biopsies were grouped as post-reperfusion (n=10), stable function (n=10), subclinical (n=10) or acute rejection (n=17), or calcineurin inhibitor nephrotoxicity (n=9) based on clinical presentation and histopathological assessment. Using quantitative real-time polymerase chain reaction analysis, chemokine transcripts were assessed relative to transcript levels in pre-procurement biopsies from live donor kidneys (n=15).
Results
Transcripts from several inflammatory chemokines (CCL3, CCL5, CXCL9, CXCL10 and CXCL11) and chemokine receptors (CCR5, CCR7 and CXCR3) were significantly elevated in allografts with subclinical and clinical acute rejection, indicating a strong polarization toward a TH1 effector phenotype during rejection. These transcripts also distinguished acutely rejecting allografts from allografts with non-rejection causes of renal dysfunction. Biopsies from patients with stable function without histological evidence of rejection had increased chemokine transcript levels that were qualitatively similar but quantitatively reduced compared to rejecting allografts.
Conclusions
This comprehensive evaluation of chemokines and their receptors in human renal transplantation defines associations between chemokine expression and clinical phenotypes, may have diagnostic utility, and highlights relevant pathways for therapeutic intervention.
Keywords: Chemokines, Renal Transplantation, RT-PCR
INTRODUCTION
Chemokines (chemotactic cytokines) and chemokine receptors are well-known regulators of mononuclear cell trafficking. Increased expression of multiple chemokines and their cognate receptors has been demonstrated in rejecting allografts (1–6). Additionally, donor organ ischemia-reperfusion injury (IRI) has been shown to induce chemokine expression, contributing to immune cell recruitment and subsequent acute cellular rejection (ACR) (7–11).
Several laboratories, including our own, have used real time polymerase chain reaction (RT-PCR) to clearly relate quantitative changes in transcript presence to clinical disease phenotypes and qualitative progression of clinical disease states (12–18). Previous studies have reported the upregulation of several chemokine and chemokine receptor transcripts including CCL2, CCL3, CCL5, CXCL10, CCR1, CCR2, CCR5 and CXCR3 in renal allografts during acute rejection (19–22). However, given the promiscuous binding of chemokine receptor-ligand pairs and an apparent imbalance between the number of ligands and receptors (23), and the likely role of chemokines in other allograft phenotypes, a comprehensive analysis of chemokines and their receptors would be useful to relate chemokines and their receptors to many clinical phenotypes within a single comprehensive platform.
To better define the relationships between chemokine and chemokine receptor expression, immune cell recruitment, and clinical allograft rejection, we evaluated human renal allograft biopsy-derived RNA using a comprehensive RT-PCR based panel of chemokines and chemokine receptors. Individual and pooled patient biopsies were grouped according to clearly defined clinical diagnoses based on clinical criteria and histopathological assessment using the Banff criteria (24). Herein we report the resultant comprehensive molecular profile of chemokines and chemokine receptors in human renal transplantation.
RESULTS
Leukocyte Infiltrates in the Studied Allografts
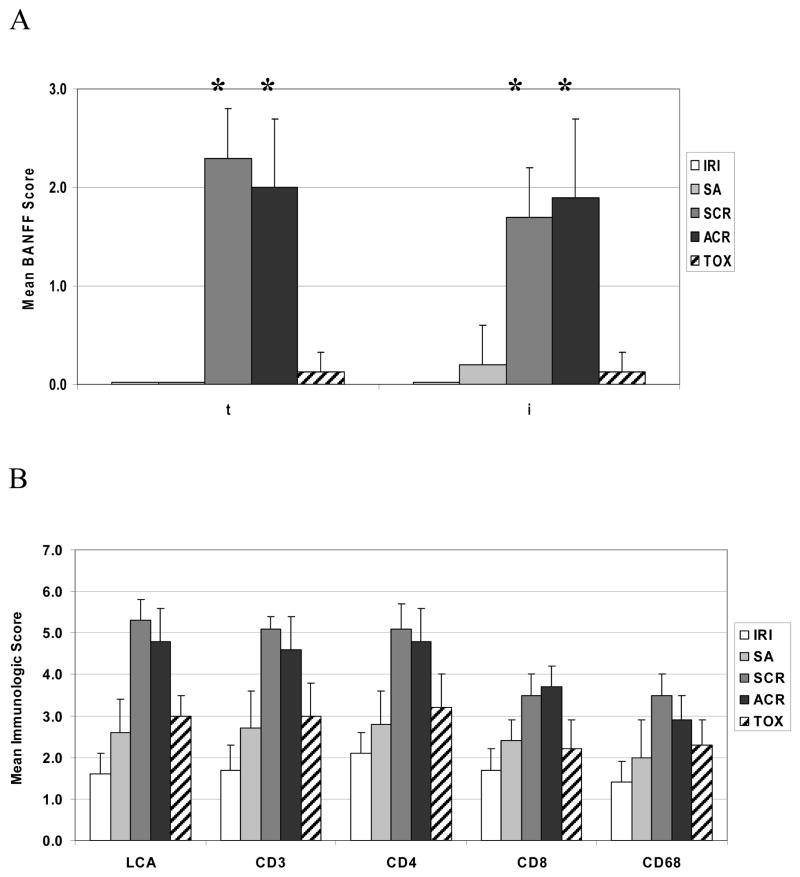
As expected, different clinical diagnoses showed characteristic and varied patterns of cellular infiltration. Normal kidney (NK) biopsies did not show ischemic injury or inflammatory cell infiltration, but did contain occasional intravascular leukocytes that were seen in all biopsies (data not shown). Mean Banff scores for tubulitis (t) and inflammation (i) for each group are shown in Fig. 1A. Interstitial or intratubular leukocytes were rare and consisted mainly of T cells. Ischemia-reperfusion injury (IRI) biopsies demonstrated mild to moderate degrees of proximal tubular necrosis and vacuolation with mild interstitial edema. There was no significant difference in score of LCA+, CD3+, CD4+, CD8+ or CD68+ cells in IRI biopsies compared to normal kidney although numerical differences existed demonstrating more than occasional inflammatory cells during IRI (Fig. 1B).
FIGURE 1.
Banff subscores and immunophenotyping of inflammatory cell infiltrates in renal allograft biopsies. (A) Post-reperfusion (IRI, open bars), stable allograft (SA, light grey bars), subclinical rejection (SCR, dark grey bars), acute cellular rejection (ACR, black bars) and toxicity (TOX, slashed bars) biopsy results are shown. The severity of Banff subscores for tubulitis (t) and inflammation (i) were significantly elevated (p<0.001) for both SCR and ACR compared to stable allografts. (B) A semi-quantitative scoring system was used to evaluate all renal allograft biopsies. Mean scores for pan T cell markers (CD3 and LCA) and specific T cell subsets (CD4 and CD8) are shown. An asterisk (*) denotes p<0.05.
Biopsies from stable allografts (SA) demonstrated modest focal interstitial infiltrates of CD3+ T cells and scattered macrophages without tubulitis, vasculitis or glomerulitis. Consistent with our previous studies(7), SA biopsies showed a greater number of LCA+, CD3+, CD4+, CD8+ and CD68+ cells (p=0.009, 0.02, <0.001, 0.03 and 0.04 respectively, Fig. 1B) compared to NK. Thus, a mild inflammatory infiltrate histologically distinguished stable allografts from normal kidney.
Subclinical rejection (SCR) and acute cellular rejection (ACR) biopsies showed a marked cellular infiltrate compared to NK or SA, with prominent tubulitis and interstitial inflammation consisting of macrophages and T cells (not shown). Despite distinct clinical presentations, neither Banff nor immunohistochemical scores differed significantly between SCR and ACR groups for those parameters tested in this study (Fig. 1A, 1B). Predictably, LCA+, CD3+, CD4+ and CD8+ T cells were significantly increased in both SCR and ACR samples compared to TOX biopsies (p=0.002, 0.004, 0.01 and 0.006, respectively, Fig. 1B). TOX biopsies displayed tubular dilation and vacuolation, cell sloughing, mild hyalinization, and had increased cellular infiltration compared to NK but were not significantly different histologically from SA despite clinical dysfunction.
Chemokines and Chemokine Receptor Transcript Profiles Distinguish Recently Reperfused and Stable Non-rejecting Allografts from Normal Kidney
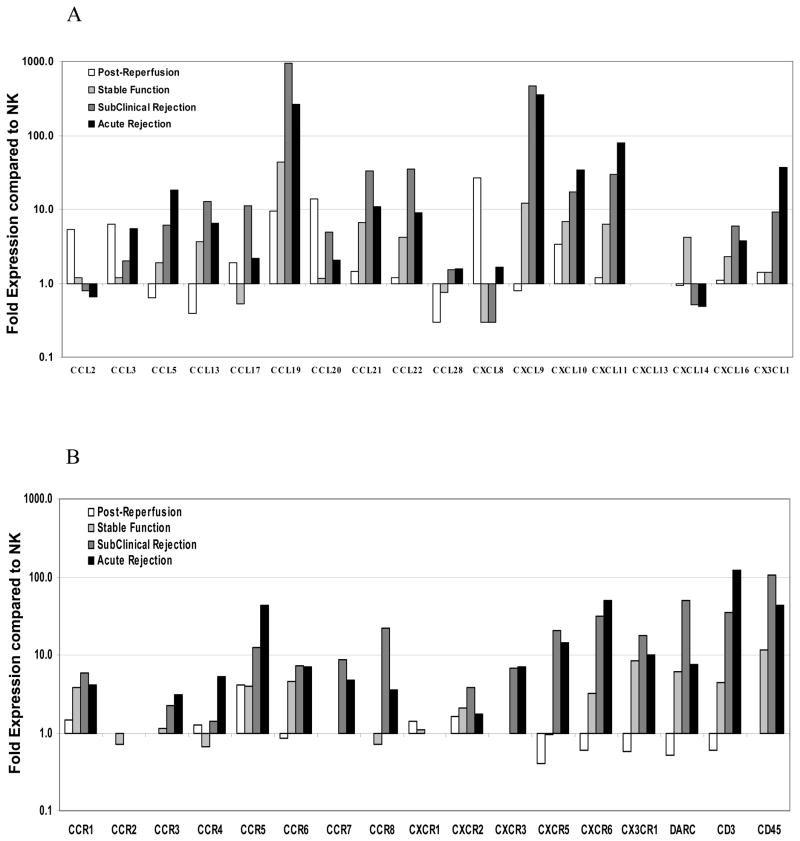
To determine whether quantitative differences in key chemokine and chemokine receptor transcripts could better discriminate clinical status post-transplantation, we pooled cDNA from patient allograft biopsies for RT-PCR analysis and compared them to normal kidneys. A comprehensive panel of chemokine and chemokine receptor targets was chosen to elucidate their relevance to monocyte and T cell recruitment and renal allograft biology (Table 1). IRI biopsies showed rapid expression of chemokine transcripts associated with early inflammation and activation of the innate immune system. CXCL8 was increased nearly 30-fold compared to normal kidneys (Fig 2A). Multiple chemokines associated with monocyte, immature dendritic cell, and natural killer cell homing (CCL2, CCL3, CCR5 receptor, CCL19 and CLL20) were also upregulated (Fig. 2A, 2B) consistent with the low level presence of monocytes within these biopsy specimens as assessed by CD68 immunohistochemistry. With the paucity of cells observed at this early time point, no associations between chemokine receptors and infiltrating cells were evident in IRI biopsies suggesting a parenchymal source of transcripts, or substantial transcription from the few cells present.
Table 1.
RT-PCR Targets: Chemokines, Chemokine Receptors, Cell Surface Molecules
| CHEMOKINES | COMMON NAME | CHEMOKINE RECEPTORS | LIGANDS |
|---|---|---|---|
| CCL2 | MCP-1 | CCR1 | CCL3, CCL5, CCL7, CCL8, CCL13, CCL14, CCL15, CCL16, CCL23 |
| CCL3 | MIP-1α | CCR2 | CCL2, CCL7, CCL8, CCL13, CCL16 |
| CCL5 | RANTES | CCR3 | CCL5, CCL7, CCL8, CCL11, CCL13, CCL15, CCL24, CCL26, CCL28, |
| CCL13 | MCP-4 | CCR4 | CCL17, CCL22 |
| CCL17 | TARC | CCR5 | CCL3, CCL4, CCL5, CCL8 |
| CCL19 | MIP-3β/ELC | CCR6 | CCL20 |
| CCL20 | MIP-3α/LARC | CCR7 | CCL19, CCL21 |
| CCL21 | 6Kine/SLC | CCR8 | CCL1 |
| CCL22 | MDC | CXCR1 | CXCL1, CXCL4, CXCL6, CXCL8 |
| CCL28 | MEC/CCK1 | CXCR2 | CXCL 1–8 |
| CXCL8 | IL-8 | CXCR3 | CXCL9, CXCL10, CXCL11 |
| CXCL9 | Mig | CXCR5 | CXCL13 |
| CXCL10 | IP-10 | CXCR6 | CXCL16 |
| CXCL11 | I-TAC | CX3CR1 | CX3CL1 |
| CXCL13 | BCA-1 | ADDITIONAL TARGETS | |
| CXCL14 | BRAK | DARC | |
| CXCL16 | CXCL16 | CD3 | |
| CX3CL1 | Fractalkine | CD45 |
FIGURE 2.
Relative gene expression of human renal allograft biopsy pools. Levels of RNA transcripts in pooled samples from post-reperfusion biopsies (IRI, open bar; n=10), stable function (SA, light grey bar; n=10), subclinical rejection (SCR, dark grey bar; n=10) and acute rejection biopsies (ACR, black bar; n=17) compared to a pool of normal donor biopsies (n=15; reference line value of 1). The relative gene expression for (A) chemokines and (B) chemokine receptors is listed on the X-axis. Where no bar is present represents no transcripts detected under our assay conditions.
Clinically stable allografts, evaluated at least one month after transplantation, demonstrated a transcriptional profile consistent with T cell recruitment and activation. Compared to IRI samples, these biopsies displayed an even greater elevation (44-fold) of CCL19 transcripts (Fig. 2A). There were elevated levels of IFNγ-inducible chemokines, CXCL9, CXCL10 and CXCL11 (12-, 7- and 6-fold respectively), compared to NK biopsies. Similarly, many T cell-associated, chemokine-associated transcripts, including CCL13, CCL21, CCL22, CX3CR1, CCR1, CCR5 and CCR6, were increased 5- to 10-fold above NK. CD3 and CD45 transcripts were also upregulated (5- and 10-fold respectively, Fig. 2B), further confirming the presence of T cell infiltrates. Although increases in both T cell-activating chemokines and T cell infiltrates were present, these renal allografts showed no evidence of clinical dysfunction.
Effector T Cell Chemokines and Chemokine Receptors are Elevated and Sustained with Ongoing Acute Rejection
There was a broad upregulation of chemokines and chemokine receptors in both SCR and ACR samples, consistent with the orchestration of an evolving alloimmune response. Similar levels of transcripts for most chemokines and receptors were detected between SCR and ACR pools. Thus, while allografts with SCR maintained clinical function comparable to stable allografts, they remained transcriptionally consistent with ACR. Transcripts associated with T cell effector function, including CXCL9, CXCL10 and CXCL11, remained significantly upregulated in both SCR and ACR compared to stable allografts (Fig. 2A), indicating strong polarization toward a TH1 effector immune response. Consistent with ongoing alloreactive T cell recruitment, CCR7 transcripts were increased and CCR7 ligands, CCL19 and CCL21, were markedly upregulated (Fig. 2A, 2B). Other inflammatory chemokines, including CCL3 and CCL5, and their receptors CCR1 and CCR5, also were increased. Interestingly, CCL17 (10- fold) and CCL22 (35-fold), ligands typically associated with TH2 and regulatory T cell responses, were upregulated in SCR biopsies compared to NK.
Chemokine Ligands for CXCR3, CCR5 and CCR7 are Evident in Allograft Rejection but not in Renal Toxicity due to Calcineurin Inhibitors
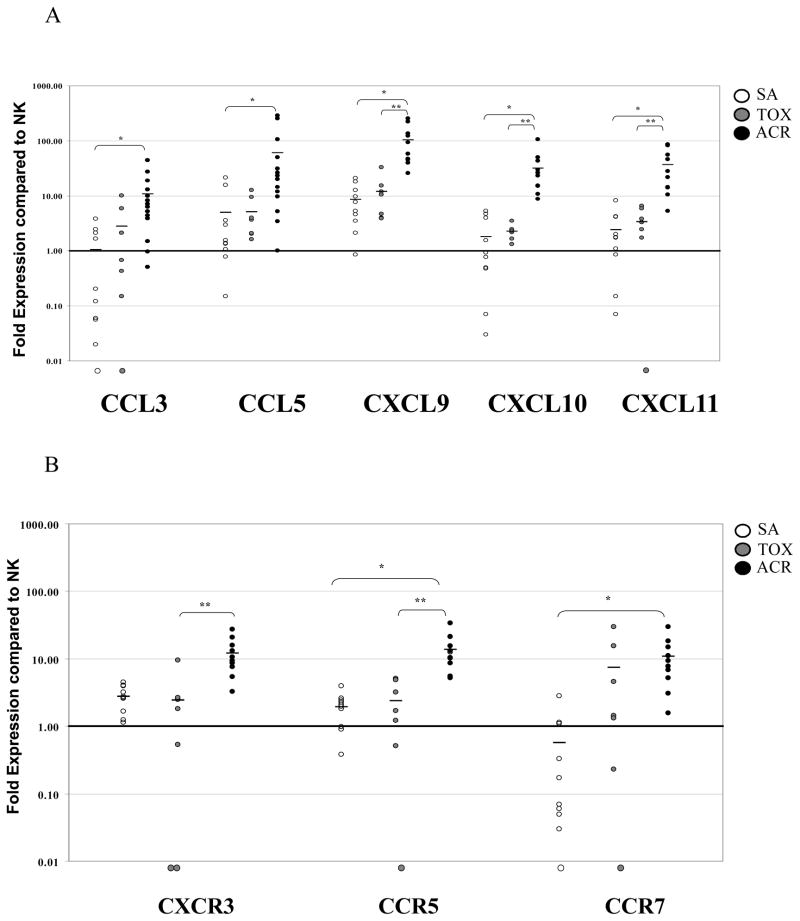
CXCR3 and CCR5 are expressed on activated CD8+ T cells, allowing them to migrate to target effector sites and mediate allograft rejection (19). CCR7 is expressed on naïve and central memory CD8+ T cells and governs recirculation through the blood and lymphoid compartments (25, 26). To more closely evaluate the role of these key CD8+-associated chemokines receptors, we compared their expression in individual patient biopsies by RT-PCR. Since there was little difference in the expression of these molecules between SCR and ACR biopsies, only ACR samples were included in this comparison (SA (n=10), ACR (n=10) and TOX (n=9)).
Both SA and TOX biopsies were generally transcriptionally quiescent, and no significant differences between these groups were observed. However, CXCL9 (p<0.001), CXCL10 (p=0.001), CXCL11 (p=0.01), and CXCR3 (p=0.006) were significantly increased in ACR compared to TOX biopsies (Fig. 3A, 3B). Additionally, transcripts for chemokine receptors CCR7 (p=0.002) and CCR5 (p=0.037), and ligands, CCL3 (p=0.016) and CCL5 (p=0.007), were elevated in ACR compared to SA. Overall, these data show that the chemokine profile associated with a CD8+ TH1 effector phenotype is specific to an alloimmune response rather than nonspecifically triggered by calcineurin inhibitor toxicity and subsequent renal dysfunction.
FIGURE 3.
Chemokine and chemokine receptor transcript levels distinguishing the nature and phenotype of infiltrating cells in human renal allografts. The relative (A) chemokine and (B) chemokine receptor transcript prevalence for each patient biopsy sample is shown for those transcripts with the most dramatic deviation from normal kidney. Individual, log-scale, quantitative fold-levels of RNA transcripts are displayed for SA (open circles), TOX (gray circles), and ACR (black circles) biopsies referenced to the pool of NK biopsies (reference line value of 1). Circles located below the graph baseline indicate undetectable levels. Bars represent the mean fold-level of RNA transcripts for each biopsy group. An asterisk (*) denotes significance between ACR and SA biopsies, (p<0.05); (**) denotes significance between ACR and TOX biopsies (p<0.05).
DISCUSSION
Chemokines are known to be critical regulators of leukocyte trafficking and activation. The initial ischemia-reperfusion injury sustained by an allograft creates a pro-inflammatory milieu that enhances allorecognition, naïve T cell priming and effector T cell recruitment, with detrimental sequelae to the allograft (27, 28). Therefore, we sought to develop distinct transcriptional profiles of chemokines and their cognate receptors that correlated with local patterns of cellular infiltration and overall clinical function. Our study profiles the changing patterns of chemokine and chemokine receptor expression in renal allografts from post-reperfusion to acute cellular rejection. It does so with a comprehensive panel allowing for relative comparisons across the entire family of chemokines to be simultaneously assessed using the same methodology, placing the magnitude of each chemokine in context.
We showed early responsiveness of the innate immune system after reperfusion through an immediate and marked upregulation of CXCL8 transcripts in post-reperfusion biopsies. Activated endothelium and monocytes release pre-formed chemokines and quickly amplify chemokine production to trigger an immune response (8, 28). CXCL8 expression during the acute reperfusion phase is commensurate with the amount of ischemia time, as allografts from deceased donors have been shown to have markedly elevated CXCL8 expression compared to allografts from live donors (29). These data highlight the early role of transcribed chemokines in addition to the known release of stored chemokines in recruiting leukocytes to the graft and suggests CXCL8 as a potential molecular therapeutic target. In a rat renal allograft model, a novel CXCL8 antagonist was shown to decrease monocyte and CD8+ T cell infiltration and limit interstitial inflammation and tubulitis (30). In this study, there was a modest elevation of CXCR1 (the receptor for CXCL8) transcripts post-reperfusion that was not statistically significant. As biopsies were performed 30–60 minutes after reperfusion, the responsive cellular infiltrate was likely missed at this early time point.
We found that transcripts of IFNγ-inducible chemokines, CXCL9, CXCL10 and CXCL11, consistent with a TH1-type immune response, were elevated early after reperfusion, and expression progressively increased in stable and rejecting allografts. The upregulation of these chemokines, commensurate with a rise in CXCR3 receptor transcripts during rejection, suggests they may play a critical role in recruiting previously activated, differentiated CXCR3+ effector cells to the graft (19, 31). In prior studies, we found that the gene targets most predictive of allograft rejection were closely associated with activated TH1 T cells, specifically T-bet and FasL (18). Quantitative differences in these gene transcripts were shown to delineate clinical progression of stable allografts through SCR and ACR. Together, these data suggest there is an immediate and ongoing recruitment of activated effector TH1 cells into the allograft, and these effector cells are crucial to the development of rejection.
The chemokine receptor CXCR3 is expressed on monocytes, TH1 lymphocytes, and natural killer cells, all critical components of tubulointerstitial inflammation (32). Increased expression of CXCR3 and its ligands has been demonstrated in clinical rejection in renal (1, 19), lung (2) and cardiac allografts (3). We observed increased levels of CXCR3 transcripts in the pooled analysis and numeric differences within the individual analyses between SA and ACR. Significant differences were evident when comparing TOX to ACR, indicating that this receptor is likely expressed by allospecific infiltrates.
CCR5 has been detected on infiltrating CD3+/CD4+ T cells during acute rejection (19, 33–34). Interestingly, naïve CD8+ T cells were also shown to upregulate CCR5, facilitating lymph node interactions with antigen-specific dendritic cell-CD4+ T cell pairs that express CCL3 and CCL4 (35). In this study, we detected elevated levels of CCR5 and CCL3 transcripts immediately after reperfusion. CCL3, CCL5 and CCR5 transcripts were also significantly elevated in ACR compared to stable allografts. The presence of this ligand-receptor pair may reflect initial T cell-APC interactions within the allograft, indicative of early T cell priming and activation.
There was also a significant upregulation of CCL19 and CCL21 transcripts in stable and rejecting allografts, most notably in allografts with subacute rejection. Both of these chemokines are CCR7 ligands and play critical roles in T cell-endothelial cell adhesion (36). Activated or injured renal endothelium may upregulate these molecules, creating a pronounced gradient for DC, macrophage, and T cell recruitment and trafficking to secondary lymphoid tissues. Mature DCs subsequently activate lymphocytes to undergo clonal expansion and gain effector function (37), and a subset of effectors upregulate CXCR3 and migrate to inflamed tissues (38). Taken together, our results profile the changing pro-inflammatory microenvironment within a renal allograft that begins at reperfusion and results in the recruitment of a progressively activated T cell compartment in stable and rejecting allografts.
Our previous studies also showed that CD152 expression was prominent in acutely rejecting allografts, suggesting that regulatory factors may be induced during a functionally significant rejection to limit the destructive process (18). Supporting evidence for this regulation is provided by the expression of CCR4 and its corresponding ligands, CCL17 and CCL22, in rejecting allografts. CCL17 and CCL22 transcripts were significantly elevated in SCR. CCR4, known to be associated with regulatory T cells, was detectable in SCR samples but only elevated in ACR biopsies, potentially reflecting a protective response to rejection.
Although many of these molecules transcriptionally defined infiltrating cell phenotypes they still did not clearly differentiate the clinical status of renal allografts. Current immunosuppressive strategies may only have modest effects on chemokine production and immune cell recruitment. It is important to acknowledge that rejection is a continuum that involves many genes related to chemotaxis, cellular activation, and effector cell maturation. It follows that macrophage and T cell chemotaxis is necessary but insufficient for rejection provided that effector T cell maturation is relatively suppressed.
Finally, there are several potential etiologies of renal allograft dysfunction that are not associated with mononuclear cell infiltration, including drug-induced nephrotoxicity, dehydration, and viral infection (39). Quantitative analysis of the chemokines and chemokine receptors that are directly related to T cell activation and chemotaxis may help differentiate cellular rejection from nonimmune causes of renal dysfunction, leading to better detection of subtle changes in the allograft that are not reflected by histology or clinical assay. Understanding the milieu of chemokines active during the different clinical stages of an allograft may help appropriately target infiltrating cell types, cellular interactions, prevent early T cell priming, and lead to improved immunosuppression strategies.
MATERIALS AND METHODS
Patients and Biopsy Acquisition
Patients were enrolled in Institutional Review Board-approved protocols following informed consent and selected for study based on clearly defined, singular diagnoses. Patients with multiple overlapping diagnoses (e.g. ACR and significant interstitial fibrosis/tubular atrophy) were excluded. Biopsies (n=71) were obtained from donors or recipients treated with standard immunosuppression: a calcineurin inhibitor (tacrolimus or cyclosporine), an antiproliferative agent (mycophenolate mofetil or sirolimus), and/or prednisone. Biopsies (18-gauge) were obtained per protocol surveillance or as clinically indicated, and assigned to one of six groups based on clinical presentation and the Banff 1997 criteria (24). Group 1 biopsies, Normal Kidney (NK, n=15), were obtained prior to vascular cross-clamp from live donors with normal renal function undergoing open donor nephrectomy. Group 2 biopsies, Ischemia-Reperfusion Injury (IRI, n=10), were procured 30–60 minutes after transplantation and revascularization. Group 3 biopsies, Stable Allograft (SA, n=10), were obtained from patients undergoing surveillance biopsy at least 1 month post-transplantation without change in renal function (serum creatinine <10% increase over baseline without pathological proteinuria), and without drug toxicity, infection, or histological evidence of rejection. Group 4 biopsies, Subclinical Rejection (SCR, n=10), were taken from patients meeting criteria for Group 3 who had a Banff score of IA or higher on biopsy. Group 5 biopsies, Acute Cellular Rejection (ACR, n=17), represent patients biopsied specifically to evaluate a rise in serum creatinine of at least 15% above baseline, who also had a Banff score of IA or higher without other demonstrable pathology. Group 6 biopsies, Toxicity (TOX, n=9), were protocol surveillance biopsies or biospies performed to evaluate an elevated creatinine and diagnosed with histological indices of calcineurin inhibitor toxicity without rejection. Patients with antibody mediated rejection or BK virus nephropathy were excluded from the study, and all biopsies were negative for SV40 viral infection. The groups did not differ with regard to patient demographics (Table 2).
Table 2.
Demographic and Clinical Data of Patient Biopsy Groups
| Group 1 NK | Group 2 IRI | Group 3 SA | Group 4 SCR | Group 5 ACR | Group 6 TOX | |
|---|---|---|---|---|---|---|
| Biopsies | 15 | 10 | 10 | 10 | 17 | 9 |
| Male/Female | 7/8 | 8/2 | 6/4 | 4/6 | 8/9 | 4/5 |
| Deceased/Living | 0/15 | 6/4 | 3/7 | 3/7 | 11/6 | 4/5 |
| SCr mg/dL* | 1.0 (0.7–1.2) | 1.3 (1.0–1.8) | 1.3 (0.7–1.6) | 1.4 (0.9–1.7) | 3.2 (1.4–12.4) | 1.7 (1.0–3.8) |
| % Change SCr* | - - - - | - - - - | −7 (−18–6.3) | 0 (−21–9.1) | 34 (19–65) | 15 (−6–27) |
| Recipient Age** | - - - - | 41 ± 13 | 39 ± 10 | 42 ± 13 | 43 ± 4 | 38 ± 6 |
| Donor Age** | 42 ± 10 | 36 ± 16 | 35 ± 11 | 37 ± 16 | 40 ± 5 | 34 ± 13 |
| HLA Match** | - - - - | 4 ± 1.6 | 3 ± 2.3 | 4 ± 1.3 | 4 ± 0.5 | 3 ± 1.6 |
| Biopsy Day | - - - - | - - - - | 360 (31–1106) | 263 (42–268) | 173 (26–1211) | 186 (4–338) |
SCr = serum creatinine,
median (range),
mean ± SD
Biopsy Preparation
Biopsies were divided at the bedside, with a 2–4mm fragment snap frozen in liquid nitrogen within 1 minute of procurement, and the remainder sent for histopathological assessment according to the Banff criteria (24). Additional sections were immunostained as previously described (7) with the following antibodies: LCA (Dako, Carpenteria, CA), CD3 (Dako), CD4 (Novocastra, Burlingame, CA), CD8 (Dako), and CD68 (BD Pharmingen, San Jose, CA). Polyomavirus infection was excluded using the anti-SV40 large T-antigen antibody PAB-416 (Oncogene Research Products, Cambridge, MA), and C4d staining was performed using the antibody Bi-RC4D (Bionet Inc, Southbridge, MA). A transplant pathologist (DK) evaluated all biopsies blinded to the clinical data. The degree of infiltration for each cell phenotype was semi-quantitatively scored from 0 to 6 based on immunohistochemical staining in the renal cortex as previously described (18). Tissue for complete immunophenotyping was available for all Group 1 biopsies, 10 cases each for Groups 2–5, and 8 cases for Group 6.
Quantification of Transcripts by Real-time Polymerase Chain Reaction (RT-PCR)
Frozen biopsy fragments were processed for cDNA and 200ng of the resulting template was used for RT-PCR as previously described (40). An extensive panel of chemokine and chemokine receptor targets was chosen for study (Table 1). Each target was analyzed in quadruplicate using 384-well, Low Density Arrays (Applied Biosystems, Inc., Foster City, CA) or individual target assays. Primers for 18s ribosomal RNA, an internal control, were analyzed simultaneously for each reaction. Reactions were amplified: 50°C for 2min, 99°C for 10min, 40 cycles at 99°C for 15sec and 60°C for 1min. Mean values of quadruplicate assays were used for analysis.
Pooled samples were prepared by combining equal amounts of cDNA from each individual within the group, and all groups were compared to a pooled sample of cDNA from Group 1 (NK). Data were analyzed using Sequence Detector Software version, 2.1 (Applied Biosystems, Inc). Quantification was derived using the comparative threshold cycle method as previously described (18, 40) and reported as an n-fold difference of the experimental sample relative to NK biopsies. No transcript associations were segregated by patient demographics or immunosuppression.
Statistics
Significance of differences between immunohistochemical staining and Banff scores was determined using the Mann-Whitney U test. Individual biopsy gene transcript data were normalized by log transformation and compared with one-way analysis of variance (ANOVA). Post-hoc inter-group comparisons were made using a Bonferroni correction to appropriately account for multiple comparisons. Significance was defined as a two-tailed p<0.05.
Acknowledgments
This study was funded in part by the Division of Intramural Research, NIDDK, NCI, NIH and in part by NIH grants RO1 AI49285-04 and 1K24 DK616962-02 (BNB) and 2P01AI044644-10 (ADK). ADK also is supported by the Georgia Research Alliance. The authors declare no conflict of interest.
ABBREVIATIONS
- ACR
Acute cellular rejection
- IRI
Ischemia-reperfusion injury
- NK
Normal Kidney
- RT-PCR
Real-time polymerase chain reaction
- SA
Stable allograft
- SCR
Subclinical rejection
- TOX
Toxicity
Footnotes
Authors’ contributions:
D. Lo and T. Weaver participated in data analysis and the preparation of this manuscript.
D. Kleiner, R. Mannon, L. Jacobson, B. Becker, S. Swanson and D. Hale participated in research performance and data analysis.
A. Kirk conceived the research design, and participated in data analysis and the preparation of this manuscript.
References
- 1.Segerer S, Cui Y, Eitner F, et al. Expression of chemokines and chemokine receptors during human renal transplant rejection. Am J Kidney Dis. 2001;37:518. [PubMed] [Google Scholar]
- 2.Agostini C, Calabrese F, Rea F, et al. CXCR3 and its ligand CXCL10 are expressed by inflammatory cells infiltrating lung allografts and mediate chemotaxis of T cells at sites of rejection. Am J Pathol. 2001;158:1703. doi: 10.1016/S0002-9440(10)64126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao DX, Hu Y, Miller GG, Luster AD, Mitchell RN, Libby P. Differential expression of the IFN-gamma-inducible CXCR3-binding chemokines, IFN-inducible protein 10, monokine induced by IFN, and IFN-inducible T cell alpha chemoattractant in human cardiac allografts: association with cardiac allograft vasculopathy and acute rejection. J Immunol. 2002;169:1556. doi: 10.4049/jimmunol.169.3.1556. [DOI] [PubMed] [Google Scholar]
- 4.Rotondi M, Rosati A, Buonamano A, et al. High pretransplant serum levels of CXCL10/IP-10 are related to increased risk of renal allograft failure. Am J Transplant. 2004;4:1466. doi: 10.1111/j.1600-6143.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- 5.Morita K, Miura M, Paolone DR, et al. Early chemokine cascades in murine cardiac grafts regulate T cell recruitment and progression of acute allograft rejection. J Immunol. 2001;167:2979. doi: 10.4049/jimmunol.167.5.2979. [DOI] [PubMed] [Google Scholar]
- 6.Miura M, Morita K, Kobayashi H, et al. Monokine induced by IFN-gamma is a dominant factor directing T cells into murine cardiac allografts during acute rejection. J Immunol. 2001;167:3494. doi: 10.4049/jimmunol.167.6.3494. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann SC, Kampen RL, Amur S, et al. Molecular and immunohistochemical characterization of the onset and resolution of human renal allograft ischemia-reperfusion injury. Transplantation. 2002;74:916. doi: 10.1097/00007890-200210150-00003. [DOI] [PubMed] [Google Scholar]
- 8.Luster AD. The role of chemokines in linking innate and adaptive immunity. Curr Opin Immunol. 2002;14:129. doi: 10.1016/s0952-7915(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 9.Hancock WW. Chemokine receptor-dependent alloresponses. Immunol Rev. 2003;196:37. doi: 10.1046/j.1600-065x.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 10.Robertson H, Kirby JA. Post-transplant renal tubulitis: the recruitment, differentiation and persistence of intra-epithelial T cells. Am J Transplant. 2003;3:3. doi: 10.1034/j.1600-6143.2003.30102.x. [DOI] [PubMed] [Google Scholar]
- 11.Fahmy NM, Yamani MH, Starling RC, et al. Chemokine and chemokine receptor gene expression indicates acute rejection of human cardiac transplants. Transplantation. 2003;75:72. doi: 10.1097/00007890-200301150-00013. [DOI] [PubMed] [Google Scholar]
- 12.Kirk AD, Bollinger RR, Finn OJ. Rapid, comprehensive analysis of human cytokine mRNA and its application to the study of acute renal allograft rejection. Hum Immunol. 1995;43:113. doi: 10.1016/0198-8859(94)00158-m. [DOI] [PubMed] [Google Scholar]
- 13.Strehlau J, Pavlakis M, Lipman M, et al. Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci. 1997;94:695. doi: 10.1073/pnas.94.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suthanthiran M. Molecular analyses of human renal allografts: differential intragraft gene expression during rejection. Kidney Int Suppl. 1997;58:S15. [PubMed] [Google Scholar]
- 15.Kirk AD, Jacobson LM, Heisey DM, Radke NF, Pirsch JD, Sollinger HW. Clinically stable human renal allografts contain histological and RNA-based findings that correlate with deteriorating graft function. Transplantation. 1999;68:1578. doi: 10.1097/00007890-199911270-00024. [DOI] [PubMed] [Google Scholar]
- 16.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 17.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panzer U, Reinking RR, Steinmetz OM, et al. CXCR3 and CCR5 positive T-cell recruitment in acute human renal allograft rejection. Transplantation. 2004;78:1341. doi: 10.1097/01.tp.0000140483.59664.64. [DOI] [PubMed] [Google Scholar]
- 19.Segerer S, Cui Y, Eitner F, et al. Expression of chemokines and chemokine receptors during human renal transplant rejection. Am J Kidney Dis. 2001;37:518. [PubMed] [Google Scholar]
- 20.Ruster M, Sperschneider H, Funfstuck R, et al. Differential expression of beta-chemokines MCP-1 and RANTES and their receptors CCR1, CCR2, CCR5 in acute rejection and chronic allograft nephropathy of human renal allografts. Clin Nephrol. 2004;61:30. doi: 10.5414/cnp61030. [DOI] [PubMed] [Google Scholar]
- 21.Mayer V, Hudkins KL, Heller F, et al. Expression of the chemokine receptor CCR1 in human renal allografts. Nephrol Dial Transplant. 2007;22:1720. doi: 10.1093/ndt/gfm007. [DOI] [PubMed] [Google Scholar]
- 22.D'Ambrosio D, Panina-Bordignon P, Sinigaglia F. Chemokine receptors in inflammation: an overview. J Immunol Methods. 2003;273:3. doi: 10.1016/s0022-1759(02)00414-3. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann SC, Hale DA, Kleiner DE, et al. Functionally significant renal allograft rejection is defined by transcriptional criteria. Am J Transplant. 2005;5:573. doi: 10.1111/j.1600-6143.2005.00719.x. [DOI] [PubMed] [Google Scholar]
- 24.Racusen LC, Solez K, Colvin RB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 25.Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat Immunol. 2008;9:970. doi: 10.1038/ni.f.213. [DOI] [PubMed] [Google Scholar]
- 26.Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A. 2008;73:975. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 27.Fischereder M, Schroppel B. The role of chemokines in acute renal allograft rejection and chronic allograft injury. Front Biosci. 2009;14:1807. doi: 10.2741/3342. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Rabb H, Womer KL. Ischemia-reperfusion and immediate T cell responses. Cell Immunol. 2007:2484. doi: 10.1016/j.cellimm.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Araki M, Fahmy N, Zhou L, et al. Expression of IL-8 during reperfusion of renal allografts is dependent on ischemic time. Transplantation. 2006;81:783. doi: 10.1097/01.tp.0000198736.69527.32. [DOI] [PubMed] [Google Scholar]
- 30.Bedke J, Nelson PJ, Kiss E, et al. A novel CXCL8 protein-based antagonist in acute experimental renal allograft damage. Mol Immunol. 2010;47:1047. doi: 10.1016/j.molimm.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Piali L, Weber C, LaRosa G, et al. The chemokine receptor CXCR3 mediates rapid and shear-resistant adhesion-induction of effector T lymphocytes by the chemokines IP10 and Mig. Eur J Immunol. 1998;28:961. doi: 10.1002/(SICI)1521-4141(199803)28:03<961::AID-IMMU961>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Daha MR, van Kooten C. Is the proximal tubular cell a proinflammatory cell? Nephrol Dial Transplant. 2000;15:41. doi: 10.1093/ndt/15.suppl_6.41. [DOI] [PubMed] [Google Scholar]
- 33.Pattison J, Nelson PJ, Huie P, et al. RANTES chemokine expression in cell-mediated transplant rejection of the kidney. Lancet. 1994;343:209. doi: 10.1016/s0140-6736(94)90992-x. [DOI] [PubMed] [Google Scholar]
- 34.Robertson H, Morley AR, Talbot D, Callanan K, Kirby JA. Renal allograft rejection: beta-chemokine involvement in the development of tubulitis. Transplantation. 2000;69:684. doi: 10.1097/00007890-200002270-00039. [DOI] [PubMed] [Google Scholar]
- 35.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding native CD8+ T cells to site of CD4+ T cell- dendritic cell interaction. Nature. 2006;440:890. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 36.Christopherson KW, Hood AF, Travers JB, Ramsey H, Hromas RA. Endothelial induction of the T-cell chemokine CCL21 in T-cell autoimmune diseases. Blood. 2003;101:801. doi: 10.1182/blood-2002-05-1586. [DOI] [PubMed] [Google Scholar]
- 37.Sallusto F, Lanzavecchia A. Understanding dendritic cell and T-lymphocyte traffic through the analysis of chemokine receptor expression. Immunol Rev. 2000;177:134. doi: 10.1034/j.1600-065x.2000.17717.x. [DOI] [PubMed] [Google Scholar]
- 38.Viola A, Molon B, Contento RL. Chemokines: coded messages for T-cell missions. Front Biosci. 2008;13:6341. doi: 10.2741/3158. [DOI] [PubMed] [Google Scholar]
- 39.Halloran PF. Call for revolution: a new approach to describing allograft deterioration. Am J Transplant. 2002;2:195. doi: 10.1034/j.1600-6143.2002.20301.x. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann SC, Pearl JP, Blair PJ, Kirk AD. Immune profiling: molecular monitoring in renal transplantation. Front Biosci. 2003;8:E444. doi: 10.2741/1167. [DOI] [PubMed] [Google Scholar]