Abstract
Export of proteins from the endoplasmic reticulum (ER) in COPII-coated vesicles occurs at defined sites, which contain the scaffolding protein Sec16. We identify TFG-1, a new conserved regulator of protein secretion that interacts directly with SEC-16 and controls the export of cargoes from the ER in C. elegans. Hydrodynamic studies indicate that TFG-1 forms hexamers, which facilitate the co-assembly of Sec16 with COPII subunits. Consistent with these findings, TFG-1 depletion leads to a dramatic decline in both SEC-16 and COPII levels at ER exit sites. The amino-terminus of human TFG was identified previously as a fusion partner of two protein kinases, creating a pair of oncogenes. We propose that fusion of these kinases to TFG relocalizes their activities to ER exit sites, where they prematurely phosphorylate substrates during ER export. Our findings provide a mechanism by which translocations involving TFG can result in cellular transformation and oncogenesis.
INTRODUCTION
The trafficking of most secretory cargoes begins with their export from the endoplasmic reticulum (ER) (1,2). In metazoans, cargoes are packaged into vesicles that emerge at defined sites on the ER and ultimately fuse with the ER-Golgi intermediate compartment (ERGIC). This process relies on the efficient recruitment of a set of soluble factors known as the COPII coat, which is composed of two multimeric protein complexes (Sec23-Sec24 and Sec13-Sec31) (3,4). A number of rare diseases have been directly linked to perturbations in COPII function, including Craniolenticulosutural dysplasia, in which a mutation in Sec23A leads to impaired collagen export from the ER (5,6). Although COPII vesicle formation has been reconstituted with purified coat proteins on synthetic membranes (7), regulators of COPII assembly remain largely unexplored.
Of the known COPII interacting proteins, the small GTPase Sar1 and the putative scaffolding protein Sec16 are among the best characterized (8). When activated by the Sec12 exchange factor, Sar1 recruits the Sec23-Sec24 complex, which forms an adaptor layer for Sec13-Sec31 lattice assembly, completing the COPII coat (9,10). Sec23 also functions as a Sar1 activating protein, which is stimulated by Sec31, leading to coat disassembly following vesicle budding (11). In contrast to Sar1, which has only been shown to bind Sec23 directly, Sec16 interacts with all COPII coat components, potentially serving as a scaffold for their recruitment (12–15). Furthermore, Sec16 may stabilize the COPII coat to prevent premature disassembly following activation of the Sec23 GAP (16). Surprisingly, few other proteins have been implicated in the regulation of COPII recruitment. Considering the necessity for controlling secretory flux during cell differentiation and development, additional factors that govern this process likely exist, and some may function via direct regulation of Sar1 or Sec16. Here, we report the identification of a new regulator of ER export, which interacts directly with Sec16 and controls COPII subunit accumulation at ER exit sites.
RESULTS
Identification of SEC-16 interacting proteins in C. elegans
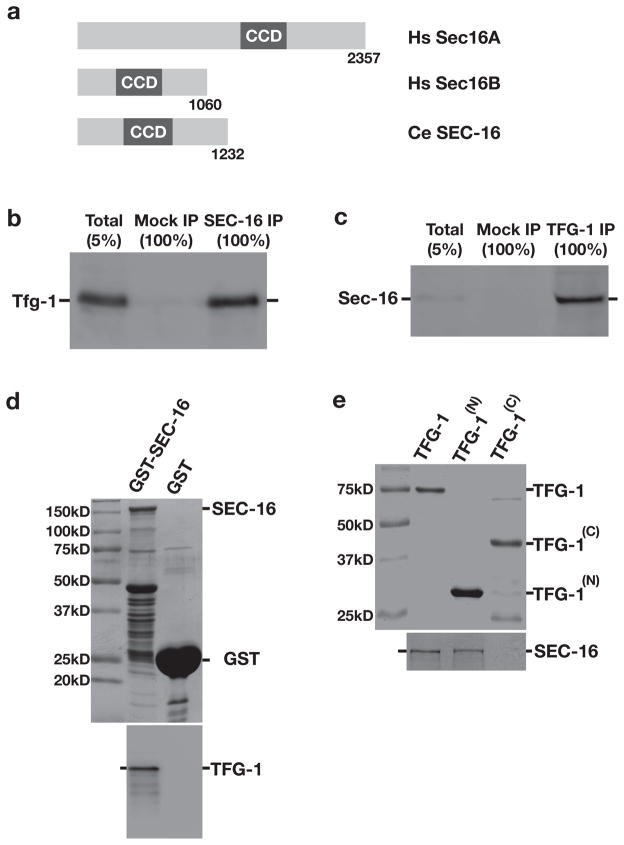
Based upon the sequence of human Sec16A, we identified a Sec16 homolog in C. elegans encoded by ZK512.5, which we named sec-16 (Fig. 1a). Antibodies directed against SEC-16 recognized a band at the appropriate molecular weight (~135 kD) following immunoblot analysis of a C. elegans lysate and were used to immunoprecipitate SEC-16 and associated proteins from detergent-solubilized embryo extracts (Supplementary Figs. S1a-S1c). We conducted solution mass spectrometry to identify 19 proteins that were reproducibly isolated by SEC-16 antibodies, which were individually depleted using RNA interference (RNAi). We found that 7 out of 19 gene products were necessary for embryo production: SEC-16, four COPII subunits (SEC-23, SEC-24.2, SEC-13, and SEC-31), the Sar1 specific GEF SEC-12, and TFG-1 (Supplementary Table 1).
Figure 1. C. elegans TFG-1 interacts with the ER exit site component SEC-16.
(a) Schematic representation of human and C. elegans Sec16 isoforms. The central conserved domain (CCD) is highlighted in each protein. (b) SEC-16 was immunoprecipitated from C. elegans embryo extract and blotted with α-TFG-1 antibodies (n=3). A mock IP was conducted in parallel using rabbit IgG. (c) TFG-1 was immunoprecipitated from C. elegans embryo extract and blotted with α-SEC-16 antibodies (n=3). A mock IP was conducted in parallel using rabbit IgG. (d) GST alone and GST-tagged full length SEC-16 were immobilized on glutathione agarose beads, which were incubated with an extract generated from E. coli expressing recombinant TFG-1. Following a series of washes, proteins were eluted using reduced glutathione, separated by SDS-PAGE, and either stained using Coomassie (top) or immunoblotted using TFG-1 antibodies (bottom). (e) Polyhistidine-tagged full length and truncated forms of TFG-1, either encoding amino acids 1–195 (TFG-1(N)) or 196–486 (TFG-1(C)), were purified from E. coli onto nickel affinity resin and incubated with freshly prepared whole worm extract (n=3). Imidazole eluted proteins were separated by SDS-PAGE, stained with Coomassie (top), and blotted with α-SEC-16 antibodies (bottom). For each figure, uncropped scans of all gels and immunoblots are provided in Supplementary Fig. S6.
We focused our attention on the interaction between SEC-16 and TFG-1. Immunoblot analysis of a C. elegans lysate using TFG-1 antibodies revealed at least two closely migrating bands immediately below the 75 kD marker, significantly larger than the predicted size of 49.8 kD (Supplementary Fig. S1d). Similarly, recombinant TFG-1 also exhibited a slow migration on SDS-PAGE (see Fig. 1e). Analysis of a SEC-16 immunoprecipitate using TFG-1 antibodies confirmed their interaction (Fig. 1b), and TFG-1 immunoprecipitation led to an enrichment of SEC-16 (Fig. 1c). We also generated recombinant forms of both TFG-1 and SEC-16 and measured their association in vitro. We found that a GST-fusion to SEC-16, but not GST alone, was capable of binding to recombinant TFG-1 (Fig. 1d). Additionally, we generated polyhistidine-tagged recombinant forms of full-length TFG-1 and two truncations of TFG-1 encoding the amino-terminus (amino acids 1–195) and carboxyl-terminus (amino acids 196–486), and measured their ability to interact with endogenous SEC-16. In a similar fashion to the full-length recombinant protein, the amino-terminal portion of TFG-1 was able to bind to SEC-16, while the carboxyl-terminus did not (Fig. 1e). These data confirm a direct biochemical association between SEC-16 and TFG-1.
TFG-1 localizes to ER exit sites with SEC-16
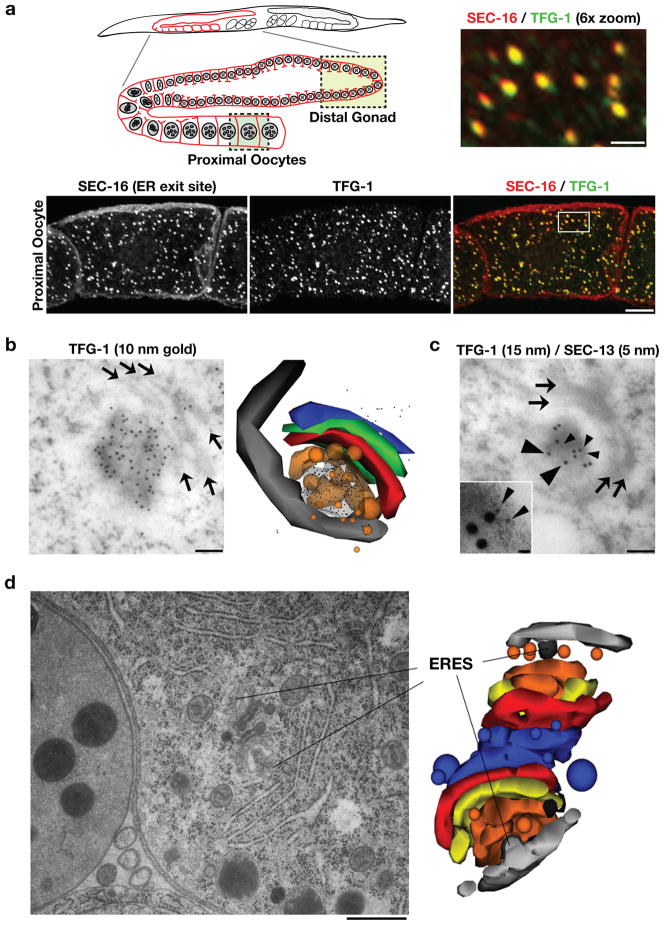
To determine whether SEC-16 interacts with TFG-1 at ER exit sites, we examined the C. elegans reproductive system, which is enriched for TFG-1 (Supplementary Fig. S1e). TFG-1 antibodies stained punctate structures throughout the cytoplasm of oocytes, 85% of which co-localized with SEC-16 on ER exit sites (Fig. 2a). In many cases, TFG-1 staining extended beyond the puncta labeled with SEC-16 (Fig. 2a, see 6× zoom). Using immuno-gold electron microscopy (EM), we further defined the localization of TFG-1 in proximal oocytes to a cloud-like region at ER exit sites that spread to the ERGIC (Fig. 2b). The high concentration of labeling observed suggested that TFG-1 forms a matrix in this region, which would correspond well to the elevated electron density seen there by EM. Dual immuno-gold labeling with TFG-1 and SEC-13 antibodies indicated that both proteins localize to an identical area next to ER exit sites, although the SEC-13 labeling was more discrete (Fig. 2c). We conclude that TFG-1 localizes to ER exit sites with both SEC-16 and COPII machinery.
Figure 2. TFG-1 localizes to ER exit sites that are juxtaposed to the Golgi.
(a) Dissected C. elegans gonads were fixed and stained using Cy2-labeled α-TFG-1 and Cy3-labeled α-SEC-16 antibodies (n=8). Both individual and merged images of proximal oocytes with TFG-1 in green and SEC-16 in red are shown (Bar, 10 μm). The upper right image is the boxed area in the panel below magnified 6× (Bar, 2 μm). Also shown is a schematic of the C. elegans reproductive system, which includes a syncytial stem cell niche in the distal gonad (boxed region in light green) and proximal oocytes that have undergone cellularization (boxed region in dark green). (b and c) Lowicryl sections of C. elegans oocytes were stained with antibodies against TFG-1 or a combination of TFG-1 and SEC-13 antibodies. Arrows highlight Golgi cisternae. Large arrowheads point out 15 nm gold particles associated with immunoreactive TFG-1, and small arrowheads highlight 5 nm gold particles associated with SEC-13. Bars, 100 nm. An inset is provided in panel c to clearly show the distribution of 5 nm particles at higher magnification (Inset bar, 15 nm). In addition, a three-dimensional reconstruction of TFG-1 immunolocalization is shown. The image was generated using the software Reconstruct from serial 50 nm thin sections. Vesicles were reconstructed using the sphere setting, and all other components (ER, ERGIC, coats, Golgi stacks) were generated using the Boissonnat surface setting. Light grey: ER; dark grey: COPII coat; orange: ER-derived transport vesicles and ERGIC; red, green and blue: Golgi cisternae; from cis to trans, respectively. (d) An electron micrograph illustrating two ER exit sites and adjacent Golgi complexes in the proximal most oocyte of an animal following high pressure freezing and freeze substitution (Bar, 500 nm). On the right is a three-dimensional reconstruction of the same pair of Golgi complexes and associated ER exit sites. The ER exit sites are surrounded by vesicles that fuse to form the ERGIC. Light grey: ER; dark grey: COPII coat; orange: ER-derived transport vesicles and ERGIC; yellow, red and blue: Golgi cisternae; from cis to trans, respectively.
The architecture of ER exit sites was also examined using EM. We found that ER exit sites range in size from 70–150 nm, and always contain coated buds emerging from smooth ER that are directed toward ERGIC and Golgi membranes (Fig. 2d; Supplementary Figs. S1f and S1g; Video S1). Electron tomography was used to confirm these findings and provide a high-resolution depiction of COPII vesicle biogenesis (Fig. 3a; Video S2). Consistent with these observations and our data showing that TFG-1 localizes specifically to ER exit sites, we failed to observe co-localization between TFG-1 and the glucuronyl transferase SQV-8, a marker of the cis-medial Golgi (Supplementary Fig. S1h). Instead, TFG-1 appeared directly adjacent to SQV-8 labeled structures, further illustrating that in C. elegans, ER exit sites are closely juxtaposed to the Golgi, forming an integrated secretion unit.
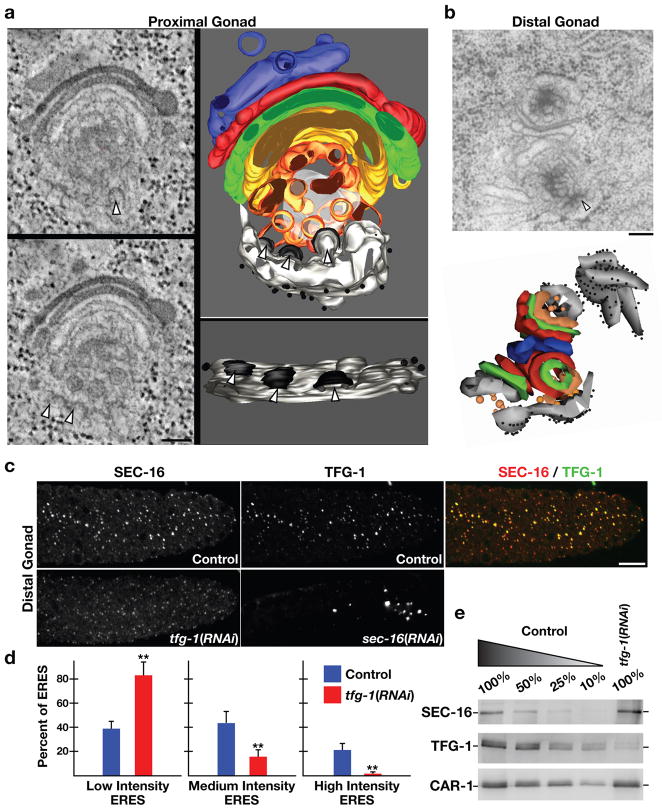
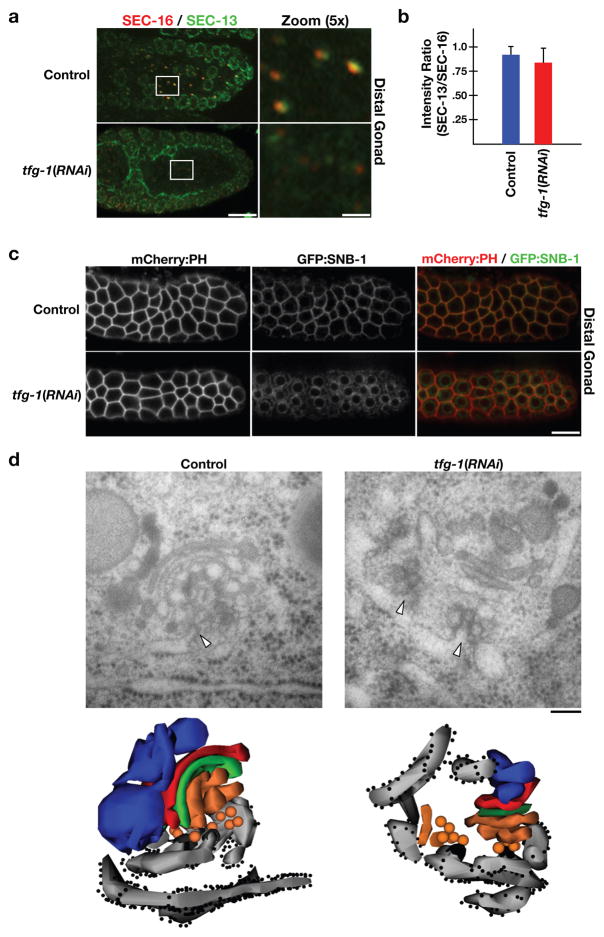
Figure 3. TFG-1 regulates SEC-16 levels on ER exit sites.
(a) In the proximal gonad, a 300 nm section of the early secretory pathway (ER exit sites, ERGIC, and Golgi) was analyzed by electron tomography. ER exit sites are highlighted by arrowheads. On the left are individual sections from the tomographic stack. On the right are two orthogonal views of the tomogram following three-dimensional reconstruction. Light grey: ER; black: COPII coat; orange and yellow: ER-derived transport vesicles and ERGIC; green, red, blue: Golgi cisternae; diffuse grey: not further resolvable matrix. Bar, 100 nm. (b) An electron micrograph illustrating ER exit sites and adjacent Golgi complexes in the distal gonad following high pressure freezing and freeze substitution. An arrowhead highlights the presence of budding vesicle from smooth ER (Bar, 100 nm). Below is a three-dimensional construction of the same Golgi complexes and associated ER exit sites. (c) Dissected gonads from control, TFG-1 depleted, and SEC-16 depleted animals were fixed and stained using Cy2-labeled α-TFG-1 and Cy3-labeled α-SEC-16 antibodies. Individual and merged images of the distal gonad with TFG-1 in green and SEC-16 in red are shown (Bar, 10 μm). (d) Fluorescence intensity of SEC-16 in the distal gonad was measured in control and TFG-1 depleted animals, and intensities were segregated into low, medium and high thresholds. To establish individual thresholds, a histogram of fluorescence intensities was equally divided into three regions, and the number of ER exit sites within each area was calculated. The bar graph indicates the percentage of all ER exit sites that fall into a specific threshold. For each condition, at least 1000 unique ER exit sites were examined. Error bars represent mean +/− SEM; 10 different animals. **p < 0.01 compared with control, calculated using a paired t test. (e) Western blots of extracts prepared from animals depleted of TFG-1 by RNAi (n=3). Serial dilutions of extracts prepared from control animals were loaded to quantify depletion levels. Blotting with α-CAR-1 antibodies was performed to control loading.
We next examined the distal region of the C. elegans germline, which constitutes a stem cell niche that constitutively requires membrane biosynthesis and secretion (17). ER exit sites in this region were morphologically similar to those seen in the proximal gonad (Fig. 3b). Immunofluorescence analysis of exit sites in the distal germline revealed a distribution of intensities for both SEC-16 and TFG-1 (Figs. 3c and 3d). However, the ratio of the fluorescence intensities between SEC-16 and TFG-1 largely did not vary among different ER exit sites, indicating that in wild-type animals, levels of SEC-16 correlated with those of TFG-1, regardless of whether they were high or low (Supplementary Fig. S2a).
To determine localization dependencies between TFG-1 and SEC-16, we depleted each protein using RNAi. Depletion of TFG-1 did not result in a significant decline in the number of SEC-16 labeled ER exit sites (Supplementary Fig. S2b). However, the distribution of SEC-16 intensities was dramatically altered under these conditions (Fig. 3d). The number of exit sites harboring a high level of SEC-16 declined significantly (approximately 30-fold), while the number of low intensity sites increased approximately 2-fold. As the stability of SEC-16 was not diminished following depletion of TFG-1 (Fig. 3e), these data suggest that TFG-1 controls the abundance of SEC-16 at ER exit sites. Depletion of SEC-16 led to nearly complete loss of puncta labeled by TFG-1, and the few structures that were observed appeared abnormally large and did not co-stain for the COPII component SEC-13 (Fig. 3c and our unpublished data). We conclude that TFG-1 requires SEC-16 for its localization, while SEC-16 requires TFG-1 to accumulate normally on ER exit sites.
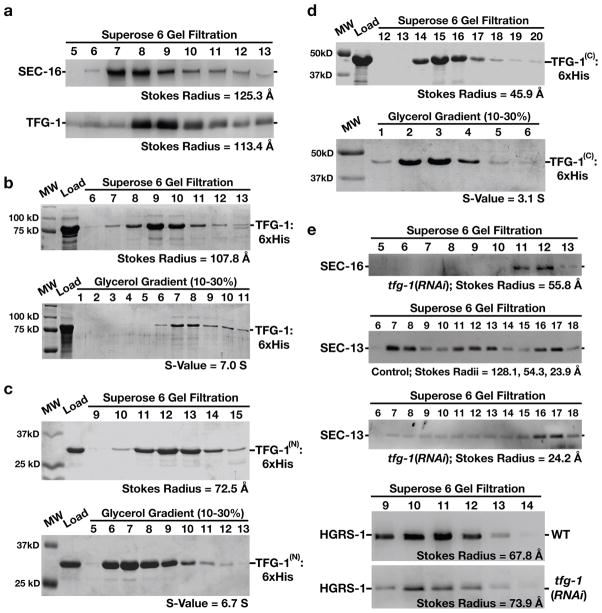
TFG-1 self-associates to form hexamers that regulate proper SEC-16 complex assembly
Based on size exclusion chromatography, native SEC-16 and TFG-1 exhibit similar elution profiles, which correspond to globular complexes larger than 800 kD (Fig. 4a). Since TFG-1 was amenable to recombinant expression, we measured its Stokes radius (107.8 Å) and sedimentation value (7.0 S) in the absence of SEC-16 (Fig. 4b). Based on these data, we calculated the native molecular weight of TFG-1 to be ~318 kD, nearly identical to the predicted mass of a TFG-1 hexamer. Furthermore, we found that the amino-terminus assembled into an octamer, while the carboxyl-terminus formed a dimer (Figs. 4c and 4d). These data suggest that the amino-terminus of TFG-1 mediates its oligomerization, while the carboxyl-terminus may restrict the full-length protein to form hexamers in solution. Since levels of TFG-1 and SEC-16 at ER exit sites co-vary in control animals and loss of TFG-1 leads to diminished accumulation of SEC-16 on ER exit sites, such hexamers of TFG-1 likely play a role in proper SEC-16 complex assembly. Consistent with this idea, depletion of TFG-1 caused a reduction in the Stokes radius of SEC-16 (by ~70 Å) isolated from whole animals (Fig. 4e). Concomitantly, loss of TFG-1 also affected the assembly of complexes containing COPII proteins. Analysis of native SEC-13 by size exclusion chromatography demonstrated that the COPII subunit is a component of a large complex (Stokes radius of 128.1 Å), similar in size to SEC-16 and TFG-1 (Fig. 4e). In addition, SEC-13 was found in a 54.3 Å complex, which likely corresponds to a SEC-13-SEC-31 heterotetramer that was described previously (10), and as a globular monomer (Stokes radius of 23.9 Å). However, following TFG-1 depletion, the majority of SEC-13 was found in a monomeric state (Stokes radius of 24.2 Å), although we continued to detect the COPII subunit in high molecular weight fractions as well. We conclude that the normal assembly of large complexes containing SEC-16 and COPII subunits is facilitated by TFG-1 hexamers.
Figure 4. The amino-terminus of TFG-1 mediates it oligomerization.
The results presented in each panel are representative of at least three individual experiments performed. In all cases, the intensities of each band were measured to identify the peak elution fraction, which was used to calculate either a Stokes radius or sedimentation value, depending on the experiment. (a) Western blots using SEC-16 antibodies (top) or TFG-1 antibodies (bottom) of C. elegans embryo extract fractionated on a Superose 6 gel filtration column. The peaks corresponding to SEC-16 and TFG-1 partially overlap. A Stokes radius was calculated for each protein based on comparison with the elution profiles of known standards. (b-d) Recombinant polyhistidine-tagged TFG-1 or fragments of TFG-1 described in Figure 1e were expressed and purified from E. coli extracts using nickel resin. A Coomassie stained gel of the peak elution fractions after fractionation of the recombinant proteins on a Superose 6 gel filtration column are shown (top). Proteins were fractionated on a 10–30% glycerol gradient (bottom), and S-values were calculated based on the location of characterized standards run on a parallel gradient. (e) Western blots of control and TFG-1 depleted C. elegans whole worm extracts fractionated on a Superose 6 gel filtration column and probed with SEC-16 antibodies (top) or SEC-13 antibodies (middle panels). Fractionation of HGRS-1, a component of the ESCRT-0 complex, was examined in both control and TFG-1 depleted conditions (bottom panels), to ensure gel filtration profiles were directly comparable. Stokes radii were calculated for each protein based on comparison with the elution profiles of known standards.
TFG-1 is required for protein secretion
We next examined the fate of COPII localization in animals with and without TFG-1. In control animals, the COPII subunit SEC-13 is found in close apposition to SEC-16 labeled exit sites and on the nuclear envelopes of proliferating germ nuclei (Fig. 5a). Following depletion of TFG-1, the intensities of both SEC-13 and SEC-16 on punctate structures within the germline declined dramatically, while localization of SEC-13 to the nuclear envelope was largely unaffected (Fig. 5a). By measuring the fluorescence intensities of SEC-13 and SEC-16 in control and TFG-1 depleted animals, we found that their ratio was not altered significantly, indicating that the remaining SEC-16 on ER exit sites can continue to recruit COPII components in the absence of TFG-1, albeit to lower levels (Fig. 5b).
Figure 5. TFG-1 is required for COPII recruitment and protein secretion.
(a) Dissected C. elegans gonads were fixed and stained using Cy2-labeled α-SEC-13 and Cy3-labeled α-SEC-16 antibodies (n=15). Merged images of the distal gonad with SEC-13 in green and SEC-16 in red are shown on the left (Bar, 10 μm). Panels to the right are magnified 5× views of the boxed area in the adjacent panel (Bar, 2 μm). (b) Bar graph showing the average ratio of SEC-13 to SEC-16 fluorescence intensities in control and TFG-1 depleted animals. For each condition, at least 250 unique ER exit sites in the distal gonad were examined. Error bars represent mean +/− SEM; 6 different animals. No statistically significant difference was observed, based on a calculation using a paired t test. (c) Swept field confocal optics were used to image anesthetized control (n=15) and TFG-1 depleted (n=15) adult animals expressing GFP:SNB-1 and mCherry:PH. Scale bar, 10 μm. (d) Electron micrographs illustrating the early secretory pathway in the proximal most oocyte of control (left) and TFG-1 depleted (right) animals following high pressure freezing and freeze substitution (Bar, 100 nm). Arrowheads highlight ER exit sites. Below each micrograph is a three-dimensional reconstruction of the same regions. Light grey: ER; dark grey: COPII coat; orange: ER-derived transport vesicles and ERGIC; green, red and blue: Golgi cisternae; from cis to trans, respectively.
To determine whether the substantially reduced levels of SEC-16 and SEC-13 were sufficient to support protein secretion following TFG-1 depletion, we examined several secreted integral membrane proteins. Under steady state conditions in the distal region of the germline, the majority of a GFP fusion to the v-SNARE synaptobrevin (GFP:SNB-1) co-localizes with a mCherry fusion to the PH domain of rat PLC1δ1 (mCherry:PH), which binds specifically to the plasma membrane (Fig. 5c; 18). However, in TFG-1 depleted animals, GFP:SNB-1 accumulated in the ER (Fig. 5c). Similarly, we found that several other transmembrane proteins, including the C. elegans caveolin-like protein CAV-1 and the glucuronyl transferase SQV-8, which normally traffic to other organelles following synthesis, accumulate in the ER in the absence of TFG-1 (Supplementary Figs. S2c and S2d).
To further examine the effect of TFG-1 depletion, we used serial section EM. In control animals, ER exit sites were juxtaposed to well-organized stacks of membranes corresponding to the ERGIC and Golgi cisternae (Fig. 5d, left). Vesicles that were 48.2 +/− 1.2 nm (n=15) in diameter were observed throughout the region between the ER and ERGIC. In contrast, subsequent to TFG-1 depletion, ERGIC and Golgi membranes appeared smaller and more poorly stacked, and the ER became fragmented (Fig. 5d, right). In addition, fewer Golgi networks were observed overall. However, consistent with our findings that reduced levels of SEC-16 at ER exit sites can continue to recruit COPII following depletion of TFG-1, we identified vesicles that were 48.8 +/− 0.7 nm (n=39) in diameter between the ER and the ERGIC. Although the sizes of vesicles were not affected by TFG-1 depletion, their distribution was altered. The majority of vesicles observed accumulated close to the ER and failed to migrate toward the ERGIC and Golgi membranes. These data suggest that TFG-1 is required for the efficient, directed movement of COPII vesicles away from the ER, but not for their initial formation.
We also generated animals that stably express a GFP fusion to the Golgi Rab-type GTPase RAB-6, which mediates retrograde transport from the Golgi to the ER, to study perturbations resulting from TFG-1 depletion. Since RAB-6 also cycles between the Golgi and early endosomes (19), we co-expressed a mCherry fusion to the early endosome marker RAB-5, to allow us to distinguish between endosome- and Golgi-associated RAB-6. In control animals, RAB-5 and RAB-6 exhibited distinct localizations in the distal region of the germline (Supplementary Fig. S2e). However, following depletion of either SEC-16 or TFG-1, RAB-6 accumulated on fewer, large punctate structures in the germline that also harbored RAB-5, suggesting that RAB-6 localization was shifted from the Golgi to enlarged early endosomes (Supplementary Fig. S2e). These data further support the idea that TFG-1 depletion leads to a defect in normal Golgi assembly.
Human TFG localizes to ER exit sites and interacts with Sec16
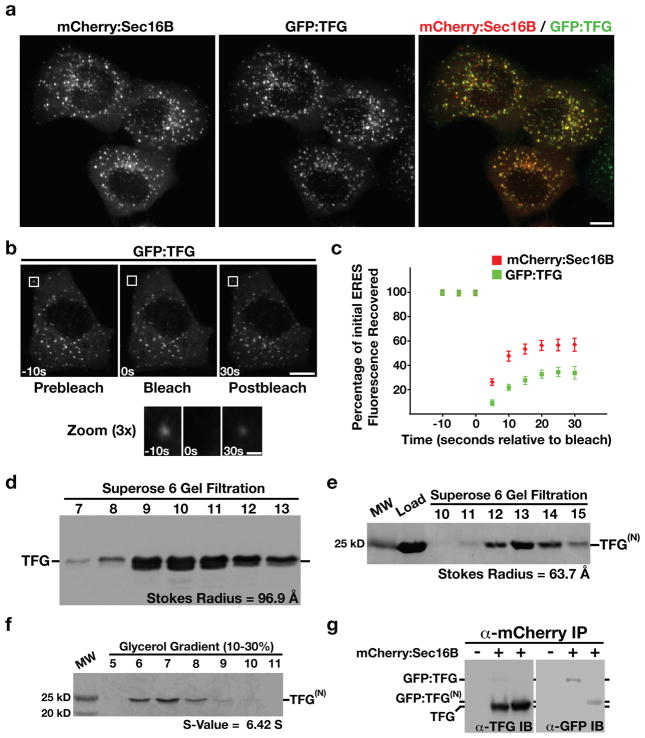
C. elegans TFG-1 has a single human homolog, previously annotated TFG (for TRK-fused gene; Supplementary Fig. S3a). We generated a GFP fusion to full-length TFG and co-transfected it into HeLa cells with a mCherry fusion to human Sec16B to measure co-localization. Analysis of cells expressing both markers indicated that greater than 90% of GFP:TFG labeled puncta also contained mCherry:Sec16B (Fig. 6a). These data demonstrate that analogous to C. elegans TFG-1, human TFG functions at ER exit sites.
Figure 6. Human TFG functions at ER exit sites and binds to Sec16.
(a) Swept field confocal optics were used to image HeLa cells that had been transiently transfected with GFP:TFG and mCherry:Sec16B (n=42). Representative color overlays of mCherry:Sec16B (red) and GFP:TFG (green) are shown. Scale bar, 10 μm. (b) Swept field confocal optics were used to monitor the recovery of GFP:TFG after photobleaching (n=15). A 3× magnified view of the boxed region where GFP:TFG was bleached is shown below. Times are in seconds relative to the bleach. Scale bars, 10 μm (top) and 1 μm (bottom). (c) Graph showing the average percentage of GFP:TFG and mCherry:Sec16B fluorescence recovered as a function of time in seconds relative to the bleach (error bars represent means +/− SEM for each time point; n=15 different cells for each fluorescent fusion protein). (d) Western blots of HeLa cell extract fractionated on a Superose 6 gel filtration column (n=3). A Stokes radius was calculated for human TFG based on comparison with the elution profiles of known standards. (e, f) A GST-tagged, truncated form of human TFG, amino acids 1–193 was expressed and purified from E. coli extracts using glutathione agarose (n=3), and the GST tag was subsequently cleaved using Prescission Protease prior to loading onto a gel filtration column or glycerol gradient. A Coomassie stained gel of the peak elution fractions after fractionation of the recombinant protein, referred to as TFG(N), on a Superose 6 gel filtration column are shown (e). The protein was also fractionated on a 10–30% glycerol gradient (f), and an S-value was calculated based on the location of characterized standards run on a parallel gradient (n=3). (g) Antibodies directed against mCherry were used to immunoprecipitate mCherry:Sec16B from HeLa cells transiently transfected with GFP:TFG or a GFP fusion to the amino-terminus of TFG referred to as GFP:TFG(N) (n=3). Isolated proteins were separated by SDS-PAGE and blotted with α-TFG (left) and α-GFP antibodies (right).
We next examined the dynamics of Sec16B relative to TFG using fluorescence recovery after photobleaching (FRAP). Similar to GFP:Sec16A (20), we found that only a fraction (56.8 +/− 6.3%) of the fluorescence of mCherry:Sec16B recovered following photobleaching, with a half-time of 5.68 +/− 0.34 seconds. In contrast, a smaller proportion of GFP:TFG recovered (33.6 +/− 5.1%), which exhibited a longer half-time of 8.22 +/− 0.64 seconds (Figs. 6b and 6c). These data indicate that Sec16B and TFG are not loaded onto exit sites simultaneously and instead suggest that they assemble onto ER exit sites via distinct mechanisms.
Similar to our findings with C. elegans TFG-1, endogenous human TFG exhibited a large Stokes radius (Fig. 6d), and a recombinant form of its amino-terminus (amino acids 1–193) assembled as an octamer with a native molecular weight of 172 kD (Figs. 6e and 6f). Thus, human TFG and C. elegans TFG-1 share multiple physical properties, which suggest they share a common function in vivo. Consistent with this idea, depletion of TFG caused a substantial delay in the secretion of VSVG(ts045)-GFP, a type I transmembrane protein used widely to study the mammalian secretory pathway (Supplementary Fig. S3b).
To determine whether human TFG binds to Sec16, we transfected cells stably expressing mCherry:Sec16B with either a GFP fusion to full length TFG or its amino-terminus, and conducted immunoprecipitations using mCherry antibodies. We found that endogenous TFG was recovered under these conditions (Fig. 6g). Additionally, we found that both GFP-tagged isoforms were able to bind to mCherry:Sec16B (Fig. 6g). These data support the idea that the amino-terminus of TFG interacts with Sec16 in an evolutionarily conserved manner.
Mistargeting of the NTRK1 kinase domain to ER exit sites causes the hyperactivation of downstream NTRK1 effectors and cell transformation
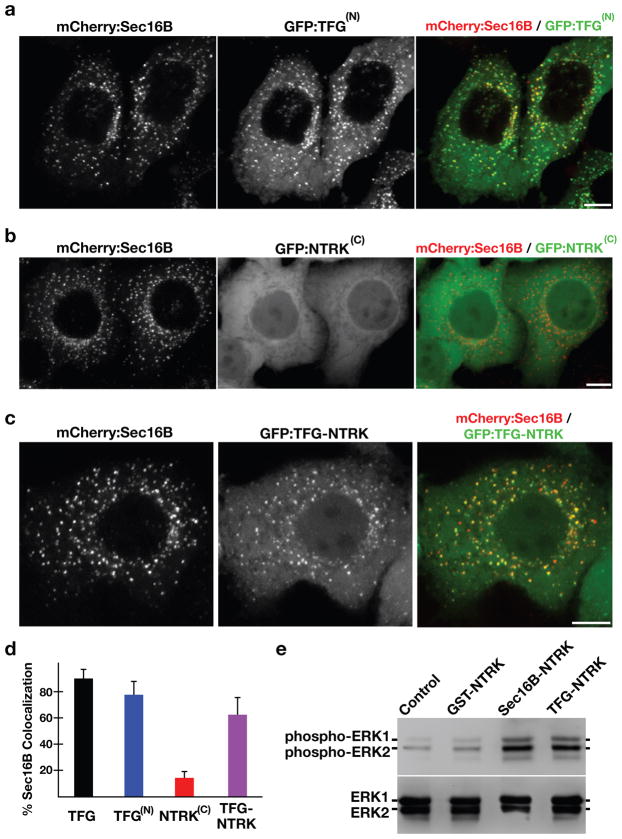
TFG was initially identified as part of a gene fusion with the receptor tyrosine kinase NTRK1 (also called TrkA), resulting in the formation of an oncogene (21). Subsequent studies have shown that TFG is also a fusion partner of the receptor tyrosine kinase ALK in some anaplastic large cell lymphomas (22). In both instances, the amino-terminus of TFG is fused to the carboxyl-terminus of the other gene, resulting in an oncogenic product. We found that the amino-terminal domain of TFG localizes to ER exit sites and is capable of redirecting the carboxyl-terminal domain of NTRK1 there as well (Figs. 7a-7d). These data suggest a role for TFG in targeting the kinase activites of NTRK1 and ALK to ER exit sites.
Figure 7. Targeting of the NTRK1 kinase domain to ER exit sites is sufficient to activate NTRK1-mediated downstream signaling.
(a–c) Swept field confocal optics were used to image HeLa cells that had been transiently transfected with mCherry:Sec16B and GFP fusions to either the amino terminus of TFG referred to as GFP:TFG(N) (n=18), the transmembrane and kinase domains of NTRK1 referred to as GFP:NTRK(C) (n=15), or a TFG(N)-NTRK1(C) fusion (n=28), which is equivalent to the oncogene characterized previously (21). Representative color overlays of mCherry:Sec16B (red) and GFP fusions (green) are shown. Scale bar, 10 μm. (d) Bar graph showing the percent co-localization between the GFP fusions described above and mCherry:Sec16B (error bars represent means +/− SEM for each condition; n=15 different cells for each condition and at least 800 unique ER exit sites were examined). (e) Extracts from hTERT-RPE1 cells stably transfected with GFP alone (Control) or various GFP fusions to the NTRK1 transmembrane and kinase domains (as indicated) were separated by SDS-PAGE and blotted using a phospho-specific ERK1-ERK2 antibody (top) and a pan-ERK1-ERK2 antibody (bottom).
NTRK1 is a high affinity receptor for nerve growth factor (NGF) (23). Binding of NGF to NTRK1 leads to its dimerization and autophosphorylation, ultimately causing the activation of several downstream signaling cascades, including the Ras-Raf-MEK-ERK pathway, to promote cell survival and growth (24). The oncogenic TFG-NTRK1 fusion protein is constitutively active (25), and expression of GFP-TFG-NTRK1 in hTERT-RPE1 cells caused an increase in the levels of phospho-ERK1 and -ERK2, as compared to control cells (Fig. 7e). To determine whether dimerization of the kinase domain is sufficient to initiate signaling downstream of NTRK1, we linked the constitutive dimer GST to NTRK1 and expressed it as a GFP fusion in hTERT-RPE1 cells. Under these conditions, levels of phospho-ERK1 and -ERK2 increased by only 2-fold as compared to control cells (Fig. 7e). These data indicate that dimerization of the NTRK1 kinase domain alone is not adequate to cause the hyperactivation of its downstream effectors as is observed following TFG-NTRK1 expression.
As suggested earlier, an alternative mechanism by which fusion to TFG may cause oncogenic NTRK1 activity is through its mislocalization to ER exit sites. Therefore, we tested the effect of targeting the NTRK1 kinase domain to exit sites independently of TFG. We fused a region of Sec16B (amino acids 1–791), which localizes to ER exit sites similarly to the full length protein, to NTRK1 and expressed it as a GFP fusion in hTERT-RPE1 cells (GFP:Sec16B-NTRK). Hydrodynamic analysis of a portion of this region in Sec16B (amino acids 34–234) indicated that it oligomerizes in vitro (Supplementary Fig. S4a). GFP:Sec16B-NTRK1 localized to ER exit sites (Supplementary Fig. S4b), and expression of this fusion protein resulted in hyperactivation of ERK1 and ERK2, similar to the effect following GFP-TFG-NTRK1 expression (Fig. 7e). We conclude that targeting of the NTRK1 kinase domain to ER exit sites is necessary for its constitutive activity in cells.
To test whether the ER exit site-targeting of NTRK1 was sufficient to transform cells, we generated NIH3T3 cell lines expressing either GFP, GFP:GST-NTRK1, GFP:TFG-NTRK1, or GFP:Sec16B-NTRK1. Cells expressing the ER exit site-targeted forms of NTRK1 produced at least 8-fold more colonies than the dimerized form of NTRK1 (Supplementary Fig. S4c). These data indicate that localization of NTRK1 to ER exit sites strongly contributes its ability to transform cells.
DISCUSSION
We have demonstrated that TFG-1 localizes to ER exit sites and is required for protein secretion in the C. elegans germline. Additionally, we have found that TFG-1 accumulates at ER exit sites in several other tissues, including the intestine, hypodermis, and muscle (Supplementary Fig. S5), suggesting a common role in multiple cell types. We further demonstrate that human TFG functions at ER exit sites and likely regulates the secretion of multiple cargoes. Consistent with this idea, overexpression of human TFG has been shown to partially rescue the trafficking defect of mutant F508del-CFTR, which aberrantly accumulates in the ER in cystic fibrosis patients (26,27). Since levels of TFG-1 at ER exit sites appear to correlate with levels of SEC-16 and COPII, additional TFG-1 may increase COPII recruitment and stimulate secretion from the ER (28–30).
Based on our data, we speculate that TFG family members generate a matrix at ER exit sites, which may serve as a molecular sink to help retain COPII components locally and facilitate efficient vesicle formation and egress. The increased membrane flux mediated by TFG-1 may also function to maintain proper Golgi organization in tissues under a high secretory demand, such as the C. elegans germline. Together, our findings reveal a new component of the early secretory pathway that regulates anterograde trafficking from the ER.
Furthermore, our studies also provide a key insight into the mechanism by which TFG functions as a proto-oncogene. In the case of the TFG-NTRK1 fusion generated by a chromosomal translocation, concentrating constitutive NTRK1 kinase activity at ER exit sites may cause to the premature stimulation of multiple effectors, including components of the ERK1-ERK2 kinase cascade, which leads to cell transformation. Notably, stimulation of ERK2 at ER exit sites may cause hyper-phosphorylation of Sec16, resulting in the formation of new exit sites that would recruit additional TFG-NTRK1 (31). Such a feed-forward mechanism may play an important role during oncogenesis.
METHODS
Antibodies
C. elegans SEC-16, TFG-1, and CYP-33E1 antibodies were raised in rabbits by immunization with GST fusions to either a fragment of SEC-16 (amino acids 813–1014), full length TFG-1, or a fragment of CYP-33E1 (amino acids 244–392) produced in E. coli. The antibodies were affinity purified from serum by binding to columns of the same antigens following removal of the GST tags by cleavage with Prescission protease. To produce SEC-13 specific monoclonal antibodies, 10 mice were each immunized with 50 μg of purified GST-SEC-13 fusion protein in PBS using complete Freund’s adjuvant. Hybridoma fusions were generated following booster injections and subsequently screened by ELISA according to standard protocols. Mouse monoclonal SEC-13 antibodies were purified via Protein-A Sepharose beads. Antibodies directed against human TFG were purchased from Bethyl Laboratories (A302-341A and A302-343A). Antibodies against ERK1 and ERK2 and phospho-ERK1 and -ERK2 were obtained from Millipore (06–182) and Invitrogen (44680G), respectively. CAR-1, SQV-8, GFP and mCherry antibodies have been described elsewhere (32–34).
Immunofluorescence, Live Imaging, and Electron Microscopy
Images were acquired on a swept field confocal microscope (Nikon Ti-E) equipped with a Roper CoolSnap HQ2 CCD camera using a Nikon 60X, 1.4NA Planapo oil objective lens. Acquisition parameters were controlled by Nikon Elements software, and image analysis was conducted using Metamorph software. Immunofluorescence of fixed gonads was performed as described previously (33) using directly labeled rabbit antibodies at a concentration of 1 μg/mL. Briefly, 60–120 Z sections at 0.2 μm steps were acquired (depending on sample thickness). The fluorescence intensity of each ER exit site was always confined to a maximum of seven Z planes, which were used to generate a maximum intensity projection. To calculate the fluorescence intensity of proteins that localize to ER exit sites, the total intensity in a box containing the ER exit site (from a maximum intensity projection) was measured and the camera background was subtracted. For live imaging of C. elegans gonads, animals were anesthetized and mounted on an agarose pad. HeLa cells were grown on 35-mm glass bottom dishes maintained at 37°C for time-lapse imaging. Photobleaching was performed using a 405-nm laser, coupled into a Nikon Photo Activation Illuminator Unit to create a single diffraction limited spot. The signal at the first post-bleach time point was subtracted from all post-bleach measurements, and the percentage of fluorescence recovered at each time point was calculated by dividing by the difference between the pre-bleach and first post-bleach measurements. Kaleidagraph software was used to fit the data and calculate the maximal fractional recovery and the half-time for recovery. To determine the percent co-localization between two proteins, ER exit sites were analyzed individually using Nikon Elements software. The VSVG(ts045)-GFP trafficking assay was conducted as described previously, with modifications as noted in the legend for Supplementary Figure S3b (35).
For high pressure freezing, a 100 μm deep aluminum platelet (Microscopy Services) was filled with 20 adult worms in a suspension of E. coli and frozen using a BalTec HPM 10. Freeze substitution was carried out in a Leica EM AFS at −90°C as described previously (36). For electron microscopy studies, 50 nm longitudinal sections of C. elegans were cut using a Leica UC6 ultramicrotome. Ribbons of sections were transferred onto Formvar-coated copper slot grids. The grids were placed in drops of 4% uranyl acetate, washed with water, and dried. They were then transferred onto lead citrate (37) and rinsed again with water. Micrographs were taken with a Proscan CCD HSS in a Zeiss EM 902A electron microscope operated in bright field mode. For 3D reconstructions, serial sections of ER exit sites were imaged, and files were aligned linearly using the software Reconstruct (38). For electron tomography, 250–300 nm EPON sections were transferred onto Formvar-coated copper slot grids and stained similarly as described for thin sections. 10 nm gold beads were applied to both sides. An orthogonal tilt series was acquired on a JEOL JEM 2100 at 200 kV from −55° to 55° (1° increments). Reconstruction was done using IMOD software (http://bio3d.colorado.edu/imod). Immuno-electron microscopy was conducted as described previously (36).
Worm Strains, RNA interference and Cell Culture
All C. elegans strains were derived from the Bristol strain N2. The generation of animals expressing fluorescent fusions with CAV-1, SNB-1 and the PH domain of rat PLC1δ were described previously (18,34). Double stranded RNA (dsRNA) was synthesized as described previously (33) from templates prepared by using primers listed in the Methods section to amplify N2 genomic DNA. For most RNAi experiments, early L4 stage hermaphrodites were soaked in dsRNA for 24 hours at 20°C. Animals were allowed to recover for 48 hours prior to analysis. For partial depletions, late L4 stage worms were soaked in dsRNA for 24 hours, followed by a 22 hour recovery period, and then analyzed. For large-scale depletion of TFG-1 necessary for gel filtration studies, L4 animals were fed bacteria expressing dsRNA directed against TFG-1 for 72 hours, prior to harvesting and extract preparation. HeLa and NIH3T3 cells were maintained in DMEM supplemented with 10% FBS (HeLa) or 10% FCS (NIH3T3), penicillin, streptomycin, and L-glutamine at 37°C in the presence of 5% CO2. hTERT-RPE1 cells were grown similarly, with the exception that DMEM/F-12 media was used. Transfections were conducted using Lipofectamine 2000 (Invitrogen), and cells were selected using puromycin (1 μg/mL for HeLa cells and 12.5 μg/mL for hTERT-RPE1 cells). Colony formation assays on monolayers of contact-inhibited NIH3T3 cells were performed as described previously (39).
Primers, dsRNAs, and plasmids used in this study
To generate dsRNAs directed against TFG-1, the following primer sets were used: (AATTAACCCTCACTAAAGGCTGCTGTGGTGGAGCATATC and TAATACGACTCACTATAGGATCTCTCGGCTCCAAAACAA), (AATTAACCCTCACTAAAGGCTTAATCTGCTCGACTTGCT and TAATACGACTCACTATAGGATGGTGCATTCAAACGGAGC), and (AATTAACCCTCACTAAAGGTTACTGCTGATACGGCGACT and TAATACGACTCACTATAGGCAGCAGCAGCAATTCGGAGC). Each dsRNA produced using these primer sets yielded similar results. To generate dsRNAs directed against SEC-16, the following primer sets were used: (AATTAACCCTCACTAAAGGGCGGTCTGCGAGTTTAGATT and TAATACGACTCACTATAGGCAAGCGGATGCAAGAAGAAT), (AATTAACCCTCACTAAAGGGCAAACCTGTAATTTAAAAT and TAATACGACTCACTATAGGCTTCTGGTTTCGATATGAGT), and (AATTAACCCTCACTAAAGGCTCCACTTCTAGCACTTCGC and TAATACGACTCACTATAGGGTAACGATCAACAACATAAC). Each dsRNA produced using these primer sets yielded similar results. For siRNA experiments, the following oligonucleotide was used to efficiently deplete TFG: 5′-CUUCUCAGCCUACUAAUUA-3′. The control siRNA used was directed against the ER exit site component TANGO1 (5′-GCAAUAACCUCAACUCUAUUU-3′), which was shown previously not to affect the trafficking of VSVG-GFP (40). To generate polyhistidine fusion constructs, DNAs were cloned by restriction digest into pRSETA, which encodes a 6× histidine tag on the amino-terminus. To generated GST fusion constructs, DNAs were cloned by restriction digest into pGEX6P-1, which encodes a GST tag on the amino-terminus, followed by a Prescission protease cleavage site. A C. elegans cDNA library was used as a template for cloning recombinant expression constructs by PCR, and cDNAs for human TFG and NTRK1 were purchased from Open Biosystems.
Mass Spectrometry Analysis and Biochemistry
Gravid adult hermaphrodites were grown synchronously in liquid culture, and embryos were isolated in buffer containing 0.6 N NaOH and 20% bleach. Extracts were generated in lysis buffer (50 mM Hepes, 1 mM EDTA, 1 mM MgCl2, 100 mM KCl and 10% glycerol) and used for immunoprecipitations as described previously (32). For mass spectrometry, proteins were precipitated using TCA. The TCA pellets were solubilized and treated with Endoproteinase Lys-C (0.1 μg/μL), followed by Trypsin (0.5 μg/μL). Following digestion, the proteins were pressure-loaded onto a fused silica capillary desalting column, placed inline with a Hewlett Packard Agilent 1100 Quaternary Pump (Version 1.4; Palo Alto, CA) and analyzed using a customized 4-step separation method (90, 120, 120, and 150 minutes respectively) (41). For each step, one full-scan mass spectrum (400–2000 m/z) occurred followed by 5 data-dependent MS/MS spectra at a 35% normalized collision energy. The spectra were searched with the SEQUEST™ algorithm (42) against the WormBase C. elegans (version 2.11 created on 01-10-2010) database.
For immunoprecipitations, HeLa or hTERT-RPE-1 cells were grown in 15 cm plates, and extracts were generated in lysis buffer, which were subjected to centrifugation at 100,000 x g prior to use. Recombinant protein expression was performed using BL21 (DE3) E. coli, and purifications were conducted using either glutathione agarose beads (for GST fusions) or nickel affinity resin (for polyhistidine-tagged proteins). For samples applied to a Superose 6 gel filtration column, the Stokes radius of each protein or protein complex was calculated from its elution volume based on the elution profiles of characterized standards. Glycerol gradients (10–30%) were poured using a Gradient Master and fractionated from the top by hand. Sedimentation values were calculated by comparing the position of the peak with that of characterized standards run on a separate gradient in parallel. To determine the native molecular weight of proteins, the following equation was used: M = 6πηNas/(1-υrho;), where M is the native molecular weight, η is the viscosity of the medium, N is Avogadro’s number, a is the Stokes radius, s is the sedimentation value, υ is the partial specific volume, and ρ is the density of the medium (43). Immunoblotting of extracts and immunoprecipitates were performed as described previously (32). To determine whether endogenous SEC-16 could interact with various fragments of recombinant TFG-1, extracts were generated from animals freshly harvested from fifteen 10-cm plates that were subsequently subjected to sonication in lysis buffer containing 1% Triton X-100. Extracts were clarified by centrifugation (100,000 x g) prior to incubation with proteins bound to nickel affinity resin. To determine the level of TFG-1 depletion following RNAi, both control and dsRNA treated animals (60 each) were hand-picked and placed into eppendorf tubes containing 100 μL of lysis buffer. Samples were sonicated in a water bath sonicator set to 80oC in the presence of 1× sample buffer for 10 min, after which they were boiled for 10 min at 100oC prior to separation by SDS-PAGE.
Statistical Analysis
Statistical significance was evaluated by performing a two-tailed Student’s t test.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the NIH (1R01GM088151-01A1) to AA and P41RR011823 to JRY. We thank Dr. Beth Weaver (UW-Madison) and Dr. Paul Bertics (UW-Madison) for use of tissue culture facilities, Dr. Robert Landick (UW Madison) for use of a Gradient Master, Dr. Karen Oegema (UC San Diego) for marker strains and antibodies, Dr. Edwin Chapman (UW-Madison) for use of Metamorph software, and Dr. Sabine Koenig for help with RNAi. We also thank Dr. David Stephens (University of Bristol, UK), Dr. Patricia Kiley (UW-Madison), and members of the Audhya lab for critically reading this manuscript. The authors declare no competing financial interests.
Footnotes
Author Contributions: Conceived and designed experiments: KW, ALS, SE, AA. Performed experiments and analyzed data: KW, ALS, AS, JH, JRM, KS, SE, AA. Contributed reagents, materials, analysis tools: SE, JRY, AA. Wrote the paper: AA.
References
- 1.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 3.Bickford LC, Mossessova E, Goldberg J. A structural view of the COPII vesicle coat. Curr Opin Struct Biol. 2004;14:147–53. doi: 10.1016/j.sbi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 5.Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, et al. Carnio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192–7. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 6.Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet. 2006;38:1198–203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–75. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 8.Miller EA, Barlowe C. Regulation of coat assembly-sorting things out at the ER. Curr Opin Cell Biol. 2010;22:447–53. doi: 10.1016/j.ceb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–8. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 10.Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–8. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 11.Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–45. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–23. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimeno RE, Espenshade P, Kaiser CA. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol Biol Cell. 1996;7:1815–23. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–6. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 15.Whittle JR, Schwartz TU. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol. 2010;190:347–61. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Supek F, Madden DT, Hamamoto S, Orci L, Schekman R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J Cell Biol. 2002;158:1029–38. doi: 10.1083/jcb.200207053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605–31. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 18.Shi A, Chen CC, Banerjee R, Glodowski D, Audhya A, Rongo C, Grant BD. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2930–43. doi: 10.1091/mbc.E10-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156:653–64. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes H, Budnik A, Schmidt K, Palmer KJ, Mantell J, Noakes C, Johnson A, Carter DA, Verkade P, Watson P, Stephens DJ. J Cell Sci. 2009;122:2924–34. doi: 10.1242/jcs.044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greco A, Mariani C, Miranda C, Lupas A, Pagliardini S, Pomati M, Pierotti MA. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol. 1995;15:6118–27. doi: 10.1128/mcb.15.11.6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, Lamant L, Falini B, Ott G, Mason, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94:3265–8. [PubMed] [Google Scholar]
- 23.Schecterson LC, Bothwell M. Neurotrophin receptors: old friends with new partners. Dev Neurobiol. 2010;70:332–8. doi: 10.1002/dneu.20767. [DOI] [PubMed] [Google Scholar]
- 24.Klesse LJ, Parada LF. Trks: signal transduction and intracellular pathways. Microsc Res Tech. 1999;45:210–6. doi: 10.1002/(SICI)1097-0029(19990515/01)45:4/5<210::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 25.Greco A, Fusetti L, Miranda C, Villa R, Zanotti S, Pagliardini S, Pierotti MA. Role of the TFG N-terminus and coiled-coil domain in the transforming activity of the thyroid TRK-T3 oncogene. Oncogene. 1998;16:809–16. doi: 10.1038/sj.onc.1201596. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Kartner N, Lukacs GL. Limited proteolysis as a probe for arrested conformational maturation of delta F508 CFTR. Nat Struct Biol. 1998;5:180–183. doi: 10.1038/nsb0398-180. [DOI] [PubMed] [Google Scholar]
- 27.Trzcinska-Daneluti AM, Ly D, Huynh L, Jiang C, Fladd C, Rotin D. High-content functional screen to identify proteins that correct F508del-CFTR function. Mol Cell Proteomics. 2009;8:780–90. doi: 10.1074/mcp.M800268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinzer S, Worz S, Kalla C, Rohr K, Weiss M. A model for the self-organization of exit sites in the endoplasmic reticulum. J Cell Sci. 2008;121:55–64. doi: 10.1242/jcs.013383. [DOI] [PubMed] [Google Scholar]
- 29.Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 2008;27:2043–54. doi: 10.1038/emboj.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes H, Stephens DJ. Assembly, organization, and function of the COPII coat. Histochem Cell Biol. 2008;129:129–51. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri HP. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J Cell Biol. 2010;189:997–1011. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JJ, III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–68. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, III, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–79. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, Grant BD. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell. 2006;17:3085–94. doi: 10.1091/mbc.E06-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–5. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 36.Rostaing P, Real E, Siksou L, Lechaire JP, Boudier T, Boeckers TM, Gertler F, Gundelfinger ED, Triller A, Marty S. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur J Neurosci. 2006;24:3463–74. doi: 10.1111/j.1460-9568.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–23. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiala JC. Reconstruct: a free editor for serial section microscopy. J Microsc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 39.Perlaky L, Valdez BC, Busch RK, Larson RG, Jhiang SM, Zhang WW, Brattain M, Busch H. Increased growth of NIH/3T3 cells by transfection with human p120 complementary DNA and inhibition by a p120 antisense construct. Cancer Res. 1992;52:428–36. [PubMed] [Google Scholar]
- 40.Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136:891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 41.Diop SB, Bertaux K, Vasanthi D, Sarkeshik A, Goirand B, Aragnol D, Tolwinski NS, Cole MD, Pradel J, Yates JR, III, Mishra RK, Graba Y, Saurin AJ. Reptin and pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 2006;9:260–6. doi: 10.1038/embor.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eng J, McCormack A, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:979–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 43.Siegel LM, Monty KJ. Determination of molecular weights and frictional rations of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.