Abstract
Cyclic AMP signals encode information required to differentially regulate a wide variety of cellular responses; yet it is not well understood how information is encrypted within these signals. An emerging concept is that compartmentalization underlies specificity within the cAMP signaling pathway. This concept is based on a series of observations indicating that cAMP levels are distinct in different regions of the cell. One such observation is that cAMP production at the plasma membrane increases pulmonary microvascular endothelial barrier integrity, whereas cAMP production in the cytosol disrupts barrier integrity. To better understand how cAMP signals might be compartmentalized, we have developed mathematical models in which cellular geometry as well as total adenylyl cyclase and phosphodiesterase activities were constrained to approximate values measured in pulmonary microvascular endothelial cells. These simulations suggest that the subcellular localizations of adenylyl cyclase and phosphodiesterase activities are by themselves insufficient to generate physiologically relevant cAMP gradients. Thus, the assembly of adenylyl cyclase, phosphodiesterase, and protein kinase A onto protein scaffolds is by itself unlikely to ensure signal specificity. Rather, our simulations suggest that reductions in the effective cAMP diffusion coefficient may facilitate the formation of substantial cAMP gradients. We conclude that reductions in the effective rate of cAMP diffusion due to buffers, structural impediments, and local changes in viscosity greatly facilitate the ability of signaling complexes to impart specificity within the cAMP signaling pathway.
Keywords: G protein-coupled receptors, adenylyl cyclase, phosphodiesterase, protein kinase A, A kinase anchoring protein
cyclic AMP is a ubiquitous second messenger that transmits information required to coordinate a wide range of cellular functions. The mechanisms by which cAMP transmits this information while ensuring signal specificity have been the subject of debate for several decades. One idea that has gained favor in recent years is that cAMP signals are localized—compartmentalized—to discrete locations within the cell (11, 18, 36). In essence, cAMP compartmentalization implies that cAMP levels in one region of the cell are high enough to activate local pools of effectors, including protein kinase A (PKA), exchange protein activated by cAMP (EPAC), and PKG, while cAMP levels in the remainder of the cell are too low to significantly activate other pools of PKA. Evidence strongly supporting the concept of compartmentalization came from a variety of studies using newly developed, single-cell cAMP probes (51, 61, 66, 67, 70, 86, 89). In essence, each of these studies provided data indicating that cAMP levels were indeed high in one region of the cell and low in other regions of the cell. To interpret these data, investigators developed mathematical descriptions of these cellular systems. Initially, simple compartmental models were used to describe the observed differences in cAMP levels in distinct subcellular locations (66, 67). These models assumed that cAMP levels rapidly equilibrated within discrete subcellular compartments, but that flux of cAMP between compartments was severely hindered. Compartmental models clearly described the observed localized cAMP signals and had predictive capabilities (61, 67). Models that included more detailed descriptions of cAMP signaling pathways in cardiac myocytes were developed (72, 73). These models required that the spatial spread of cAMP signals be slowed by buffering and restriction on diffusion. As such, these studies outlined quantitative frameworks with which we could begin to understand the flux of information through the cAMP pathway.1
More recently, two studies used elegant mathematical modeling approaches to describe the spatial spread of cAMP signals in different cellular systems (54, 56). Interestingly, both groups reached the conclusion that hindered diffusion is not required for the localization of cAMP signals. An underlying assumption in each of these studies was that the concentrations of adenylyl cyclase (AC) and phosphodiesterase (PDE) were two or more orders of magnitude higher than would be expected in most cell types. This highlights an overall limitation in the studies described above, i.e., the difficulty in estimating values of key parameters such as the subcellular localizations and activities of enzymes.
The goal of our study was to better understand how altering the subcellular locations and magnitudes of AC and PDE activities, cellular shape, buffering, and other factors that lower the effective cAMP diffusion coefficient influence the spatial spread of cAMP. We have chosen to use pulmonary microvascular endothelial cells (PMVECs) as a model system because data strongly suggest that the location of cAMP synthesis is a critical determinant of whether cAMP signals strengthen or disrupt endothelial barrier integrity (76, 77, 82). Specifically, these studies demonstrated that cAMP produced at the plasma membrane enhances endothelial barrier integrity whereas cAMP produced within the cytosol disrupts barrier integrity. Of particular relevance is a study in which Sayner et al. (76) demonstrated that cAMP produced by a heterologously expressed, forskolin-activatable, soluble AC construct (soluble CI/CII) triggered barrier disruption; however, when this construct was redirected to the plasma membrane, cAMP production did not disrupt the barrier (76).
We have developed mathematical models describing the spatial spread of cAMP signals in PMVECs using the “Virtual Cell” modeling environment (http://www.vcell.org). The models described here incorporate a realistic geometry of cultured PMVECs. Other geometries were examined to better understand how cell shape contributes to cAMP gradients. In most simulations, AC and PDE activities were limited to physiological levels by constraining the simulated rates of cAMP accumulation to match experimental measurements of total cAMP accumulation in the presence and absence of PDE inhibitors. The effects of altering the spatial distribution of AC and PDE activities, as well as the effects of altering cAMP buffering capacity and the effective cAMP diffusion coefficient were examined. The results from these simulations indicate that no single factor (e.g., the subcellular localization of AC or PDE activities, buffering, effective cAMP diffusion coefficient) is primarily responsible for localizing cAMP signals. Specifically, in cultured PMVECs, neither the subcellular localization of AC and PDE activities nor cell shape was sufficient to compartmentalize cAMP signals. Thus, other factors that slow the spatial spread of cAMP signals are likely to contribute to compartmentalization in PMVECs. These factors may include cAMP buffering, structural impediments to cAMP movement such as organelles, clustered proteins, cytoskeletal networks, and the F-actin cortical rim, and local changes in cytosolic viscosity.
MATERIALS AND METHODS
Cell culture.
Rat PMVECs were isolated as described previously (81) by the cell culture core of the Center for Lung Biology at the University of South Alabama. Procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of South Alabama. PMVECs were maintained in Dulbecco's modified Eagle's medium (DMEM, Life Technologies) supplemented with 10% vol/vol fetal bovine serum (Gemini), 100 μg/ml streptomycin, and 100 U/ml penicillin, pH 7.0. Cells were grown in 100-mm culture dishes at 37°C in a humidified atmosphere of 95% air-5% CO2.
Measurement of cAMP.
Cyclic AMP levels were assessed by standard radioimmunoassay (Biomedical Technologies, Stoughton, MA). Briefly, experiments were performed on confluent monolayers of PMVECs grown in six-well dishes. The cells were serum starved in DMEM supplemented with 1% vol/vol fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin, pH 7.0 for 12–14 h before experiments. The cells were treated with vehicle (DMSO) or 10 μM rolipram (a PDE4-specific inhibitor) for 10 min before treatment with vehicle or 10 μM forskolin (an AC activator) for 0 to 50 min as indicated. Reactions were stopped by the addition of HCl (1 N final concentration) and neutralized with NaOH. cAMP content was estimated from standard curves and normalized to both protein level (BCA assay, Pierce) and total cell volume (cell count × average cell volume) to estimate total cellular cAMP concentration.
Measurement of PDE activity.
Cyclic AMP PDE activity was measured as detailed previously (68, 87, 88, 99). Briefly, cells were harvested and homogenized in ice-cold hypotonic buffer containing (in mM) 20 Tris·HCl, 5 MgCl2, 0.5 EDTA, pH 7.4, with protease inhibitors: 10 μM TLCK, 2 μM leupeptin, 2 μM pepstatin A, 10 μM benzamidine, 2,000 units aprotinin/ml. Cells were homogenized and aliquots were assayed for PDE activity using 0.01–5 μM [3H]cAMP as a substrate. Protein concentrations were determined using protein assays (Bio-Rad) with BSA as a standard.
Confocal microscopy.
PMVECs were cultured to confluence on 25-mm glass coverslips. Cells were labeled with calcein green AM (4 μM, 20 min), wheat agglutinin Alexa Fluor 647 (10 μg/ml, 5 min) and DRAQ5 (50 μM, 20 min) and washed twice with phosphate-buffered saline before imaging. A series of images were captured at 0.2-μm intervals (z-stack) at excitation and emission wavelengths of 488 and 510 nm (calcein green), and 647 and 700 nm (wheat agglutinin 647 and DRAQ5) using either a Leica TCS SP2 or Nikon A1R confocal microscope.
Model description.
Simulations of cAMP signaling within PMVECs were run using the Virtual Cell environment. The Virtual Cell environment offers a structured platform for the development of mathematical descriptions of biological systems. Here we have utilized this environment to model the synthesis, hydrolysis, diffusion, and buffering of cAMP within the geometry of a cultured PMVEC from a confluent monolayer as well as in other idealized geometries, including a geometry based on measurements of cell thickness and aspect ratios from ex vivo pulmonary endothelial cells (30). Geometric parameters are listed in Table 1. The spatial distribution of cAMP within these geometries was governed by a set of differential equations that were solved using the finite volume solver available within the Virtual Cell environment (38, 78, 79). The models described in the present study, “PMVEC cAMP model” and “PMVEC cAMP model with point source,” are accessible at the Virtual Cell web site, www.vcell.org/vcell_models/published_models.html, under share models/wfeinstein.
Table 1.
Parameters describing cellular geometries used for simulations
| Spherical Cell | PMVEC | IPEC | ExPEC | |
|---|---|---|---|---|
| Maximum length, μm | 18.68 | 38 | 52 | 370 |
| Maximum width, μm | 18.68 | 25 | 22 | 41.5 |
| Maximum height, μm | 18.68 | 9 | 2 | 6.15 |
| Cellular surface area, μm2 | 1,093 | 2,090 | 1,880 | 23,009 |
| Cytosolic volume, μm3 | 2,801 | 2,806 | 255 | 2,804 |
| Nuclear surface area, μm2 | 346 | 505 | 210 | 839 |
| Nuclear volume, μm3 | 608 | 612 | 79 | 590 |
| S/V, μm−1 | 0.39 | 0.74 | 7.37 | 8.21 |
| Mesh nodes | 50 × 50 × 50 | 110 × 80 × 40* | 100 × 50 × 50 | 300 × 50 × 130 |
PMVEC, cultured pulmonary microvascular endothelial cell. Idealized pulmonary endothelial cell (IPEC) geometry is based on images of ex vivo vascular preparations. Expanded pulmonary endothelial cell (ExPEC) geometry is an expanded version of the idealized pulmonary cell such that its volume is similar to that of the PMVEC.
Initial conditions were obtained by running simulations to equilibrium with a basal AC activity level of 60 cAMP·min−1·AC−1 and PDE activity as described below. Under these conditions, basal cAMP levels were ∼50 nM. Forskolin-mediated stimulation of cAMP synthesis was treated as a step increase in AC activity from basal levels to activated levels of 1,200 cAMP·min−1·AC−1. In all simulations, AC was localized to the plasma membrane with a cellular concentration of 7.06 nM, unless otherwise specified. This level of AC activity within a cell was estimated from the rate of forskolin-induced cAMP accumulation as described in results.
PDE was treated as a Michaelis-Menten enzyme with a Km of 2 μM and a maximal cAMP turnover rate of 100 s−1, unless indicated otherwise. To examine the effects of subcellular localization of PDE activity on the spatial spread of cAMP, PDE activity was uniformly distributed within either the subplasmalemmal region, the cytosol, or the perinuclear region, as indicated. Sequestering PDE activity to three distinct distributions allowed us to investigate potential contributions of localized PDE activity to cAMP compartmentalization. In a subset of simulations, AC or PDE activity was concentrated at discrete subcellular locations.
Buffering of cAMP (e.g., cAMP binding to PKA) was described by a mass action equation with k1 = 0.013 μM/s and k−1 = 0.006 s-1, yielding a KD ∼ 0.46 μM or k1 = 0.06 μM/s and k−1 = 0.006 s-1, yielding a KD of 0.1 μM. This allowed us to examine the effects of different buffer affinities on the spatial distribution of cAMP signals. cAMP buffers were uniformly distributed in the cytosol at indicated concentrations. A complete list of parameter values and parameter descriptions as well as relevant references is given in Table 2. The maximal time step was 0.1 s. To ensure simulation accuracy, simulation results were compared with those run with finer mesh size and smaller time steps. Mesh size, maximal time step, and tolerances were adjusted as necessary. All results are from simulations run in three dimensions.
Table 2.
Model parameters and the ranges of model parameters used
| Variable | Cellular Concentration | Kinetic Parameter | Subcellular Location | Reference No. |
|---|---|---|---|---|
| AC | 7.06 nM | cAMP production: | Subplasmalemmal region | (21)* |
| basal, 1.0 nM/s | ||||
| forskolin, 20 nM/s | ||||
| PDE | 2.95 nM | Km = 2 μM; | Subplasmalemmal, cytosolic, or perinuclear region, as indicated | (17, 32)* |
| Kcat = 100 cAMP/s | ||||
| cAMP | Basal [cAMP] ∼ 50 nM | Effective diffusion coefficient = 300, 30, 3, 0.3 μm2/s, as indicated | Diffusible | (7, 15, 61, 66, 71)* |
| cAMP buffers | 0 to 100 μM, as indicated | KD =100 or 460 nM, as indicated | Uniformly distributed within cytosol | (7, 27, 71)* |
AC, adenylyl cyclase; PDE, phosphodiesterase.
Indicated parameter was manually fit to describe data in Fig. 2 or published work, as described in text.
RESULTS
Recent studies have deduced that specificity within the cAMP signaling pathway is achieved, at least in part, by the compartmentalization of cAMP signals to discrete regions of the cell (8, 11, 18, 33, 36, 51, 66, 67, 70, 76, 77, 89). Several cellular mechanisms have been proposed to contribute to cAMP compartmentalization including the following: localization of PDE activity, localization of AC activity, cell shape, cAMP buffering, and restrictions on the effective cAMP diffusion coefficient caused by the local cellular environment (36, 37, 54, 56, 66, 71). Here, we utilize mathematical models of cAMP production, hydrolysis, buffering, and diffusion to investigate potential contributions of these mechanisms to cAMP compartmentalization within PMVECs. Model details are described in materials and methods.
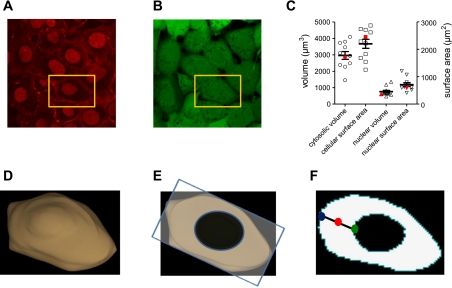
We initially determined the geometry of individual PMVECs from largely confluent monolayers for use in subsequent simulations. The geometry of cultured PMVECs was estimated by three-dimensional reconstruction of image stacks obtained using confocal microscopy. The plasma membrane, nucleus, and cytosolic space were labeled with wheat agglutinin Alexa Fluor 647, DRAQ5, and calcein green AM, respectively (Fig. 1, A and B). Cytosolic volume, cellular surface area, nuclear volume, and nuclear surface area were estimated based upon reconstructions of image stacks (Fig. 1). The PMVEC chosen for subsequent simulations had approximately average cytosolic volume (2,806 μm3), cellular surface area (2,090 μm2), nuclear volume (612 μm3), and nuclear surface area (505 μm2), yielding a surface area-to-volume (S/V) ratio of 0.74 μm−1 (Fig. 1C). Figure 1D shows that the cell geometry chosen for subsequent simulations had the expected “fried egg” appearance of cultured PMVECs.
Fig. 1.
Geometry of cultured pulmonary microvascular endothelial cells (PMVECs). Geometries were based on measurements of a series of slices (z-stacks, confocal microscopy). A: wheat agglutinin Alexa Fluor 647 and DRAQ5 were used to label plasma membranes and nuclei of confluent PMVEC monolayers. B: calcein green was used to label the intracellular space. C: the distribution of dimensions for individual PMVECs. Cytosolic volume was estimated as the intracellular volume minus the nuclear volume. D: three-dimensional geometry of the cultured PMVEC used in subsequent simulations (cell indicated by gold rectangle in A). This cell had average volume, surface area, nuclear volume, and nuclear surface area (indicated in red symbols in C). E: an individual slice through this cell from which results of subsequent simulations are displayed. All simulations were run in three dimensions. F: to simplify the representation of four-dimensional data, we present the time course of cAMP signals at three distinct subcellular locations within the plane depicted in E: at the plasma membrane, within the cytosol, and in the perinuclear space, indicated by the blue, red, and green circles, respectively.
Results presented herein are from simulations of cAMP signaling in a three-dimensional cell. However, it is difficult to portray four-dimensional data (X, Y, Z, and time). Here, we have used two approaches to represent simulation results. In the first approach, simulation results are presented as the time course of cAMP signals at three distinct subcellular locations (at the plasma membrane, within the bulk cytosol, and in the perinuclear space, indicated by blue, red, and green circles, respectively) from a horizontal plane of the cell (Fig. 1, E and F). This approach simplifies the display of four-dimensional data while allowing accurate assessment of the time course and amplitudes of cAMP signals at different locations within the cell. In the second approach, simulation results are presented as the difference between the maximum and minimum cAMP levels within the cell, normalized to the maximum cAMP level (e.g., Fig. 4). This approach allows convenient display of spatial differences in agonist-induced cAMP levels caused by systematic changes in model parameters (e.g., the Km of PDE activity).
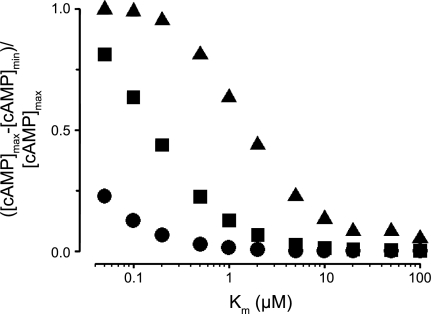
Fig. 4.
Decreased Km of PDE activity potentiates formation of cAMP gradients. Normalized differences between maximum and minimum cAMP levels ([cAMP]max and [cAMP]min) from simulations with effective cAMP diffusion coefficients of 300 (●), 30 (■), and 3 μm2/s (▴) are shown. Simulations were evaluated at 5 min following activation of AC. PDE and AC activities were uniformly distributed within the cytosol and along the plasma membrane, respectively. Km values for PDE4, the predominant PDE isoform in PMVECs, were typically between 1 and 5 μM.
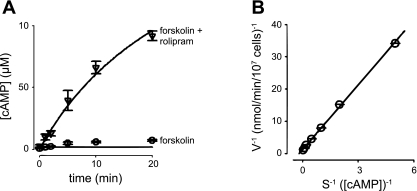
We next measured the time course of forskolin (10 μM)-induced cAMP accumulation in PMVECs pretreated for 10 min with either vehicle control (DMSO) or the PDE4-specific inhibitor rolipram (10 μM) because PMVECs primarily express PDE4 (99). In the absence of PDE inhibitors, forskolin-induced cAMP accumulation reached a plateau within 10 min. However, in the presence of rolipram, cAMP continued to accumulate for 20 min. (Fig. 2A). Similar results were observed in cells pretreated with both rolipram (10 μM) and the nonspecific PDE inhibitor IBMX (500 μM, data not shown). The time frame of cAMP accumulation is consistent with the time frame in which PKA phosphorylates near-membrane targets in several cellular systems (8, 68, 74). To constrain subsequent models, we estimated the total PDE activity in PMVECs (Fig. 2B). Data fits were made using standard curve fitting approaches and the least squared error criteria. On the basis of these fits, we estimated the maximal PDE4 activity in PMVECs to be ∼26 μM/min, consistent with previous studies (99). This result is similar to our estimate of the maximal endogenous PDE activity in human embryonic kidney-293 (HEK-293) cells (∼30 μM/min, not shown). This level of PDE activity is consistent with the PDE concentration used in subsequent simulations (2.95 nM, unless otherwise indicated). With this level of PDE activity, 7.06 nM AC with a turnover number of 20 cAMP·AC−1·s−1 were required to approximate the time course of cAMP accumulation. The parameters in this model are not unique. Twofold increases or decreases in both AC and PDE activities had little effect on the quality of the fit or intracellular cAMP gradients unless the effective cAMP diffusion coefficient was substantially decreased (see Figs. 4 and 5).
Fig. 2.
cAMP accumulation and phosphodiesterase (PDE) activity in PMVECs. A: time course of intracellular cAMP accumulation triggered by addition of 10 μM forskolin following 10 min of pretreatment with vehicle (○) or 10 μM rolipram (a PDE4 inhibitor, ▿). The basic model used to describe the spatial spread of cAMP (solid line) adequately fits overall cAMP accumulation. Simulations including cAMP buffers better fit the time course of cAMP accumulation in the absence of rolipram (not shown). B: cAMP PDE activity measured in PMVECs as described in materials and methods (○). Solid lines represent curve fits to data. On the basis of the fits, Km and Vmax of PDE activity were estimated to be 4.5 μM and 0.71 nmol·min−1·10−7 cells. Data are presented as means ± SE.
Fig. 5.
Effects of increasing AC and PDE activities on cAMP signals. A: the kinetics of total cellular cAMP accumulation become faster as the levels of AC and PDE activities are increased. B: normalized differences between maximum and minimum cAMP levels within the cell determined 5 min following activation of AC. The open circle depicts estimated AC and PDE activities from cultured PMVECs. In these simulations, AC activity was uniformly distributed along the plasma membrane, PDE activity was distributed throughout the cytosol, and the effective cAMP diffusion coefficient was 300 μm2/s. AC and PDE activities are as indicated.
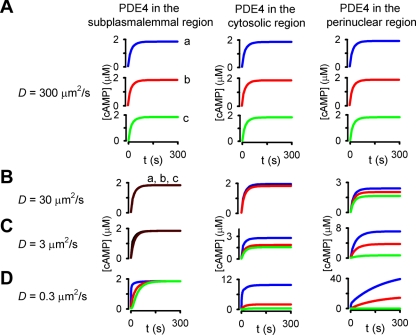
We then modeled the effects of altering the subcellular distribution and kinetics of PDE activity on cAMP gradients. These simulations were conducted with an effective cAMP diffusion coefficient of 300 μm2/s, which is the effective diffusion coefficient of cAMP in bulk cytoplasm (2, 15, 47). Initial simulations used PDE activity with a Km of 2 μM and a maximal cAMP turnover rate of 100 s−1. PDE activity was uniformly distributed within either the subplasmalemmal region, the cytosolic region, or the perinuclear region (Fig. 3A). Regardless of the subcellular distribution of PDE activity, the time course of cAMP signals was nearly identical throughout the cell and little or no cAMP gradients were observed. These simulations indicate that in the absence of other contributing factors, the levels of PDE4 activity observed in PMVECs are unlikely to facilitate substantial cAMP gradients.
Fig. 3.
Simulations depicting the effects of subcellular PDE distribution on spatial spread of cAMP signals. The time course of agonist-induced cAMP accumulation at three subcellular locations, the plasma membrane (blue line, a), the cytosol (red line, b), and the perinuclear region (green line, c), as depicted in Fig. 1F, is shown. In these simulations, adenylyl cyclase (AC) activity was uniformly distributed along the plasma membrane; PDE activity was distributed uniformly in either the subplasmalemmal, cytosolic, or perinuclear regions (left, center, and right columns, respectively); and the effective cAMP diffusion coefficient (D) was 300, 30, 3, and 0.3 μm2/s (A, B, C, and D, respectively). Decreasing the effective cAMP diffusion coefficient D from 300 to 0.3 μm2/s facilitated the formation of cAMP gradients regardless of the subcellular distribution of PDEs.
Several studies have presented data that are consistent with the hypothesis that the effective diffusion coefficient of small molecules (including cAMP) is lower than would be expected in cytosol (5, 9, 20, 42, 44, 48, 49, 55, 61, 66). Thus, we examined the effects of altering the subcellular distribution of PDE activity in simulations in which the effective diffusion was reduced 10- to 1,000-fold (Fig. 3, B–D). When PDE activity was constrained to the subplasmalemmal region, relatively small cAMP gradients were observed, even when the effective diffusion coefficient was reduced 1,000-fold. The cAMP gradients that were observed were primarily due to a reduction in the effective diffusion coefficient, not membrane-localized PDE activity. This result was not surprising given that PDE was effectively colocalized with AC. However, when PDE activity was distributed throughout the cytosol or constrained to the perinuclear region, and the effective cAMP diffusion coefficient was reduced 100-fold, substantial differences in cAMP levels were observed at the three indicated subcellular locations, indicating the formation of substantial cAMP gradients.
We next sought to assess the effects of altering the affinity of PDEs for cAMP on cAMP gradients. The magnitude of cAMP gradients is presented as the maximum cAMP level minus the minimum cAMP level normalized to the maximum cAMP level 5 min following activation of AC (Fig. 4). We observed that increasing the affinity of PDEs for cAMP had little effect on cAMP gradients when the effective diffusion coefficient was 300 μm2/s, but it had substantial effects on cAMP gradients at effective diffusion coefficients of 3 and 0.3 μm2/s. These simulations indicate that without other contributing factors, the activities of high-affinity PDEs such as PDE3 and PDE7 are not sufficient to generate physiologically significant cAMP gradients in pulmonary endothelial cells.
Computational studies have demonstrated that high PDE levels can contribute to cAMP gradients when the effective cAMP diffusion coefficient is ∼300 μm2/s (40, 54, 56, 73). These computational studies are conceptually consistent with experimental studies that demonstrated that high levels of PDE activity generate cGMP gradients in retinal rod outer segments (12). Thus, we sought to determine whether our models of PMVECs were consistent with previous models. As would be expected, increasing either PDE or AC activities alone significantly altered the amplitude of total cAMP accumulation (not shown). Increasing both AC and PDE activities in parallel caused a substantial acceleration of the overall kinetics of the cAMP response (Fig. 5A) and an increase in cAMP gradients evaluated 5 min after AC activation (Fig. 5B). However, the levels of AC and PDE required to generate biologically significant gradients, i.e., cAMP levels high enough to activate PKA in one region of the cell without activating PKA in the rest of the cell, were only achieved at AC and PDE activities 100- to 500-fold greater than those measured in PMVECs.
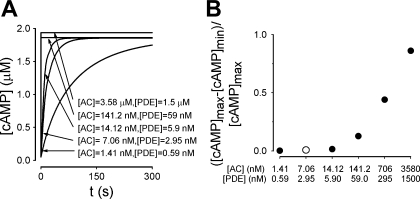
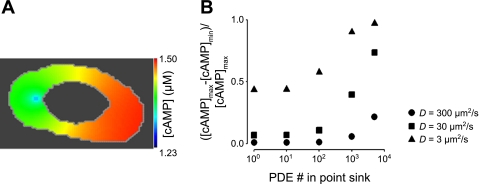
To further explore potential contributions of localized PDE activity to the generation of cAMP gradients, we modeled PDE activity at a discrete point within the cell. This is analogous to the binding of PDE4 to scaffolding domains such as A kinase anchoring proteins (AKAPs) (22, 35, 95). In these simulations, the total number of PDEs (and therefore the maximal PDE activity) remained constant, and only the distribution changed. For example, if 100 PDEs were localized within the point source, then 4,900 PDEs were uniformly distributed within the cytosol; if 1,000 PDEs were localized to a single location, then 4,000 were uniformly distributed within the cytosol. AC activity was uniformly distributed along the plasma membrane, and the effective cAMP diffusion coefficient was 300 μm2/s. Interestingly, when all of the PDE activity was localized into one point, the minimum cAMP level was only 21% lower than the maximum level 5 min after AC activation (Fig. 6). When the localized PDE activity was further increased or the effective diffusion coefficient was lowered, localized PDE activity was able to generate substantial cAMP gradients (Fig. 6B).
Fig. 6.
Effects of localized PDE activity on cAMP gradients. In these simulations a subset of PDE activity was concentrated at a specific subcellular location (a point sink) while total cellular PDE activity remained constant. For example, if 100 PDEs were localized within the point source, then 4,900 PDEs were uniformly distributed within the cytosol; if 1,000 PDEs were localized to a single location, then 4,000 were uniformly distributed within the cytosol. AC activity was evenly distributed along the plasma membrane. A: simulated distribution of cAMP within a PMVEC when the total cellular PDE activity (295 nM/s) was sequestered into one location. D = 300 μm2/s. B: effects of increasing localized PDE activity (while total cellular PDE activity remained constant) on normalized differences between maximum and minimum cAMP levels at effective cAMP diffusion coefficients of 300 (●), 30 (■), and 3 μm2/s (▴). Simulations were evaluated 5 min following AC activation. Minimum cAMP levels for simulations run at each effective diffusion coefficient (t = 5 min) were as follows: D = 300 μm2/s: 1.36 ≤ [cAMP]min ≤ 1.41 μM; D = 30 μm2/s: 1.02 ≤ [cAMP]min ≤ 1.38 μM; D = 3 μm2/s: 0.26 ≤ [cAMP]min ≤ 1.16 μM.
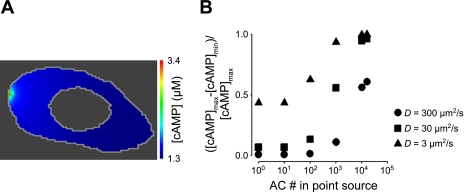
In the simulations presented above, AC activity was assumed to be uniformly distributed throughout the plasma membrane. However, PMVECs primarily express AC6 (16, 82), a Ca2+-inhibitable AC that localizes within caveolae (25, 26, 58, 62, 74). Caveolae are flask-shaped invaginations of the plasma membrane ranging in diameter from 50 to 150 nm (3) that allow clustering of proteins such as G protein-coupled receptors and AC to discrete domains within the plasma membrane (39, 58). Thus, we sought to simulate the effects of localizing AC activity to discrete regions of the plasma membrane. In these simulations, basal AC activity was uniformly distributed throughout the plasma membrane (as in the simulations described above), and agonist-stimulated AC activity was treated as an isolated point source and uniformly distributed along the plasma membrane. The maximal AC activity was held constant (e.g., 1,000 AC localized to the point source and 11,000 uniformly distributed along the plasma membrane). PDE activity was uniformly distributed throughout the cytosol, and the effective cAMP diffusion coefficient was 300 μm2/s. When 100 ACs (∼0.7% of the AC activity that was estimated in PMVECs) were localized within a 100-nm square region of the plasma membrane—similar to the packing of acetylcholine receptors within the electric organ of Torpedo marmarota (31)—only small cAMP gradients were produced within 5 min of AC activation (Fig. 7B). As the rate of cAMP production from the point source was increased, larger cAMP gradients were observed. At the point when all of the AC activity in the cell was localized to a single point, large cAMP gradients were observed within the cell. Reduction in the effective cAMP diffusion coefficient increased the magnitude of cAMP gradients at all levels of AC activity (Fig. 7B).
Fig. 7.
Effect of localized AC activity on the spatial spread of cAMP signals. AC activity was concentrated within a single location on the plasma membrane while total cellular AC activity remained constant. PDE activity was evenly distributed throughout the cytosol. A: the spatial distribution of cAMP signals 5 min following activation of AC. Total cellular AC activity (141.2 nM/s) was sequestered at one location. D = 300 μm2/s. B: effect of increasing localized AC activity on normalized differences between maximum and minimum cAMP levels with effective cAMP diffusion coefficients of 300 (●), 30 (■), and 3 (▴) μm2/s. Minimum cAMP levels for simulations run at each effective diffusion coefficient (time, t, = 5 min) were as follows: D = 300 μm2/s, 1.33 ≤ [cAMP]min ≤ 1.41 μM; D = 30 μm2/s: 0.96 ≤ [cAMP]min ≤ 1.38 μM; D = 3 μm2/s: 0.033 ≤ [cAMP]min ≤ 1.16 μM.
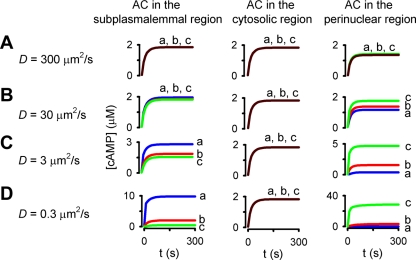
The simulations presented in Fig. 7 examine the effects of localizing AC activity to discrete points at the plasma membrane. While this is a reasonable approximation for the distribution of endogenous AC6 (16, 82), it is not representative of the endogenous soluble AC (sAC) or the pathological situation in which cells exposed to Pseudomonas aeruginosa are intoxicated with the secreted exotoxic AC, ExoY (77). To better describe the contributions of sAC constructs to cAMP compartmentalization, we ran a series of simulations in which AC was uniformly distributed at the plasma membrane, in the cytosol, or in the perinuclear space. In each case, total AC activity remained constant and PDE activity was uniformly distributed within the cytosol. Regardless of AC localization, little or no cAMP gradients were observed when the effective cAMP diffusion coefficient was 300 or 30 μm2/s (Fig. 8, A and B). However, when the effective diffusion coefficient was reduced to 3 or 0.3 μm2/s, substantial cAMP gradients were observed (when AC was localized to either the plasma membrane or the perinuclear space) (Fig. 8, C and D). Thus, simulations presented to this point indicate that differential localization of AC and PDE activities can indeed contribute to cAMP gradients, but only when either high levels of AC and PDE activity are present or when the effective cAMP diffusion coefficient is at least 100-fold lower than that measured in salt water.
Fig. 8.
Simulations depicting the effects of subcellular AC distribution on spatial spread of cAMP signals. The time course of agonist-induced cAMP accumulation at three subcellular locations, the plasma membrane (blue line, a), the cytosol (red line, b), and the perinuclear region (green line, c), as depicted in Fig. 1F, is shown. In these simulations, AC activity was uniformly distributed along the plasma membrane, within the cytosol, or in the perinuclear region (left, center, and right columns, respectively); PDE activity was distributed uniformly in the cytosol; and the effective cAMP diffusion coefficient was 300, 30, 3, and 0.3 μm2/s (A, B, C, and D, respectively). Decreasing the effective cAMP diffusion coefficient D from 300 to 0.3 μm2/s facilitated the formation of cAMP gradients when AC activity was located at the plasma membrane or in the perinuclear region.
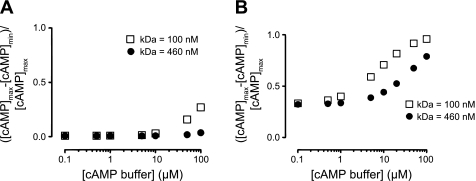
Several mechanisms can contribute to a reduced effective cAMP diffusion coefficient. They include the buffering of cAMP by binding proteins (e.g., PKA), structural impediments (e.g., diffusion through a cytoskeletal meshwork), and changes in the viscosity of cytoplasm. Here we separate the effects of buffering from other impediments to diffusion. Specifically, we have included known concentrations of a fixed buffer (e.g., PKA bound to AKAPs) with cAMP affinities of 100 or 460 nM uniformly distributed within the cytosol. AC activity was either uniformly distributed along the plasma membrane (Fig. 9A) or localized to a single point source at the plasma membrane (Fig. 9B), and PDE activity was uniformly distributed within the cytosol. The simulations clearly demonstrate that when the effective cAMP diffusion coefficient was 300 μm2/s and AC activity was distributed along the plasma membrane, little or no cAMP gradients were generated with high- or low-affinity buffers, even with 100 μM cAMP binding protein distributed throughout the cell (Fig. 9A). When the effective diffusion coefficient was reduced to 3 μm2/s, cAMP gradients were augmented by buffer concentrations >10 μM (not shown). These results may seem surprising given the extensive literature describing the effects of Ca2+ buffering on the formation of Ca2+ gradients (e.g., see Refs. 53, 94, and 98). However, one must consider that Ca2+ channels allow Ca2+ fluxes on the order of 103 to 105 ions/s, whereas AC produces cAMP at rates on the order of 60 cAMP/s (see Ref. 66). Thus we considered the case in which 100 ACs were clustered (e.g., a reasonable upper limit of AC clustering within caveolae) and the effective cAMP diffusion coefficient was 300 μm2/s. Under these conditions, cAMP gradients were augmented by buffer concentrations >10 μM, with the higher-affinity buffers having a greater effect. Cyclic AMP gradients were further augmented at lower effective diffusion coefficients (not shown).
Fig. 9.
Effect of cAMP buffering on the spatial spread of cAMP signals. Normalized differences between maximum and minimum cAMP levels were evaluated in simulations that included cAMP buffering proteins, with KD = 460 nM (●) or KD = 100 nM (□). Buffering proteins were fixed and uniformly distributed within the cytosol. A: cAMP differences occurring when AC activity was uniformly distributed along the plasma membrane. B: cAMP differences occurring when AC activity was localized to a discrete location. PDE activity was evenly distributed in the cytosol, and the effective diffusion coefficient was 300 μm2/s. Simulations were evaluated 1 min following AC activation.
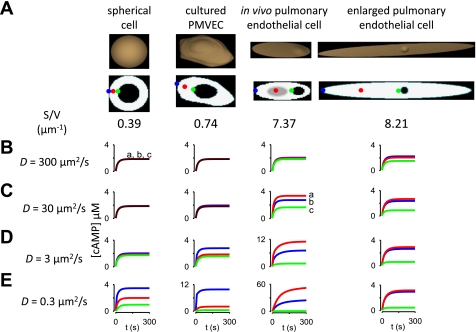
While the localization of AC and PDE activities, cAMP buffering, and reduction in the effective cAMP diffusion coefficient are likely to contribute to the generation of cAMP gradients, other factors may also be involved. Previous experimental and theoretical studies have deduced that cell shape may slow the spatial spread of second messenger signals such as Ca2+ and cAMP in dendrites and dendritic spines (54, 84, 85). Here we sought to determine whether cell shape, and more specifically S/V ratio, contributed to the generation of cAMP gradients in PMVECs. We examined the effects of cell shape in four different geometries (Fig. 10A): 1) a spherical cell with a S/V ratio = 0.39 μm−1, 2) a cultured PMVEC with a S/V ratio = 0.74 μm−1 (described above), 3) an idealized pulmonary endothelial cell based on ex vivo volume and surface area measurements from intact segments of the pulmonary vasculature with a S/V ratio = 7.37 μm−1 (30), and 4) an idealized pulmonary endothelial cell that was expanded to the volume of cultured PMVECs, with S/V ratio = 8.21 μm−1. The idealized pulmonary endothelial cell has an exquisitely thin region (Fig. 10A, indicated in gray) that represents observed changes in endothelial cell thickness in ex vivo lung preparations (30). In these simulations, AC and PDE activities were uniformly distributed along the plasma membrane and throughout the cytosol, as described above.
Fig. 10.
Time course of cAMP signals at different subcellular locations in different cellular geometries. A: cellular geometries and two-dimensional slice from a spherical cell (first column), a cultured PMVEC (second column), an idealized pulmonary endothelial cell (third column), and an enlarged endothelial cell that has a similar volume to a cultured PMVEC (fourth column). Surface area to volume (S/V) ratios are as indicated. The gray region in the idealized pulmonary endothelial cell (third column) represents the location of a 30 nm thick region within the cell. This exquisitely thin region represents observed changes in endothelial cell thickness in ex vivo preparations (30). B–E: time course of cAMP accumulation at three subcellular locations [a (plasma membrane), b (cytosol), and c (perinuclear); blue, red, green lines, respectively] in simulations in which the effective cAMP diffusion coefficient was set to 300 (B), 30 (C), 3 (D), and 0.3 (E) μm2/s.
In cells with a low S/V ratio (the spherical cell and cultured PMVEC), no substantial cAMP gradients were observed when the effective cAMP diffusion coefficient was 300, 30, or 3 μm2/s (Fig. 10, B–D). However, when the effective cAMP diffusion coefficient was lowered to 0.3 μm2/s, cAMP gradients were apparent (Fig. 10E). In cells with higher S/V ratios, substantial cAMP gradients were observed when the effective cAMP diffusion coefficient was 30 μm2/s. This was particularly notable in the thin region of the idealized pulmonary endothelial cell. In this region the S/V ratio was increased, thus increasing the ratio of cAMP synthesis to cAMP hydrolysis (AC activity/PDE activity) and local cAMP concentration. The magnitude of cAMP gradients further increased as the effective diffusion coefficient was reduced. These results are consistent with previous studies indicating that cell shape (and S/V ratio) can impact the localization of cAMP signals (54).
The simulations presented thus far were designed to look at the effects of specific components that could underlie compartmentalized cAMP signals in PMVECs. On the basis of these simulations and measured values of total cellular cAMP accumulation and PDE activity, we propose a plausible model for the spatial spread of cAMP signals in cultured PMVECs. In this model, AC activity was sequestered into 100 point sources (analogous to 100 caveolae); PDE activity was distributed between the plasma membrane (30%), cytosol (50%), and perinuclear region (20%); and 1 μM high-affinity cAMP buffer (KD = 100 nM) was uniformly distributed in the cytosol. Simulations of this model suggest that substantial cAMP gradients begin to form when the effective cAMP diffusion coefficient is <3 μm2/s (Fig. 11), consistent with the simulations presented above. On the basis of these simulations, we feel that a reasonable effective cAMP diffusion coefficient would be ∼2 μm2/s. Under these conditions, cAMP gradients would be generated in response to activation of AC (Fig. 12, A and C). These gradients would lead to activation of PKA (or EPAC) in the near-membrane region without significant activation of PKA in the cytosolic or perinuclear regions. As such, these results are consistent with previous studies indicating that the production of cAMP in the near-membrane space strengthens endothelial barrier function (16, 76, 77). Simulations also indicate that inhibition of PDE4 activity with 10 μM rolipram [inhibition constant (Ki) = 0.1 μM] would allow cAMP levels to increase throughout the cell (Fig. 12, B and D). These results are consistent with previous studies demonstrating that activation of AC with forskolin and inhibition of PDE4 activity with 10 μM rolipram (or a dominant negative PDE4D construct) allow cAMP-mediated regulation of cytosolic (microtubule associated) proteins and subsequent endothelial barrier disruption (19, 74).
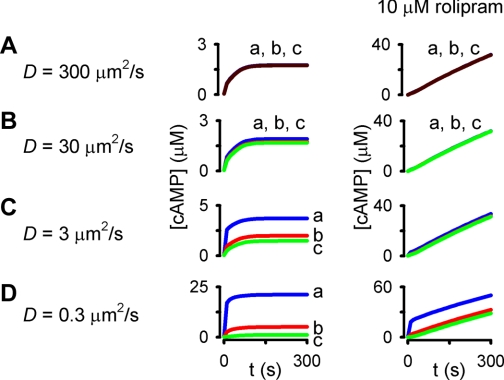
Fig. 11.
Effects of altering the effective cAMP diffusion coefficient on a plausible model of cAMP signaling in PMVECs. Simulations are analogous to those presented in Fig. 10: simulations of a cell in which 12,000 ACs were sequestered into 100 locations at the plasma membrane. PDE activity was distributed at the plasma membrane (30%), within the cytosol (50%), and in the perinuclear space (20%). A total of 1 μM of a fixed buffer (KD = 100 nM) was present in the cell. A–D: the effective diffusion coefficient was varied from 300 to 0.3 μm2/s as indicated. The right column depicts simulations in which 10 μM rolipram (a PDE4 inhibitor with Ki = 0.1 μM) was present.
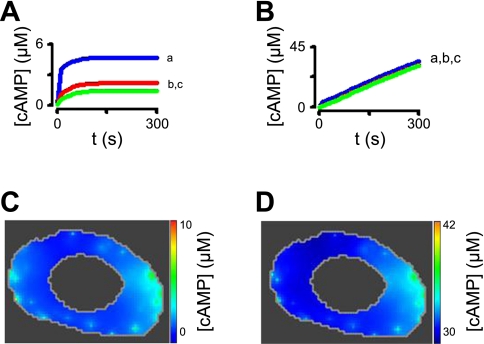
Fig. 12.
Plausible description of the spatial spread of cAMP signals in PMVECs. A and C: simulations of a cell in which a total of 12,000 ACs were sequestered into 100 locations at the plasma membrane. PDE activity was distributed at the plasma membrane (30%; a), within the cytosol (50%; b), and in the perinuclear space (20%; c). A total of 1 μM of a fixed buffer (KD = 100 nM) was present in the cell, and the effective diffusion coefficient was 2 μm2/s. Under these conditions, activation of AC initiated the generation of substantial cAMP gradients. B and D: simulations depicting the effects of treatment with 10 μM of a PDE inhibitor with a Ki of 0.1 μM (e.g., rolipram). Pretreatment with a PDE inhibitor allowed cAMP to accumulate throughout the cell. The spatial spread of cAMP 5 min following activation of AC is depicted in C and D.
DISCUSSION
Since the discovery of cAMP, investigators have speculated about the nature of mechanisms that underlie specificity within the cAMP signaling pathway (63, 64, 83). Experimental evidence supporting the hypothesis that cAMP signals are compartmentalized was provided nearly 20 years later (11, 18). It was another 10 years before cGMP gradients were measured in retinal rod outer segments (12, 29), and 15 years before cAMP gradients were deduced from the differential activation of discrete pools of PKA within single cells (36). Even then, the magnitude of restrictions or limits on the spatial spread of cAMP signals was not well understood until quantitative methods for assessing local cAMP concentrations were developed (66, 67). In the past decade, a variety of studies have provided evidence that both cAMP and cGMP signals are compartmentalized (8, 37, 51, 66, 67, 70, 76, 86, 89, 91). More recently, data have suggested that gradients in PKA activity may occur at the leading edge of migrating cells (60). On the basis of work presented in these and other studies, investigators have proposed several mechanisms that may contribute to the compartmentalization of cAMP signals: high cellular levels of AC and PDE activities, localization of AC and PDE activities to discrete regions of the cell, restrictions on cAMP diffusion, cAMP buffering, and cellular shape. However, conceptual models of signaling systems often have underlying assumptions that are not well described. This has hindered our understanding of specificity within the cAMP signaling pathway. Here, we describe mathematical models developed to evaluate the potential contributions of each of these mechanisms to cAMP compartmentalization. Simulations suggest that localization of AC and PDE activities, buffering by PKA, and cell geometry are by themselves insufficient to generate cAMP gradients in cultured PMVECs. Using these models we have examined the interplay between these mechanisms and other potential restrictions on the spatial spread of cAMP. On the basis of both experimental studies of cAMP signaling in PMVECs and model simulations, we conclude that restrictions on the spatial spread of cAMP signals by high concentrations of cAMP buffers, changes in cytosolic viscosity, or structural impediments may facilitate the formation of cAMP gradients. This is in stark contrast to recent modeling studies in which the authors concluded that restrictions on cAMP diffusion are not required for the formation of cAMP gradients (54, 56). Below we discuss how differences in model assumptions contribute to the disparate conclusions.
Localized AC and PDE activities are unlikely to be solely responsible for cAMP gradients in PMVECs.
Recent mathematical descriptions of cAMP signals have relied on high levels of AC and PDE activities to describe cAMP gradients (40, 54, 56). The authors of these studies in turn concluded that restrictions on cAMP diffusion were not required for the generation of cAMP gradients. The simulations presented here suggest that without high AC and PDE activities, the spatial spread of cAMP must be constrained by other mechanisms.
The assumption of high enzyme concentrations has been justified, at least in part, by the high guanylyl cyclase (GC) and PDE concentrations in rod outer segments (45, 50, 96). In this system, high enzyme concentrations are capable of generating cGMP gradients (12, 29). Cyclic GMP gradients in retinal rods are further accentuated by a 10- to 100-fold reduction in the longitudinal cGMP diffusion coefficient, due in part to buffering by the noncatalytic cGMP binding site of PDE6, hindered diffusion or increased tortuosity caused by the structure of the rod outer segment, and possibly, altered viscosity of cytoplasm (57, 92). While high GC and PDE concentrations have clearly been demonstrated in rod outer segments, they do not necessarily exist in other cell types. For example, estimates of maximal AC and PDE activities in rat ventricular myocytes were ∼25 and 12 μM/min, respectively (62, 65). These levels are comparable with our estimates of maximal AC and PDE activities in PMVECs of ∼8.5 and 26 μM/min, respectively. Similarly, we estimate that the maximal PDE activity in HEK-293 cells is ∼30 μM/min. However, Oliveira et al. (56) assumed a Vmax for PDE4 activity 500- to 1,000-fold higher than our measured value, with a similar Km, in their model of cAMP signaling in HEK-293 cells. The use of high AC and PDE activities allowed the formation of cAMP gradients without substantial contributions from buffering or slowed cAMP diffusion. The models presented here also indicate that high AC and PDE activities could form cAMP gradients. It should be noted that phosphorylation of PDE4 by PKA can lead to an increase in PDE activity. We have measured a PKA-mediated, two- to threefold increase in PDE4 activity in both HEK-293 cells and PMVECs (68, 93, 99). In PMVECs, PKA-mediated phosphorylation also may inhibit endogenous AC6 activity (16). However, based on our in vitro measurements of cAMP accumulation and PDE activity, it is unlikely that AC and (PKA-stimulated) PDE levels are high enough in PMVECs to generate substantial cAMP gradients in the absence of diffusional restrictions.
High AC and PDE concentrations may occur in other systems, especially in specialized cellular domains such as neuronal processes. Neves et al. (54) recently proposed a model of cAMP signals within hippocampal neurons in which the PDE activity within the processes was set to 480 μM/min. It is possible that high levels of PDE activity could be reached within the confined spaces of neuronal processes, although the actual localized enzymatic activity within these spaces has not been measured.
Subcellular localization of AC and PDE activities.
Early studies indicated that ACs are localized within spatially isolated regions of cells such as olfactory cilia and neuronal synapses (52, 59). These observations led to the suggestion that the subcellular localization of AC activity is an important component in signaling specificity. Subsequent studies have demonstrated that AC activity is both physically and functionally sequestered within distinct regions of the plasma membrane, e.g., caveolae (16, 24, 58, 74, 80). Recent evidence indicates that ACs are also localized via binding to protein scaffolds such as A kinase anchoring proteins (AKAPs) (23). The functional importance of AC localization has been clearly demonstrated in PMVECs (74, 76, 77). In these cells, activation of either endogenous or heterologously expressed, plasma membrane resident ACs facilitates endothelial barrier enhancement. In contrast, activation of soluble ACs—whether endogenous sAC, bacterial exotoxin ExoY, or heterologously expressed, forskolin-activated, soluble AC—triggers endothelial barrier disruption (75–77). These observations are analogous to the segregated cGMP signals observed upon activation of soluble or particulate GC (13, 61, 100). The simulations presented here demonstrate that localized AC activity can indeed facilitate the generation of cAMP gradients. However, the gradients only become physiologically relevant when other factors, such as buffering and additional restrictions on cAMP diffusion, contribute to the localization of cAMP signals.
Similarly, specific PDE subtypes bind to different scaffold proteins, have distinct subcellular localizations, and regulate specific cellular functions (1, 4, 8, 22, 46, 51, 69, 70, 86, 95). As such, PDE activity is likely a critical factor in determining specificity within the cAMP pathway. Yet, the simulations presented herein indicate that the maximal PDE activities in PMVECs are unable to generate physiologically meaningful cAMP gradients without other contributing factors. This observation held true even when all cellular PDE activity was concentrated at a single location.
Hindered cAMP diffusion.
Several studies have provided data consistent with the hypothesis that the concentrations of small diffusible molecules, including cAMP, cGMP, ATP, and Na+, are significantly different in the near-membrane space than in other cellular locations (5, 9, 20, 44, 61, 66, 67). This reduction in the effective diffusion coefficient is far greater than had been assessed to steric hindrance due to intracellular structures (48, 49, 55). The reasons for these differences have not yet been determined. However, on the basis of the activities of the enzymes involved, it seems likely that the effective diffusion coefficients of these molecules are lower than would be expected. Reduction in the effective diffusion coefficient may occur due to several factors, including structural impediments to diffusion, local changes in viscosity, electrostatic effects, and buffering (discussed below). Structural impediments that may hinder the spatial spread of small molecules include organelles, clustered proteins, cytoskeletal networks, and the F-actin cortical rim. Structural components may also alter the local electrostatic environment and local viscosity, e.g., F-actin gelation (10, 34, 97). The extent to which structural elements reduce the effective cAMP diffusion coefficient is not known and may well depend on cell type (42, 48, 49, 55).
In the models presented here we have assumed that the effective cAMP diffusion coefficient is uniform throughout the cell. This allowed an assessment of mechanisms that may affect the spatial spread of cAMP. However, it should be noted several studies have concluded that reductions in the effective diffusion coefficient may be more prominent between the near-membrane space and bulk cytosol than within the bulk cytosol (33, 66, 67, 73, 89). This may explain the success of compartmental models in describing the physiological effects of localized cAMP signals (33, 61, 66, 67, 71, 73).
Cyclic AMP buffering.
While contributions of both structural impediments and changes in cytosolic viscosity to the overall reduction in the effective cAMP diffusion coefficient have been difficult to estimate, the contributions of cAMP buffering by known buffering proteins are more easily assessed. Cyclic AMP is believed to bind primarily to effector proteins including PKA, protein kinase G (PKG), exchange protein activated by cAMP (EPAC), noncatalytic binding sites of PDEs, cyclic nucleotide-gated channels, as well as other ion channels. In most cell types, these proteins are low affinity (KD > 5 μM), low abundance (cellular concentrations < 1 μM), or both. Of these, the regulatory subunit of PKA is the most likely to contribute significantly to buffering due to its relatively high cellular concentration of cAMP binding sites, typically 1 to 2 μM, and apparent cAMP binding affinity, typically between 0.1 and 1 μM (7, 27). On the basis of the simulations presented in Figs. 9 and 12, PKA buffering has little effect on cAMP gradients without contributions from other factors. As such, these simulations are consistent with our previous conclusion that buffering by known cAMP effectors is insufficient to account for the slow spatial spread of cAMP within cells (66, 67). These simulations are also consistent with the simulations of cAMP signaling in cardiac myocytes presented by Saucerman et al. (73), in which both cAMP buffering by PKA and reductions in the effective diffusion coefficient were required to describe functional differences between isoproterenol- and prostaglandin-induced cAMP signals.
Cellular shape.
The relationship between cellular S/V ratio and enzyme activity (higher S/V ratios allow greater concentrations of plasma membrane-associated enzymes with respect to cell volume) has been well established (e.g., see Ref. 90). Recent studies have found that this is also true for neuronal processes including retinal rod outer segments, olfactory cilia, and dendritic spines (28, 43, 50, 84, 85, 96). More recently, Neves et al. (54) investigated the effects of cell shape, including S/V ratio, on information flux through signaling cascades in dendrites. Their simulations demonstrate that as the S/V ratio is decreased, due to an increase in the diameter of an idealized dendrite, signal gradients collapsed. These studies highlight the importance of overall cell shape. However, local cell shape may also impact second messenger gradients. For example, the sequestration of signaling proteins such as AC may not be the only effect caveolae have on compartmentalized cAMP signals. The shape of caveolae, especially when in clusters, may facilitate signal localization by reducing the space into which cAMP can diffuse. Similarly, neuronal processes may form a type of diffusional hindrance, in effect forcing cAMP to diffuse in one dimension (e.g., through the dendrite). The simulations presented here also demonstrate that the S/V ratio does indeed affect the ability of cAMP to accumulate in localized regions of the cell. This was particularly true in the idealized pulmonary endothelial cell, a cell with a region of reduced thickness (30 nm) in which cAMP accumulated to higher levels than in the rest of the cytosolic space (Fig. 10).
Comparison of model simulations with experimental observations.
It has been observed that cAMP produced in different subcellular compartments of PMVECs causes distinct physiological effects. Cyclic AMP produced at the plasma membrane triggers an increase in endothelial barrier integrity; whereas, cAMP produced in the cytosol triggers a reduction in endothelial barrier integrity in both cultured endothelial monolayers and intact lung vasculature (76, 77, 82). In addition, activation of endogenous, plasma membrane-localized AC leads to phosphorylation of near-membrane cytoskeletal proteins such as filamin, but not phosphorylation of cytosolic proteins, such as tau. Conversely, activation of soluble ACs leads to phosphorylation of cytosolic proteins such as tau, but not near-membrane proteins such as filamin (19, 74). These experimental observations support our simulation results, suggesting that activation of plasma membrane-localized AC leads to formation of cAMP gradients that trigger near-membrane PKA activity without substantial increases in PKA activity in other regions of the cell only when the effective cAMP diffusion coefficient is below ∼3 μm2/s (Figs. 3, 8, 11, and 12). Conversely, our model suggests that activation of AC localized in the perinuclear region would trigger cytosolic PKA activity, but not near-plasma membrane PKA activity (Fig. 8).
It has also been observed that, in PMVECs, inhibition of PDE4 activity allows cAMP produced at the plasma membrane to disrupt endothelial barrier integrity and to trigger tau phosphorylation. This is presumably due to the inappropriate spatial spread of cAMP from the near-membrane space into the bulk cytosol. These observations also support the models presented here (Figs. 11 and 12). Interestingly, simulations of these models suggest that while PDE activity alone is insufficient to generate cAMP gradients in PMVECs, PDE activity is required for the formation of cAMP gradients. As such, these models are consistent with observations demonstrating the importance of PDE activity in the localization of cyclic nucleotide signals (6, 8, 19, 36, 61, 66, 67, 70).
Recent studies have demonstrated that disease states such as acute lung injury and asthma cause changes in enzyme localization, enzyme activity, and cell shape. Prolonged treatment with pharmaceutical agents can similarly cause multiple changes to cellular systems. Mathematical models offer the unique ability to isolate the effects of individual parameters on a specific outcome—here we describe the effects of enzyme activity, enzyme localization, cellular shape, and effective diffusion coefficient on cAMP gradients—in a manner that is not readily possible experimentally. For example, it is difficult to predictably alter the subcellular localization of PDE4 without impacting a multitude of cellular processes. The situation becomes more complicated when altering the effective cAMP diffusion coefficient. We have attempted to do this using cytochalasin D to disrupt the F-actin cortical rim. In these experiments, we observed that the lowest concentration of cytochalasin D that would disrupt the cortical rim would also cause cell rounding, lower both AC and PDE activities, and most likely alter AC and PDE localization as well as the effective cAMP diffusion coefficient. These observations illustrate that pharmacological manipulations of enzyme activity often trigger multiple specific and nonspecific cellular responses.
A limitation of most models describing signaling systems, including the models presented here, is that the experimental observations used to constrain models of cellular signaling systems are based on in vitro cultured cell preparations. While it is often necessary to measure enzyme kinetics and localization as well as the signals themselves in vitro, it is often unclear whether the basic conclusions of experimental or modeling studies can be extrapolated to in vivo systems. It is possible, even likely, that enzyme activity, localization, and even isoforms can vary between in vitro and in vivo systems. Cell shape can also be markedly different (see Fig. 10). And the differences do not end there. For example, the models presented here assume that cell shape and effective cAMP diffusion coefficient are constant within the time frame of the signals. These assumptions are unlikely true in vivo. Recent studies by Fredberg and colleagues have suggested that, in response to stretch, both smooth muscle and endothelial cells undergo rapid fluidization of the F-actin network followed by slower resolidification (14, 41). Fluidization of the F-actin network was accompanied by increased macromolecular mobility. Thus, in cells of the lung, which undergo cyclic stretch, both cell shape and viscosity are likely to change with time.
However, even with these limitations, the mathematical models presented here provide a framework with which we can interpret both in vitro and in vivo data. The models suggest that while the differential localizations of AC and PDE activities are required to generate cAMP gradients, they are by themselves insufficient to generate cAMP gradients (except when cyclase and PDE activities are high, e.g., rod outer segments). Taken together, the results presented here also suggest that the localization of AC, PDE, PKA, and target proteins (e.g., the L-type Ca2+ channel) alone is insufficient to ensure signaling specificity within cells. Cell shape, buffering, and other factors that slow the spatial spread of cAMP are necessary to localize the cAMP signals themselves.
GRANTS
This work was supported by National Institutes of Health Grants R01HL094455, P01H066299, and S10RR027535 and American Heart Association Grant 11GRNT7430039.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
W.P.F. and B.Z. performed the experiments; W.P.F., B.Z., S.J.L., S.L.S., and T.C.R. analyzed the data; W.P.F., S.J.L., S.L.S., and T.C.R. interpreted the results of the experiments; W.P.F. prepared the figures; W.P.F. drafted the manuscript; S.J.L., S.L.S., and T.C.R. conception and design of the research; S.J.L., S.L.S., and T.C.R. edited and revised the manuscript; S.J.L., S.L.S., and T.C.R. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank B. M. Slepchenko and L. Loew for valuable advice on the implementation of mathematical models in the Virtual Cell modeling environment. We also thank Drs. J. W. Karpen, T. Stevens, and W. Wagner for valuable discussions about this research and helpful comments on the manuscript.
Footnotes
This article is the topic of an editorial focus by Fiona Murray (52a).
REFERENCES
- 1. Abi-Gerges A, Richter W, Lefebvre F, Mateo P, Varin A, Heymes C, Samuel JL, Lugnier C, Conti M, Fischmeister R, Vandecasteele G. Decreased expression and activity of cAMP phosphodiesterases in cardiac hypertrophy and its impact on beta-adrenergic cAMP signals. Circ Res 105: 784–792, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allbritton NL, Meyer T, Stryer L. Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science 258: 1812–1815, 1992 [DOI] [PubMed] [Google Scholar]
- 3. Anderson RG, Jacobson K. a role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296: 1821–1825, 2002 [DOI] [PubMed] [Google Scholar]
- 4. Ariga M, Neitzert B, Nakae S, Mottin G, Bertrand C, Pruniaux MP, Jin SL, Conti M. Nonredundant function of phosphodiesterases 4D and 4B in neutrophil recruitment to the site of inflammation. J Immunol 173: 7531–7538, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Arnon A, Hamlyn JM, Blaustein MP. Ouabain augments Ca2+ transients in arterial smooth muscle without raising cytosolic Na+. Am J Physiol Heart Circ Physiol 279: H679–H691, 2000 [DOI] [PubMed] [Google Scholar]
- 6. Barnes AP, Livera G, Huang P, Sun C, O'Neal WK, Conti M, Stutts MJ, Milgram SL. Phosphodiesterase 4D forms a cAMP diffusion barrier at the apical membrane of the airway epithelium. J Biol Chem 280: 7997–8003, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Beavo JA, Bechtel PJ, Krebs EG. Activation of protein kinase by physiological concentrations of cyclic AMP. Proc Natl Acad Sci USA 71: 3580–3583, 1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackman BE, Heimann J, Horner K, Wang D, Richter W, Rich TC, Conti M. PDE4D and PDE4B function in distinct subcellular compartments in mouse embryonic fibroblasts. J Biol Chem 286: 12590–12601, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bridge JH, Smolley JR, Spitzer KW. The relationship between charge movements associated with ICa and INa-Ca in cardiac myocytes. Science 248: 376–378, 1990 [DOI] [PubMed] [Google Scholar]
- 10. Brotschi EA, Hartwig JH, Stossel TP. The gelation of actin by actin-binding protein. J Biol Chem 253: 8988–8993, 1978 [PubMed] [Google Scholar]
- 11. Brunton LL, Hayes JS, Mayer SE. Functional compartmentation of cAMP and protein kinase in heart. Adv Cyclic Nucleotide Res 14: 391–397, 1981 [PubMed] [Google Scholar]
- 12. Cameron DA, Pugh EN., Jr The magnitude, time course and spatial distribution of current induced in salamander rods by cyclic guanine nucleotides. J Physiol 430: 419–439, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castro LR, Verde I, Cooper DM, Fischmeister R. Cyclic guanosine monophosphate compartmentation in rat cardiac myocytes. Circulation 113: 2221–2228, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C, Krishnan R, Zhou E, Ramachandran A, Tambe D, Rajendran K, Adam RM, Deng L, Fredberg JJ. Fluidization and resolidification of the human bladder smooth muscle cell in response to transient stretch. PLoS One 5: e12035, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen CH, Nakamura T, Koutalos Y. Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophys J 76: 2861–2867, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cioffi DL, Moore TM, Schaack J, Creighton JR, Cooper DM, Stevens T. Dominant regulation of interendothelial cell gap formation by calcium-inhibited type 6 adenylyl cyclase. J Cell Biol 157: 1267–1278, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem 76: 481–551, 2007 [DOI] [PubMed] [Google Scholar]
- 18. Corbin JD, Sugden PH, Lincoln TM, Keely SL. Compartmentalization of adenosine 3′:5′-monophosphate and adenosine 3′:5′-monophosphate-dependent protein kinase in heart tissue. J Biol Chem 252: 3854–3861, 1977 [PubMed] [Google Scholar]
- 19. Creighton J, Zhu B, Alexeyev M, Stevens T. Spectrin-anchored phosphodiesterase 4D4 restricts cAMP from disrupting microtubules and inducing endothelial cell gap formation. J Cell Sci 121: 110–119, 2008 [DOI] [PubMed] [Google Scholar]
- 20. Despa S, Bers DM. Na/K pump current and Nai in rabbit ventricular myocytes: local Nai depletion and Na buffering. Biophys J 84: 4157–4166, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dessauer CW, Gilman AG. The catalytic mechanism of mammalian adenylyl cyclase equilibrium binding and kinetic analysis of P-site inhibition. J Biol Chem 272: 27787–27795, 1997 [DOI] [PubMed] [Google Scholar]
- 22. Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437: 574–578, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Efendiev R, Samelson BK, Nguyen BT, Phatarpekar PV, Baameur F, Scott JD, Dessauer CW. AKAP79 interacts with multiple adenylyl cyclase (AC) isoforms and scaffolds AC5 and -6 to alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptors. J Biol Chem 285: 14450–14458, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fagan KA, Mahey R, Cooper DMF. Functional co-localization of transfected Ca2+-stimulated adenylyl cyclases with capacitative Ca2+ entry sites. J Biol Chem 271: 12438–12444, 1996 [DOI] [PubMed] [Google Scholar]
- 25. Fagan KA, Mons N, Cooper DMF. Dependence of the Ca2+-inhibitable adenylyl cyclase of C6–2B glioma cells on capacitative Ca2+ entry. J Biol Chem 273: 9297–9305, 1998 [DOI] [PubMed] [Google Scholar]
- 26. Fagan KA, Smith KE, Cooper DMF. Regulation of the Ca2+-inhibitable adenylyl cyclase type VI by capacitative Ca2+ entry requires localization in cholesterol-rich domains. J Biol Chem 275: 26530–26537, 2000 [DOI] [PubMed] [Google Scholar]
- 27. Francis S, Corbin JD. Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit Rev Clin Lab Sci 36: 275–328, 1999 [DOI] [PubMed] [Google Scholar]
- 28. Gold GH. Controversial issues in vertebrate olfactory transduction. Annu Rev Physiol 61: 857–871, 1999 [DOI] [PubMed] [Google Scholar]
- 29. Gray-Keller M, Denk W, Shraiman B, Detwiler PB. Longitudinal spread of second messenger signals in isolated rod outer segments of lizards. J Physiol 513: 679–692, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haies DM, Gil J, Weibel ER. Morphometric study of rat lung cells. I. Numerical and dimensional characteristics of parenchymal cell population. Am Rev Respir Dis 123: 533–541, 1981 [DOI] [PubMed] [Google Scholar]
- 31. Heuser JE, Salpeter SR. Organization of acetylcholine receptors in quick-frozen, deep-etched, and rotary-replicated Torpedo postsynaptic membrane. J Cell Biol 82: 150–173, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Houslay MD, Sullivan M, Bolger GB. The multienzyme PDE4 cyclic adenosine monophosphate-specific phosphodiesterase family: intracellular targeting, regulation, and selective inhibition by compounds exerting anti-inflammatory and antidepressant actions. Adv Pharmacol 44: 225–342, 1998 [DOI] [PubMed] [Google Scholar]
- 33. Iancu RV, Jones SW, Harvey RD. Compartmentation of cAMP signaling in cardiac myocytes: a computational study. Biophys J 92: 3317–3331, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Isenberg G, Wohlfarth-Bottermann KE. Transformation of cytoplasmic actin. Importance for the organization of the contractile gel reticulum and the contraction–relasation cycle of cytoplasmic actomyosin. Cell Tissue Res 173: 495–528, 1976 [DOI] [PubMed] [Google Scholar]
- 35. Jin SLC, Bushnik T, Lan L, Conti M. Subcellular localization of rolipram-sensitive, cAMP phosphodiesterases. J Biol Chem 273: 19672–19678, 1998 [DOI] [PubMed] [Google Scholar]
- 36. Jurevicius J, Fischmeister R. cAMP compartmentation is responsible for a local activation of cardiac Ca2+ channels by beta-adrenergic agonists. Proc Natl Acad Sci USA 93: 295–299, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jurevicius J, Skeberdis VA, Fischmeister R. Role of cyclic nucleotide phosphodiesterase isoforms in cAMP compartmentation following beta2-adrenergic stimulation of ICa,L in frog ventricular myocytes. J Physiol 551: 239–252, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kapustina M, Vitriol E, Elston TC, Loew LM, Jacobson K. Modeling capping protein FRAP and CALI experiments reveals in vivo regulation of actin dynamics. Cytoskeleton 67: 519–534, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kenworthy A. Peering inside lipid rafts and caveolae. Trends Biochem Sci 27: 435–438, 2002 [DOI] [PubMed] [Google Scholar]
- 40. Kholodenko BN, Kolch W. Giving space to cell signaling. Cell 133: 566–567, 2008 [DOI] [PubMed] [Google Scholar]
- 41. Krishnan R, Park CY, Lin YC, Mead J, Jaspers RT, Trepat X, Lenormand G, Tambe D, Smolensky AV, Knoll AH, Butler JP, Fredberg JJ. Reinforcement versus fluidization in cytoskeletal mechanoresponsiveness. PLoS One 4: e5486, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kushmerick MJ, Podolsky RJ. Ionic mobility in muscle cells. Science 166: 1297–1298, 1969 [DOI] [PubMed] [Google Scholar]
- 43. Lagnado L, Baylor DA. Signal flow in visual transduction. Neuron 8: 995–1002, 1992 [DOI] [PubMed] [Google Scholar]
- 44. Leblanc N, Hume JR. Sodium current-induced release of calcium from cardiac sarcoplasmic reticulum. Science 248: 372–376, 1990 [DOI] [PubMed] [Google Scholar]
- 45. Leskov IB, Klenchin VA, Handy JW, Whitlock GG, Govardovskii VI, Bownds MD, Lamb TD, Pugh EN, Jr, Arshavsky VY. The gain of rod phototransduction: reconciliation of biochemical and electrophysiological measurements. Neuron 27: 525–537, 2000 [DOI] [PubMed] [Google Scholar]
- 46. Liu H, Maurice DH. Phosphorylation-mediated activation and translocation of the cyclic AMP-specific phosphodiesterase PDE4D3 by cyclic AMP-dependent protein kinase and mitogen-activated protein kinases. A potential mechanism allowing for the coordinated regulation of PDE4D activity and targeting. J Biol Chem 274: 10557–10565, 1999 [DOI] [PubMed] [Google Scholar]
- 47. Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature 366: 283–286, 1993 [DOI] [PubMed] [Google Scholar]
- 48. Luby-Phelps K, Lanni F, Taylor DL. Probing the structure of cytoplasm. J Cell Biol 102: 2015–2022, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mastro AM, Babich MA, Taylor WD, Keith AD. Diffusion of a small molecule in the cytoplasm of mammalian cells. Proc Natl Acad Sci USA 81: 3414–3418, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Molday RS. Photoreceptor membrane proteins, phototransduction, and retinal degenerative diseases: The Friedenwald Lecture. Invest Ophthalmol Vis Sci 39: 2493–2513, 1998 [PubMed] [Google Scholar]
- 51. Mongillo M, McSorley T, Evellin S, Sood A, Lissandron V, Terrin A, Huston E, Hannawacker A, Lohse MJ, Pozzan T, Houslay MD, Zaccolo M. Fluorescence resonance energy transfer-based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ Res 95: 67–75, 2004 [DOI] [PubMed] [Google Scholar]
- 52. Mons N, Harry A, Dubourg P, Premont RT, Iyengar R, Cooper DMF. Immunohistochemical localization of adenylyl cyclase in rat brain indicates a highly selective concentration at synapses. Proc Natl Acad Sci USA 92: 8473–8477, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a. Murray F. The interplay of multiple molecular and cellular components is necessary for compartmentalization of cAMP. Focus on “Assessment of cellular mechanisms contributing to cAMP compartmentalization in pulmonary microvascular endothelial cells.” Am J Physiol Cell Physiol (January 11, 2012). doi:10.1152/ajpcell.00012.2012 [DOI] [PubMed] [Google Scholar]
- 53. Neher E, Augustine GJ. Calcium gradients and buffers in bovine chromaffin cells. J Physiol 450: 273–301, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, Taubenfeld SM, Alberini CM, Schaff JC, Blitzer RD, Moraru I, Iyengar R. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell 133: 666–680, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Novak IL, Kraikivski P, Slepchenko BM. Diffusion in cytoplasm: effects of excluded volume due to internal membranes and cytoskeletal structures. Biophys J 97: 758–767, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oliveira RF, Terrin A, Di Benedetto G, Cannon RC, Koh W, Kim M, Zaccolo M, Blackwell KT. The role of type 4 phosphodiesterases in generating microdomains of cAMP: large scale stochastic simulations. PLoS One 5: e11725, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Olson A, Pugh EN., Jr Diffusion coefficient of cyclic GMP in salamander rod outer segments estimated with two fluorescent probes. Biophys J 65: 1335–1352, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ostrom RS, Liu X, Head BP, Gregorian C, Seasholtz TM, Insel PA. Localization of adenylyl cyclase isoforms and G protein-coupled receptors in vascular smooth muscle cells: expression in caveolin-rich and noncaveolin domains. Mol Pharmacol 62: 983–992, 2002 [DOI] [PubMed] [Google Scholar]
- 59. Pace U, Hanski E, Salomon Y, Lancet D. Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature 316: 255–258, 1985 [DOI] [PubMed] [Google Scholar]
- 60. Paulucci-Holthauzen AA, Vergara LA, Bellot LJ, Canton D, Scott JD, O'Connor KL. Spatial distribution of protein kinase A activity during cell migration is mediated by A-kinase anchoring protein AKAP Lbc. J Biol Chem 284: 5956–5967, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Piggott LA, Hassell KA, Berkova Z, Morris AP, Silberbach M, Rich TC. Natriuretic peptides and nitric oxide stimulate cGMP synthesis in different cellular compartments. J Gen Physiol 128: 3–14, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Post SR, Hilal-Dandan R, Urasawa K, Brunton LL, Insel PA. Quantification of signalling components and amplification in the beta-adrenergic-receptor-adenylate cyclase pathway in isolated adult rat ventricular myocytes. Biochem J 311: 75–80, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rall TW. Introduction. Adv Cyclic Nucleotide Res 5: 1–2, 1975 [Google Scholar]
- 64. Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem 232: 1065–1076, 1958 [PubMed] [Google Scholar]
- 65. Reeves ML, Leigh BK, England PJ. The identification of a new cyclic nucleotide phosphodiesterase activity in human and guinea-pig cardiac ventricle. Implications for the mechanism of action of selective phosphodiesterase inhibitors. Biochem J 241: 535–541, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rich TC, Fagan KA, Nakata H, Schaack J, Cooper DMF, Karpen JW. Cyclic nucleotide-gated channels colocalize with adenylyl cyclase in regions of restricted cAMP diffusion. J Gen Physiol 116: 147–161, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rich TC, Fagan KA, Tse TE, Schaack J, Cooper DMF, Karpen JW. A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc Natl Acad Sci USA 98: 13049–13054, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rich TC, Xin W, CM , Hassell KA, Piggott LA, Le X, Karpen JW, Conti M. Cellular mechanisms underlying prostaglandin-induced transient cAMP signals near the plasma membrane of HEK-293 cells. Am J Physiol Cell Physiol 292: C319–C331, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Richter W, Jin SL, Conti M. Splice variants of the cyclic nucleotide phosphodiesterase PDE4D are differentially expressed and regulated in rat tissue. Biochem J 388: 803–811, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rochais F, Abi-Gerges A, Horner K, Lefebvre F, Cooper DMF, Conti M, Fischmeister R, Vandecasteele G. A specific pattern of phosphodiesterases controls the cAMP signals generated by different Gs-coupled receptors in adult rat ventricular myocytes. Circ Res 98: 1081–1088, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saucerman JJ, Brunton LL, Michailova AP, McCulloch AD. Modeling beta-adrenergic control of cardiac myocyte contractility in silico. J Biol Chem 278: 47997–48003, 2003 [DOI] [PubMed] [Google Scholar]
- 72. Saucerman JJ, McCulloch AD. Mechanistic systems models of cell signaling networks: a case study of myocyte adrenergic regulation. Prog Biophys Mol Biol 85: 261–278, 2004 [DOI] [PubMed] [Google Scholar]
- 73. Saucerman JJ, Zhang J, Martin JC, Peng LX, Stenbit AE, Tsien RY, McCulloch AD. Systems analysis of PKA-mediated phosphorylation gradients in live cardiac myocytes. Proc Natl Acad Sci USA 103: 12923–12928, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sayner S, Balczon R, Frank D, Cooper D, Stevens T. Filamin A is a phosphorylation target of plasma membrane but not cytosolic adenylyl cyclase activity. Am J Physiol Lung Cell Mol Physiol 301: L117–L124, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sayner S, Kunstadt R, Daigle J. Bicarbonate disruption of the pulmonary endothelial cell barrier by activation of the endogenous soluble adenylyl cyclase. Am J Respir Crit Care Med 183: A4178, 2011 [Google Scholar]
- 76. Sayner SL, Alexeyev M, Dessauer CW, Stevens T. Soluble adenylyl cyclase reveals the significance of cAMP compartmentation on pulmonary microvascular endothelial cell barrier. Circ Res 98: 675–681, 2006 [DOI] [PubMed] [Google Scholar]
- 77. Sayner SL, Frank DW, King J, Chen H, VandeWaa J, Stevens T. Paradoxical cAMP-induced lung endothelial hyperpermeability revealed by Pseudomonas aeruginosa ExoY. Circ Res 95: 196–203, 2004 [DOI] [PubMed] [Google Scholar]
- 78. Schaff J, Fink CC, Slepchenko B, Carson JH, Loew LM. A general computational framework for modeling cellular structure and function. Biophys J 73: 1135–1146, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Slepchenko BM, Schaff JC, Carson JH, Loew LM. Computational cell biology: spatiotemporal simulation of cellular events. Annu Rev Biophys Biomol Struct 31: 423–441, 2002 [DOI] [PubMed] [Google Scholar]
- 80. Smith KE, Gu C, Fagan KA, Hu B, Cooper DMF. Residence of adenylyl cyclase type 8 in caveolae is necessary but not sufficient for regulation by capacitative Ca2+ entry. J Biol Chem 277: 6025–6031, 2002 [DOI] [PubMed] [Google Scholar]
- 81. Stevens T, Creighton J, Thompson WJ. Control of cAMP in lung endothelial cell phenotypes. Implications for control of barrier function. Am J Physiol Lung Cell Mol Physiol 277: L119–L126, 1999 [DOI] [PubMed] [Google Scholar]
- 82. Stevens T, Nakahashi Y, Cornfield DN, McMurtry IF, Cooper DMF, Rodman D. Ca2+-inhibitable adenylyl cyclase modulates pulmonary artery endothelial cell cAMP content and barrier function. Proc Natl Acad Sci USA 92: 2696–2700, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem 232: 1077–1092, 1958 [PubMed] [Google Scholar]
- 84. Svoboda K, Tank DW, Denk W. Direct measurement of coupling between dendritic spines and shafts. Science 272: 716–719, 1996 [DOI] [PubMed] [Google Scholar]
- 85. Takechi H, Eilers J, Konnerth A. A new class of synaptic response involving calcium release in dendritic spines. Nature 396: 757–760, 1998 [DOI] [PubMed] [Google Scholar]
- 86. Terrin A, Di Benedetto G, Pertegato V, Cheung YF, Baillie G, Lynch MJ, Elvassore N, Prinz A, Herberg FW, Houslay MD, Zaccolo M. PGE1 stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: role of compartmentalized phosphodiesterases. J Cell Biol 175: 441–451, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thompson WJ, Appleman MM. Multiple cyclic nucleotide phosphodiesterase activities from rat brain. Biochemistry 10: 311–316, 1971 [PubMed] [Google Scholar]
- 88. Thompson WJ, Ashikaga T, Kelly JJ, Liu L, Zhu B, Vemavarapu L, Strada SJ. Regulation of cyclic AMP in rat pulmonary microvascular endothelial cells by rolipram-sensitive cyclic AMP phosphodiesterase (PDE4). Biochem Pharmacol 63: 797–807, 2002 [DOI] [PubMed] [Google Scholar]
- 89. Warrier S, Ramamurthy G, Eckert RL, Nikolaev VO, Lohse MJ, Harvey RD. cAMP microdomains and L-type Ca2+ channel regulation in guinea-pig ventricular myocytes. J Physiol 580: 765–776, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Weiss RL, Kukora JR, Adams J. The relationship between enzyme activity, cell geometry, and fitness in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 72: 794–798, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Willoughby D, Wong W, Schaack J, Scott JD, Cooper DMF. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J 25: 2051–2061, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu Q, Chen C, Koutalos Y. Longitudinal diffusion of a polar tracer in the outer segments of rod photoreceptors from different species. Photochem Photobiol 82: 1447–1451, 2006 [DOI] [PubMed] [Google Scholar]
- 93. Xin W, Tran TM, Richter W, Clark RB, Rich TC. Functional roles of GRK and PDE activities in the regulation of beta2 adrenergic signaling. J Gen Physiol 134: 349–364, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xu T, Naraghi M, Kang H, Neher E. Kinetic studies of Ca2+ binding and Ca2+ clearance in the cytosol of adrenal chromaffin cells. Biophys J 73: 532–545, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yarwood SJ, Steele MR, Scotland G, Houslay MD, Bolger GB. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J Biol Chem 274: 14909–14917, 1999 [DOI] [PubMed] [Google Scholar]
- 96. Yau KW. Phototransduction mechanism in retinal rods and cones: The Friedenwald Lecture. Invest Ophthalmol Vis Sci 35: 9–32, 1994 [PubMed] [Google Scholar]
- 97. Yin HL, Zaner KS, TPS Ca2+ control of actin gelation. Interaction of gelsolin with actin filaments and regulation of actin gelation. J Biol Chem 255: 9494–9500, 1980 [PubMed] [Google Scholar]
- 98. Zhou Z, Neher E. Mobile and immobile calcium buffers in bovine adrenal chromaffin cells. J Physiol 469: 245–273, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhu B, Kelly J, Vemavarapu L, Thompson WJ, Strada SJ. Activation and induction of cyclic AMP phosphodiesterase (PDE4) in rat pulmonary microvascular endothelial cells. Biochem Pharmacol 68: 479–491, 2004 [DOI] [PubMed] [Google Scholar]
- 100. Zolle O, Lawrie AM, Simpson AWM. Activation of the particulate and not the soluble guanylate cyclase leads to the inhibition of Ca2+ extrusion through localized elevation of cGMP. J Biol Chem 275: 25892–25899, 2000 [DOI] [PubMed] [Google Scholar]