Insulin biosynthesis involves the efficient folding of a single polypeptide-chain precursor, with concomitant formation of three disulfides, to give proinsulin and the subsequent enzymatic removal of the C-peptide to give mature insulin.[1,2] A proinsulin- or mini-proinsulin-based approach is currently used in the recombinant production of human insulin.[3,4] However, recombinant production of insulin analogues is effectively limited to the creation of mutants from the twenty genetically encoded amino acids. In contrast to this, total chemical synthesis of insulin would in principle enable the incorporation of a wide range of non-natural amino acids and other chemical modifications into the molecule,[5] and would thus enable the full exploration of the medicinal chemistry of this important therapeutic molecule. Until now, however, we have lacked an efficient approach to the chemical synthesis of human insulin.[5] This has impeded development of next-generation insulin analogues containing non-standard side chains, D-amino acids[6,7] or other novel chemical structural features.
Current chemical methods for insulin synthesis are limited by inefficient chain combination to give the three native disulfide bonds. Early chemical syntheses of insulin relied on inefficient folding/disulfides formation from separate A- and B-chains, which were prepared by solution[8-10] or solid phase peptide synthesis.[11] More recently, it has been found that optimal folding/disulfides formation requires a 2-to-3-fold stoichiometric excess of A-chain over the B-chain, and gives only a ∼12% folding yield based on the limiting amount of B chain[12]; because of the excess A-chain used, only ∼7% of the total weight of A- and B-chains ends up as final insulin product. Total synthesis with chemically directed formation of the disulfide bonds has been reported,[13,14] but has not found widespread use.
An alternative approach to high-yield folding/disulfide formation of the insulin molecule is to use a chemical tether to mimic the effect of covalently linking the A- and B-chains as occurs in proinsulin and mini-proinsulin precursors. Most of the previously studied chemically tethered insulin precursors have involved covalent linking of the N-terminal of the insulin A chain to the side chain of LysB29, near the C-terminal of the insulin B-chain.[15-20] With the goal of an efficient total synthesis of human insulin and analogues, a variety of different length chemical tethers between these two functionalities has been explored, using both non-cleavable[15,16] and cleavable[17-20] tethers. The shortest tether reported to be effective in promoting high yield folding/disulfide formation contained 8 carbon atoms.[15]
Recently, there have been attempts to extend the chemical tether approach to provide a more effective total chemical synthesis of insulin.[21,22] We reported a proof-of-principle synthesis of human insulin via a chemically synthesized ‘mini-proinsulin’ prepared by oxime-forming ligation.[22] A temporary ‘chemical tether’ that linked the N-terminus of the A chain to LysB28 near the C-terminal of the B chain enabled us to fold/form disulfides with high efficiency. However, our approach involved a relatively long and complex chemical tether, which made the synthesis laborious. In addition, it was necessary to remove the chemical tether enzymatically in a subsequent step, as was the case for a similar ‘mini-proinsulin’ chemical synthesis approach to insulin(desB30).[21] Thus, the strategy was not practical for the efficient generation of insulin chemical analogues.
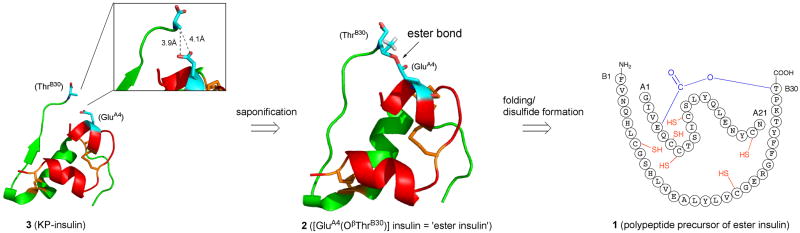
Ideal features of an optimal chemically tethered mini-proinsulin would include: straightforward preparation by existing synthetic methods; efficient folding/disulfides formation; and, ready chemical conversion to mature insulin. A model target molecule is provided by Insulin Lispro® ([LysB28,ProB29]-human insulin (herein designated “KP-insulin”), the active ingredient of Humalog® (Eli Lilly and Co.), a first-generation rapid-acting insulin analogue widely used for the treatment of diabetes mellitus.[23] In examining the three-dimensional structure of KP-insulin,[24] we noticed that the β-hydroxyl group of ThrB30 was in close proximity to and virtually in contact with the side-chain carboxyl group of GluA4 (Figure 1). This suggested to us that a covalently linked molecule in which the A- and B-chains of insulin are directly connected via an ester bond (i.e. with no additional auxiliary moiety as a tether) between the β-hydroxyl group of ThrB30 and the γ-carboxyl group of GluA4 (Figure 1) might serve as a surrogate proinsulin, in order to promote efficient folding/disulfide formation in an insulin precursor molecule. We set out to make such a molecule and explore its folding properties, and its chemical conversion to insulin.
Figure 1. Retrosynthetic analysis for the preparation of human KP-insulin by the ‘ester insulin’ strategy.
KP-insulin is a fast acting form of human insulin in which the native ProB28-LysB29 sequence is inverted to LysB28-ProB29.[23] KP-insulin coordinates are from Protein Databank entry 1LPH.[24] Insulin A-chain is shown in red, B-chain in green, and GluA4 and ThrB30 in cyan. The γ-CH3 and β-OH of ThrB30 are not visible due to disorder in the crystal structure; however, the possible positions of the β-OH of ThrB30 can be accurately inferred since the position of the ThrB30 β-carbon is known. The desired ester linked molecule (1) was prepared by native chemical ligation[25] of [PheB1-ValB18]-αthioester and the ester linked CysB19-[A1-GluA4(OβThrB30)-A21] peptide as described in the Supporting Information.
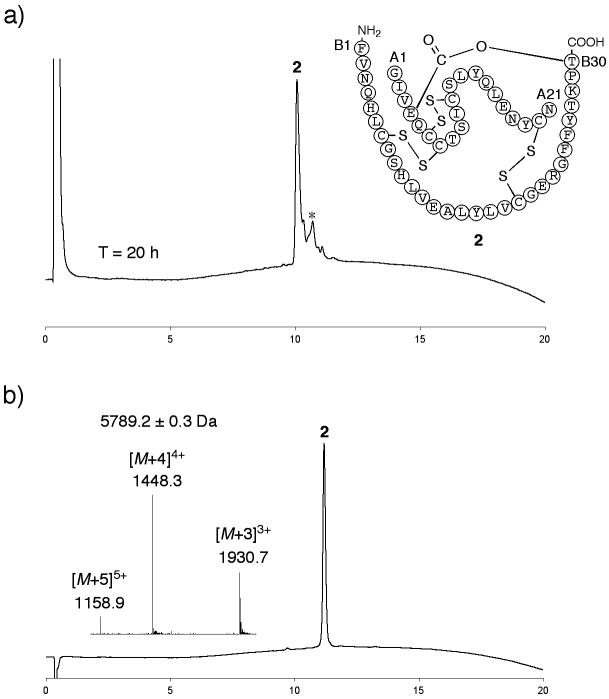
A retrosynthetic analysis of a route to KP-insulin via the ester insulin precursor (1) is shown in Figure 1. The folding properties of purified (1) were examined under the following conditions: ∼ 0.3 mg mL−1 1, 20 mM Tris, 8 mM Cys, 1 mM cystine, 1.5 M GnHCl, pH = 7.3 (Figure S3). Folding was complete in 20 h with the formation of folded ester insulin 2 as the predominant product (Figure 2a). The observed mass of compound 2 decreased by 5.8 ± 0.2 Da compared to that of the reduced polypeptide 1, consistent with the formation of three disulfide bonds in 2. We estimated that the HPLC yield of folded ester insulin 2 from 1 was ∼70%. The excellent folding profile of 2 demonstrates that the ThrB30-GluA4 ester linkage made the molecule as favorable for folding/disulfides formation as does the C-peptide (35 amino acids long) in the proinsulin molecule.[26] The folding yield of 2 was also similar to that previously observed for oxime-linked ‘mini-proinsulin’ (∼60%)[22] where the N-terminal of A-chain was connected to LysB28 near the C-terminal of B-chain. Folded ester insulin 2 was isolated after HPLC purification (Figure 2b).
Figure 2. Folding/disulfides formation to give ester insulin.
a) Folding of the ester insulin precursor 1 to form ester insulin 2 was monitored by LC after 20 h (UV profiles at 214 nm are shown). Essentially similar data were obtained at T = 1 h. Folding conditions were 1: ∼0.3 mg mL−1, Tris: 20 mM, Cys: 8 mM, cystine: 1 mM, GnHCl: 1.5 M, pH = 7.3, *Cys adducts; b) purified ester insulin 2. (Inset) On-line ESI-MS spectra taken at the top of the main peak in chromatogram. The chromatographic separations were performed using a linear gradient (5-65%) of buffer B in buffer A over 15 min (buffer A = 0.1% TFA in water; buffer B = 0.08% TFA in acetonitrile) (UV profiles at 214 nm). Columns with different reverse phase packings were used for a) and b).
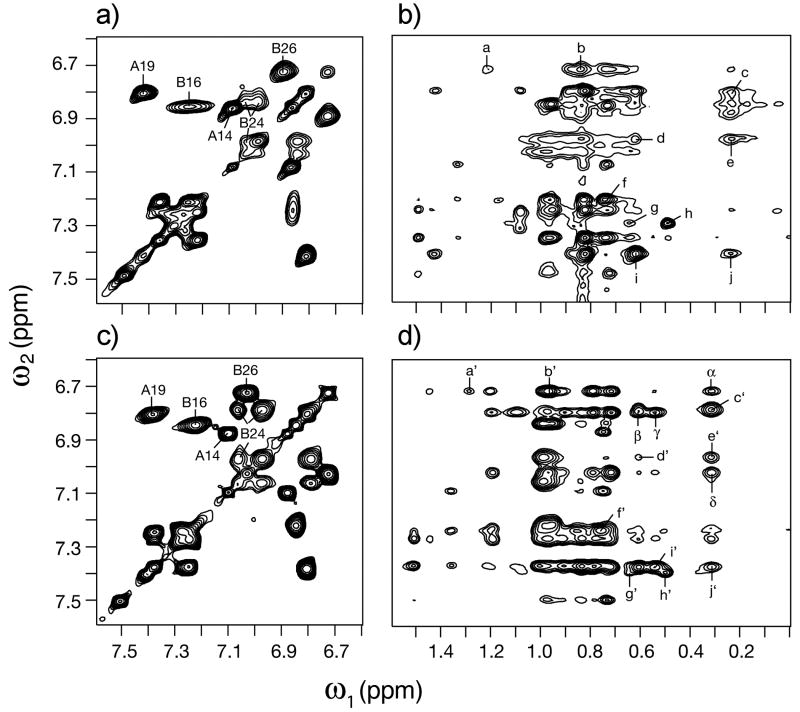
In order to investigate the folded conformation of ester insulin, 2D 1H NMR studies were conducted at pH 3.0 in 10 mM deuterioacetic acid at 25 and 37 °C. Use of acidic pH was chosen to retard the rate of hydrolysis of the ester during the 7 days of NMR data acquisition in H2O and D2O. Under these conditions the solution structure of KP-insulin (Q.X.H. and M.A.W., manuscript in preparation) is essentially identical to its crystal structure (T-state protomer).[24] Because KP-insulin (like wild-type insulin) contains multiple aromatic side chains at key positions in the structure (4 Tyr, 3 Phe, and 2 His), its aromatic spectrum provides a fingerprint of the folded structure. Comparison of TOCSY 1H spectra of ester insulin and KP-insulin demonstrated retention of a native-like pattern of aromatic chemical shifts (Figure 3a versus 3c). In particular, the large and corresponding secondary chemical shifts of TyrA19 and TyrB16 suggest that the α-helical moiety of the structure is retained, and the upfield shifts of PheB24 and TyrB26 suggest that ester insulin retains a native-like αβ U-turn involving the C-terminal segment of the tethered B-chain. The B24 spin system is anomalously broadened. Similar broadening is also observed involving non-aromatic resonances in the segments B27-B30 and A1-A5 (comparison of NOESY spectra in Figure 3b versus 3d). We ascribe such broadening to constrained millisecond motions in the ester insulin molecule, leading to incomplete averaging of chemical shifts.
Figure 3. Tertiary structure of ester insulin and its relation to mature insulin, by 2D 1H-NMR.
(Panels a and b) Spectra of ester insulin. Aromatic TOCSY spin systems are shown in panel a). Assignments of Tyr and Phe spin systems are as labeled; NOEs between aormatic protons (vertical axis) and aliphatic protons (horizontal axis) are shown in panel b). (Panels c and d) Corresponding spectra of KP-insulin. Aromatic TOCSY spin systems are shown in panel c) with related NOEs in panel d). Assignment of NOE cross-peaks a-j (a‘-j’) are reported in the Supporting Information. Samples were dissolved in 10 mM deuterioacetic acid (pH 3.0). Spectra were acquired at 25°C and 700 MHz.
Despite limitations of the NMR analysis near the ester moiety itself, complete resonance assignments were obtained elsewhere and enabled a detailed comparison between inter-residue nuclear Overhauser effects (NOEs) in ester insulin and the corresponding KP-insulin. Diagnostic long-range NOEs characteristic of the tertiary structure of insulin are retained in ester insulin. Of particular note are native-like patterns of chemical shifts and NOEs involving the three cystines, evidence of native disulfide pairing. In addition, ester insulin exhibits selected inter-residue NOEs in the A6-A12 segment that are in accord with insulin crystal structures but attenuated or not seen in the NMR spectrum of KP-insulin.
Interestingly, ester insulin itself had less than 1% activity (>6 nM) in the insulin receptor binding assay as compared to KP-insulin (0.044 nM) in the same assay. It has previously been shown that an Ala replacement at either ThrB30 or GluA4 did not cause any loss of the binding affinity,[5] suggesting that the inactive nature of ester insulin may arise from conformational restriction caused by the ester bond (see NMR studies, above) in accord with the low activities of chemically tethered insulin analogs[27] and single-chain analogs.[28,29]
Next, we investigated the chemical conversion of ester insulin to KP-insulin. Saponification of ester insulin 2 was performed under the following conditions: ∼0.12 mg mL−1 2, 25 mM sodium hydroxide, 25% acetonitrile (in water), 4 °C. As shown in Figure 4a, after 24 h reaction the desired KP-insulin molecule 3 was obtained in an HPLC yield of ∼95%. A few percent of individual A- and B-chains were observed as side products probably because of disruption of disulfide bonds under these basic conditions. Following saponification, the mass increased by 18.5 ± 0.5 Da compared to that of 2, consistent with the addition of the elements of water in the formation of 3. We obtained pure KP-insulin 3 after final HPLC purification (Figure 4b); the isolated yield of KP-insulin 3 from ester insulin 2 was 93%. The reverse-phase HPLC retention time of synthetic KP-insulin 3 was identical to that of an authentic sample of biosynthetic KP-insulin (extracted from Humalog®). The synthetic KP-insulin 3 was also characterized by measurement of the relative binding affinity to the insulin receptor (Figure 4c). Within experimental uncertainty, the activity of synthetic KP- insulin was the same as that of an authentic sample of Humalog. This further confirmed the formation of the correct disulfide bonds in the folded ester insulin molecule, and their retention in the isolated synthetic KP-insulin after saponification.
Figure 4. Conversion of ester insulin to native KP-insulin.
a) saponification of ester insulin 2 to give KP-insulin 3. Conditions were 2: 0.12 mg mL−1, NaOH: 25 mM, acetonitrile-H2O (2.5:7.5), 4 °C. Reaction mixture at T = 2 h (upper panel) and T = 24 h (lower panel), #unrelated column contaminant, *derived from A-chain, **B-chain; b) purified KP-insulin 3. (Inset) On-line ESI-MS spectra taken at the top of the main peak. Chromatographic separations were performed as described in Figure 2 legend; c) binding affinities of synthetic KP-insulin 3 and authentic KP-insulin (purchased from Eli Lilly and Co) to the insulin receptor.
In order to evaluate the utility of ester insulin for the efficient chemical synthesis of insulin analogues, we prepared the ester-containing polypeptide precursor of [Gly23D-Ala]KP-insulin. The protein diastereomer [Gly23D-Ala]KP-insulin was designed to investigate the contribution of the GlyB23 residue to receptor recognition. As in ester insulin, the polypeptide precursor of [Gly23D-Ala] ester insulin was efficiently folded with concomitant formation of three disulfides, and the oxidized[Gly23D-Ala] ester insulin was saponified in a similar manner to give the desired [Gly23D-Ala]KP-insulin (Figure S4). The receptor binding affinity of the [Gly23D-Ala]KP-insulin was 0.021±0.004 nM, a 2-fold higher activity than that of KP-insulin (0.044±0.007 nM). [Gly23D-Ala] ester insulin had 100-fold worse activity (2.1±0.3 nM) compared with the [Gly23D-Ala]KP-insulin mature form (Figure S4C). The 2-fold higher activity of the [Gly23D-Ala] analog suggests that the GlyB23 residue of the KP-insulin contributes to the receptor recognition by maintenance of the positive phi angle at B23, as was previously suggested by studies of DKP-insulin.[6]
In conclusion, we have designed and synthesized [GluA4(OβThrB30)]insulin (ester insulin 2) as a surrogate proinsulin with a ‘zero length’ chemical tether moiety, and explored the potential utility of this novel molecule as an intermediate for the total chemical synthesis of human insulins. The reduced ester insulin precursor folded efficiently (∼70% HPLC yield) under standard redox conditions with concomitant formation of the three native disulfides. Thus, the ThrB30-GluA4 ester linkage made folding the precursor molecule as favorable as does the 35 residue C-peptide in proinsulin. Finally, saponification of ester insulin gave the native folded insulin molecule in near-quantitative yield. Synthetic KP-insulin produced by the ester insulin route had full receptor-binding activity.
With suitable optimization of its preparation, ester insulin may prove to be the key to a simple and effective route to the total chemical synthesis of insulin.[5] The ester insulin precursor polypeptide could be made by any of several synthetic routes, including the hybrid solution-solid phase methodology used for the cost effective large-scale manufacture of long peptides.[30] We believe that ester insulin will be a useful intermediate for the efficient generation of insulin analogues in the research laboratory and for cost-effective chemical manufacture of human insulins.
Footnotes
This research was supported by NIH grant RO1 GM075993 to S.B.H.K.. Y. S. is grateful for a JSPS Postdoctoral Fellowship for Research Abroad. We thank Salih Özcubukcu (University of Chicago) for his support in NMR characterization of new compounds.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Youhei Sohma, Department of Biochemistry and Molecular Biology, Department of Chemistry, Institute for Biophysical Dynamics, The University of Chicago, Chicago IL 60637 (USA)
Dr. Qing-Xin Hua, Department of Biochemistry, Case Western Reserve University, Cleveland Ohio 44106 (USA)
Prof. Jonathan Whittaker, Department of Biochemistry, Case Western Reserve University, Cleveland Ohio 44106 (USA)
Prof. Michael A. Weiss, Department of Biochemistry, Case Western Reserve University, Cleveland Ohio 44106 (USA)
Prof. Stephen B. H. Kent, Email: skent@uchicago.edu, Department of Biochemistry and Molecular Biology, Department of Chemistry, Institute for Biophysical Dynamics, The University of Chicago, Chicago IL 60637 (USA).
References
- 1.Oyer PE, Cho S, Peterson JD, Steiner DF. J Biol Chem. 1971;246:1375–1386. [PubMed] [Google Scholar]
- 2.Kemmler W, Peterson JD, Steiner DF. J Biol Chem. 1971;246:6786–6791. [PubMed] [Google Scholar]
- 3.Frank BH, Chance RE. MMW Munch Med Wochenschr. 1983;125(Suppl 1):14–20. [PubMed] [Google Scholar]
- 4.Thim L, Hansen MT, Norris K, Hoegh I, Boel E, Forstrom J, Ammerer G, Fiil NP. Proc Natl Acad Sci USA. 1986;83:6766–6770. doi: 10.1073/pnas.83.18.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer JP, Zhang F, DiMarchi RD. Biopolymers (Peptide Science) 2007;88:687–713. doi: 10.1002/bip.20734. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa SH, Hua QX, Hu SQ, Jia W, Wang S, Katsoyannis PG, Weiss MA. J Biol Chem. 2006;281:22386–22396. doi: 10.1074/jbc.M603547200. [DOI] [PubMed] [Google Scholar]
- 7.Zhao M, Wan ZL, Whittaker L, Xu B, Phillips NB, Katsoyannis PG, Ismail-Beigi F, Whittaker J, Weiss MA. J Biol Chem. 2009;284:32178–32187. doi: 10.1074/jbc.M109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meienhofer J, Schnabel E, Bremer H, Brinkhoff O, Zabel R, Sroka W, Klostermeyer H, Brandenburg D, Okuda T, Zahn H. Z Naturforsch. 1963;18b:1120–1121. [PubMed] [Google Scholar]
- 9.Katsoyannis PG, Fukuda K, Tometsko A, Suzuki K, Tilak M. J Am Chem Soc. 1964;86:930–932. [Google Scholar]
- 10.Du YC, Jiang RQ, Tsou CL. Sci Sin. 1965;14:229–236. [Google Scholar]
- 11.Marglin B, Merrifield RB. J Am Chem Soc. 1966;88:5051–5052. doi: 10.1021/ja00973a068. [DOI] [PubMed] [Google Scholar]
- 12.Hua Q, Chu Y, Jia W, Phillips N, Wang R, Katsoyannis P, Weiss M. J Biol Chem. 2002;277:43443–43453. doi: 10.1074/jbc.M206107200. [DOI] [PubMed] [Google Scholar]
- 13.Sieber P, Kamber B, Hartmann A, Jöhl A, Riniker B, Rittel W. Helv Chim Acta. 1977;60:27–37. doi: 10.1002/hlca.19770600105. [DOI] [PubMed] [Google Scholar]
- 14.Akaji K, Fujino K, Tatsumi T, Kiso Y. J Am Chem Soc. 1993;115:11384–11392. [Google Scholar]
- 15.Brandenburg D, Wollmer A. Hoppe-Seyler's Z Physiol Chem. 1973;354:613–627. doi: 10.1515/bchm2.1973.354.1.613. [DOI] [PubMed] [Google Scholar]
- 16.Wollmer A, Brandenburg D, Vogt HP, Schermutzki W. Hoppe-Seyler's Z Physiol Chem. 1974;355:1471–1476. [PubMed] [Google Scholar]
- 17.Brandenburg D, Schermutzki W, Zahn H. Hoppe-Seyler's Z Physiol Chem. 1973;354:1521–1524. [PubMed] [Google Scholar]
- 18.Geiger R, Obermeier R. Biochem Biophys Res Commun. 1973;55:60–66. doi: 10.1016/s0006-291x(73)80059-2. [DOI] [PubMed] [Google Scholar]
- 19.Obermeier R, Geiger R. Hoppe-Seyler's Z Physiol Chem. 1975;356:1631–1634. [PubMed] [Google Scholar]
- 20.Busse WD, Carpenter FH. Biochemistry. 1976;15:1649–1657. doi: 10.1021/bi00653a010. [DOI] [PubMed] [Google Scholar]
- 21.Tofteng AP, Jensen KJ, Schäffer L, Hoeg-Jensen T. ChemBioChem. 2008;9:2989–2996. doi: 10.1002/cbic.200800430. [DOI] [PubMed] [Google Scholar]
- 22.Sohma Y, Kent SBH. J Am Chem Soc. 2009;131:16313–16318. doi: 10.1021/ja9052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson JH, Jr, Brunelle RL, Keohane P, Koivisto VA, Trautmann ME, Vignati L, DiMarchi R. Arch Intern Med. 1997;157:1249–1255. [PubMed] [Google Scholar]
- 24.Ciszak E, Beals JM, Frank BH, Baker JC, Carter ND, Smith GD. Structure. 1995;3:615–622. doi: 10.1016/s0969-2126(01)00195-2. PDB code 1LPH. [DOI] [PubMed] [Google Scholar]
- 25.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 26.Winter J, Lilie H, Rudolph R. Anal Biochem. 2002;310:148–155. doi: 10.1016/s0003-2697(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa SH, Tager HS. J Biol Chem. 1989;264:272–279. [PubMed] [Google Scholar]
- 28.Markussen J, Jørgensen KH, Sørensen AR, Thim L. Int J Peptide Protein Res. 1985;26:70–77. [PubMed] [Google Scholar]
- 29.Hua QX, Hu SQ, Jia W, Chu YC, Burke GT, Wang SH, Wang RY, Katsoyannis PG, Weiss MA. J Mol Biol. 1998;277:103–118. doi: 10.1006/jmbi.1997.1574. [DOI] [PubMed] [Google Scholar]
- 30.Bray BL. Nature Rev Drug Discovery. 2003;2:587–593. doi: 10.1038/nrd1133. [DOI] [PubMed] [Google Scholar]