Abstract
Although T cells have been shown to play a direct role in kidney ischemia-reperfusion injury (IRI), little is known about the underlying mechanisms. We hypothesized that studying the transcriptional responses in kidney-infiltrating T cells would help elucidate novel therapeutic targets for kidney IRI. Unilateral renal pedicle clamping for 45 min was performed in male C57BL/6 mice, and CD3+ T cells were isolated from the kidney and purified. Transcriptional activities of T cell were measured by array-based PCR compared between ischemic kidneys and contralateral nonischemic kidneys. Among total of 89 genes analyzed, 24, 22, 24, and 37 genes were significantly changed at 6 h, day 3, day 10, and day 28 after IRI. Genes associated with cytokines, chemokines, and costimulatory molecules were upregulated. Pathway analysis identified CC motif chemokine receptor 5 (CCR5) as a candidate pathophysiological pathway. CCR5 upregulation was validated at the protein level, and CCR5 blockade improved renal function after kidney IRI. Using discovery techniques to identify transcriptional responses in purified kidney-infiltrating cells enabled the elucidation of novel mechanisms and therapeutic targets for IRI.
Keywords: acute kidney injury, T lymphocyte, array-based QRT-PCR, chemokine receptor 5
ischemia-reperfusion injury (IRI) is the most common cause of acute kidney injury (AKI) in both native kidneys and transplant allografts (34). Despite advances in renal replacement therapy, the mortality in patients with native kidney IRI and morbidity in patients with kidney transplantation remain high, and there is no specific therapy (44). Unveiling the pathophysiology of kidney IRI will allow us to elucidate novel therapeutic targets. Inflammation has been established to contribute substantially to the pathogenesis of IRI (12, 42, 45), and previous studies have demonstrated that T cells are important mediators of IRI (7, 17–18, 36). T cell-deficient mice (nu/nu) showed renal protection from IRI, and adoptive T cells transfer with the CD4+ T cells subset restored early renal damage following IRI (11, 15). However, the underlying mechanisms by which T cells infiltrate and mediate kidney IRI are largely unclear. Transcription analysis has helped discover biomarkers and mechanistic pathways and new candidate therapeutic approaches directed toward organ injury (8, 20). Although many studies have examined whole kidney transcriptional responses during kidney IRI (20, 29, 46), transcriptional studies on kidney-infiltrating immune cells during IRI has not been performed. We hypothesized that transcriptional response of T cells infiltrating into the postischemic kidney during IRI would help us to understand T cell responses and lead to discovery of novel molecular targets for modulating kidney IRI.
We performed unilateral kidney ischemia followed by reperfusion in male C57BL/6 mice, isolated kidney mononuclear cells (KMNCs) with our advanced techniques, and sorted for pure CD3+ T cells with microbeads. We examined transcriptional responses in these isolated and purified kidney-infiltrating T cells. We found that kidney IRI induced significant changes in large numbers of genes, and these changes included cytokine/chemokine signaling and a costimulatory pathway even 4 wk after IRI. Pathway analysis for main biological functions of genes with significant changes identified CC motif chemokine receptor 5 (CCR5) as a candidate mediator of IRI. CCR5 was one of the highest upregulated genes following IRI, and it was one component of significant functional pathways at all time points. To test the hypothesis that CCR5 modulated kidney IRI, we first validated upregulation of CCR5 in CD3+ infiltrating T cells at the protein level by flow cytometry. We then blocked CCR5 with a neutralizing antibody in mice undergoing bilateral kidney ischemia-reperfusion. We found that blockade of CCR5 in vivo protected mice from kidney IRI. Modifying discovery techniques to study small populations of kidney-infiltrating cells during IRI can lead to new insights into mechanisms of kidney IRI.
METHODS
Animal and experimental protocols.
Male C57BL/6J mice were purchased from The Jackson Laboratory. All mice were 7- to 10-wk-old males and were housed in a specific pathogen-free barrier animal facility. The Johns Hopkins University Animal Care and Use Committee approved all studies. Mice were anesthetized with an intraperitoneal injection of pentobarbital sodium (75 mg/kg). Following an abdominal midline incision, left renal pedicles were bluntly dissected and clamped with a microvascular clamp (Roboz Surgical Instrument, Gaithersburg, MD) for 45 min. This ischemic time was chosen based on our preliminary experiments, in which 30 min of ischemia led to only a mild kidney histological response on days 10 and 28 and 60 min of ischemia led to severe histological injury. During the procedures, mice were kept well hydrated with warm (37°C) sterile saline. After the clamps were removed, the wounds were sutured and the mice were allowed to recover with free access to chow and water. Randomly selected mice were euthanized at 6 h, on day 2, day 10, and day 28 after surgery. Both postischemic kidneys and contralateral kidneys were collected and compared. In a CCR5 blockade experiment, a 30-min bilateral renal pedicle-clamping model was applied to assess early renal dysfunction.
KMNC extraction and CD3+ T cell purification.
KMNCs were isolated according to the method previously described (4). Briefly, harvested kidneys were immersed in RPMI buffer (Mediatech, Manassas, VA) containing 5% fetal bovine serum and disrupted mechanically using a Stomacher 80 Biomaster (Seward, UK). The disrupted kidney tissues were meshed and strained through a cell strainer (70 μm). The strained suspension was then centrifuged, and the cell pellet was washed and then suspended in 36% Percoll (Amersham Pharmacia Biotech, Piscataway, NJ) followed by gentle overlaying onto 72% Percoll. After centrifugation at 1,000 g for 30 min at room temperature, KMNCs were collected from the Percoll interface, washed twice, and counted on a hemocytometer using trypan blue exclusion. Then, KMNCs were reacted with rat anti-CD3 antibody (BD Bioscience, San Jose, CA), and CD3+ T cells were isolated from KMNCs using magnetic bead separation (Goat anti-rat magnetic beads; Miltenyi Biotec, Auburn, CA). The purity of the population was confirmed by flow cytometric analysis and reached >90% (data not shown).
Purification and preparation of RNA.
Total RNA was extracted from the CD3+ T cells population using the TRIzol reagent method (Invitrogen, Carlsbad, CA). The quality of total RNA samples was assessed using Nanodrop (Thermoscientific, Waltham, MA), an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), and RT2 RNA QC PCR Array (SABiosciences, Gaithersburg, MD).
Superarray RT-PCR (quantitative RT-PCR) analysis.
RT was performed on total RNA isolated from CD3+ T cells extracted from kidneys and processed (Applied Biosystems, Foster City, CA). A High-Capacity cDNA Archive kit first-strand synthesis system for RT-PCR was used according to the manufacturer's protocol. Quantitative real-time PCR (QRT-PCR) was performed using an RT2 Profiler PCR Array from SuperArray (SABiosciences). RT2 Profiler PCR Arrays are designed for relative QRT-PCR based on SybrGreen detection and performed on a 1-sample/1-plate, 96-well format using primers for a preset list of genes corresponding to a particular biological pathway. The specific array types included here were mouse Th1-Th2-Th3 PCR Arrays (PAMM-034). In brief, cDNA volumes were adjusted to ∼2.5 ml with SuperArray RT2 Real-Time SYBR Green/ROX PCR 2X Master Mix (PA-012). Twenty-five microliters of cDNA mix was added to all wells. The PCR plate was sealed, spun at 1,500 rpm × 4 min, and real-time PCR was performed on an Applied Biosystems 7300 Real Time PCR System. ABI instrument settings include setting reporter dye as “SYBR” and passive reference as “ROX,” deleting “UNG Activation,” and adding “Dissociation Stage.”
Relative gene expressions were calculated by using the 2−ΔΔCt method, in which Ct indicates cycle threshold, the fractional cycle number where the fluorescent signal reaches detection threshold (26). The normalized ΔCt value of each sample is calculated using up to a total of five endogenous control genes (18S rRNA, HPRT1, RPL13A, GAPDH, and ACTB). Fold-change values are presented as average fold-change = 2−(average ΔΔCt) for genes in treated relative to control samples (NCBI tracking system no. 15831202; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23747).
Global functional analysis and biological functional networking.
The functional analysis that identifies the biological functions that were significantly associated with identified candidate genes was conducted using the Ingenuity Pathways Knowledge Base tool (http://www.ingenuity.com). The significance value of each function is a measurement of how likely it is that a group of the candidate genes is involved in a represented (x-axes) function. Fisher's exact test was used to calculate a P value determining the probability of each biological function assigned to our candidate genes. The Ingenuity-modified network for biological functional pathways was built between genes which showed significant changes (listed in Table 1) based on the literature.
Table 1.
Lists of genes showing significant changes after IRI at each time point
| Upregulated Genes | |||||||
|---|---|---|---|---|---|---|---|
| 6 h |
Day 3 |
Day 10 |
Day 28 |
||||
| Symbol | FC | Symbol | FC | Symbol | FC | Symbol | FC |
| CD80 | 9.9 | SOCS3 | 9.1 | CCL7 | 11.2 | CCL7 | 49.8 |
| CCL7 | 8.6 | TNF | 6.0 | CCR2 | 8.3 | CTLA4 | 12.3 |
| CCR2 | 8.4 | TLR4 | 5.9 | SOCS3 | 7.6 | TNFSF4 | 10.3 |
| SOCS3 | 7.9 | CCR5 | 5.5 | CCR5 | 7.0 | IL10 | 9.6 |
| CD40 | 7.7 | CCR3 | 5.1 | PTPRC | 5.3 | IL2 | 8.0 |
| CCR3 | 6.8 | CCL7 | 4.7 | JUNB | 5.0 | TLR6 | 7.0 |
| TLR4 | 6.7 | CCR2 | 3.5 | TNF | 4.9 | CCR5 | 6.8 |
| TLR6 | 6.0 | CD80 | 3.1 | CCR3 | 4.3 | TNFSF8 | 6.7 |
| IGSF6 | 6.0 | Spp1 | 2.6 | IGSF6 | 4.3 | CXCR3 | 6.4 |
| IL17a | 5.7 | PTPRC | 2.3 | CD40 | 4.1 | CD40 | 6.2 |
| CCR5 | 5.6 | CD86 | 4.0 | CCR3 | 6.2 | ||
| PTPRC | 5.6 | TLR4 | 3.6 | IL1R1 | 6.1 | ||
| TNF | 5.1 | TLR6 | 3.5 | CCR2 | 6.1 | ||
| IL4Ra | 4.6 | Spp1 | 3.3 | TNF | 5.5 | ||
| SOCS1 | 3.9 | CD4 | 2.8 | TLR4 | 5.4 | ||
| CD86 | 3.2 | TNFSF4 | 2.8 | CD86 | 5.3 | ||
| IL18 | 3.1 | CEBPB | 2.7 | GFI1 | 5.3 | ||
| IL13Ra1 | 2.9 | CXCR3 | 2.7 | SOCS3 | 4.7 | ||
| NFkB1 | 2.8 | IRF1 | 2.4 | IL27 | 4.4 | ||
| CEBPB | 2.8 | CCL5 | 2.4 | CCL5 | 4.3 | ||
| CCL5 | 2.5 | IL2Ra | 4.2 | ||||
| NFATC2 | 2.1 | CD40Ig | 4.1 | ||||
| CD80 | 3.4 | ||||||
| PTPRC | 3.4 | ||||||
| ICOS | 2.6 | ||||||
| CD28 | 2.4 | ||||||
| Downregulated genes | |||||||
| MAF | −2.7 | IL12RB2 | −2.0 | BCL6 | −2.3 | MAPK9 | −2.1 |
| TCFCP2 | −2.8 | CREBBP | −2.2 | MAPK9 | −2.3 | NFATC2ip | −2.1 |
| NFATC3 | −2.5 | IL15 | −2.7 | IL9 | −2.2 | ||
| INHA | −2.6 | SOCS5 | −3.1 | TMED1 | −2.3 | ||
| IL18bp | −2.8 | IL18bp | −2.6 | ||||
| CD40Ig | −3.2 | MAF | −3.2 | ||||
| MAPK8 | −3.4 | MAPK8 | −3.9 | ||||
| MAPK9 | −3.4 | BCL6 | −4.0 | ||||
| IL15 | −3.7 | CCR10 | −6.2 | ||||
| CD27 | −3.8 | IL15 | −7.7 | ||||
| SOCS5 | −4.2 | TCFCP2 | −11.2 | ||||
| TCFCP2 | −12.8 | ||||||
IRI, ischemia-reperfusion injury; FC, fold-change.
Genomic clustering and signature analyses.
Hierarchical clustering was performed using the MeV (MultiExperiment Viewer) component (http://www.tm4.org/mev.html) of the TM4 system for microarray data management and analysis. Genes significantly affected by IRI after 45 min of ischemia were combined and clustered using MeV software. Fold-change values (log2) were calculated by subtracting the average of their corresponding controls (n = 5–8) from individual gene expression values of each biological replicate. The analysis across time points was conducted using supervised or unsupervised clustering, respectively, with application of an uncentered Pearson correlation and average linkage algorithm.
Flow cytometry analysis of KMNCs.
Surface and intracellular staining of KMNCs was performed as previously described (4). Isolated KMNCs were preincubated with anti-CD16/CD32 Fc receptor blocking antibody for 10 min to minimize nonspecific antibody binding. Cells were then incubated with monoclonal antibodies (anti-mouse CD3, CD4, CD8, CD11c, CD19, CD40, CD69, CD80, TCR-β, NK1.1, F4/80, Mac-1, Ly-6G and MHC class II; all from BD Biosciences except anti-mouse F4/80 antibody and anti-mouse CCR5 antibody, which were from eBioscience) for 25 min at 4°C, washed with FACS buffer, and fixed with a 1% paraformaldehyde solution. Four-color immunofluorescence staining was analyzed using a FACSCalibur instrument (BD Biosciences) and FCS Express V3 (De Novo Software, Los Angeles, CA). Each assay included at least 10,000 gated events.
Intracellular cytokine staining.
One million KMNCs were suspended in 200 μl of RPMI 1640 medium with 10% of FBS, l-glutamine, and penicillin/streptomycin and incubated at 37°C in the presence of PMA (5 ng/ml), ionomycin (500 ng/ml; both Sigma-Aldrich), and monensin (BD Biosciences). After 5-h culture, cells were washed and stained for surface markers using anti-TCR-β-allophycocyanin antibodies. These cells were then permeabilized with Cytofix/Cytoperm solution (BD Biosciences) for 20 min and washed twice with perm/wash buffer. Cells were then incubated with anti-TNF-α-FITC, washed with perm/wash buffer, and analyzed with a FACSCalibur instrument (BD Biosciences).
CCR5 blockade.
To study the mechanistic role of CCR5 in T cells infiltrating into the kidney after IRI, 25 mg/kg of anti-CCR5 antibody and IgG2B isotype control antibody (R&D Systems) diluted with saline were administered to 7-wk-old male C57BL/6J mice (ip) 2 days before renal IRI (n = 16–18/group). Thirty minutes of bilateral renal pedicle clamping were applied to both groups, and they were maintained at a constant temperature (37°C). This bilateral clamping model was chosen because bilateral renal pedicle clamping causes acute kidney injury with an increase in serum creatinine concentration so that the latter could be used as a marker for early functional changes in the kidney, which cannot be performed with a unilateral clamp model. The same dosage of anti-CCR5 and an isotype control antibody was also injected into the intraperitoneal cavity immediately after the start of reperfusion. The antibody injection protocol was based on a previous study (40), and CCR5 binding was verified with flow cytometry (data not shown).
Assessment of renal function.
Blood samples were obtained from the tail vein before and at 24, 48, and 72 h after renal IRI. Serum creatinine levels (mg/dl) were measured with a Cobas Mira plus autoanalyzer (Roche Diagnostics, Indianapolis, IN).
Kidney histological analysis.
Mice were euthanized at 72 h after renal IRI. All kidneys were harvested after exsanguination. Tissue samples were fixed with 10% buffered formalin followed by paraffin embedding, and then renal sections were stained with hematoxylin and eosin (H&E). Renal tubular damage was scored in a blinded fashion by a renal pathologist.
Measurement of chemokines by ELISA.
CCL3 and CCL5 protein levels at days 0 and 3 after IRI were measured in whole-kidney protein extracts in each experimental group with an ELISA kit (Ray Biotech, Norcross, GA) according to the manufacturer's recommended protocol.
Statistics.
All data are expressed as means ± SE. Group means were compared using Mann-Whitney analysis and ANOVA followed by Newman-Keuls post hoc analysis using GraphPad Prism version 4 (GraphPad Software, La Jolla, CA). The survival rate was evaluated using the Kaplan-Meier method. Statistical significance was considered when a P value was <0.05.
RESULTS
Gene expression changes in kidney-infiltrating CD3+ T cells after IRI.
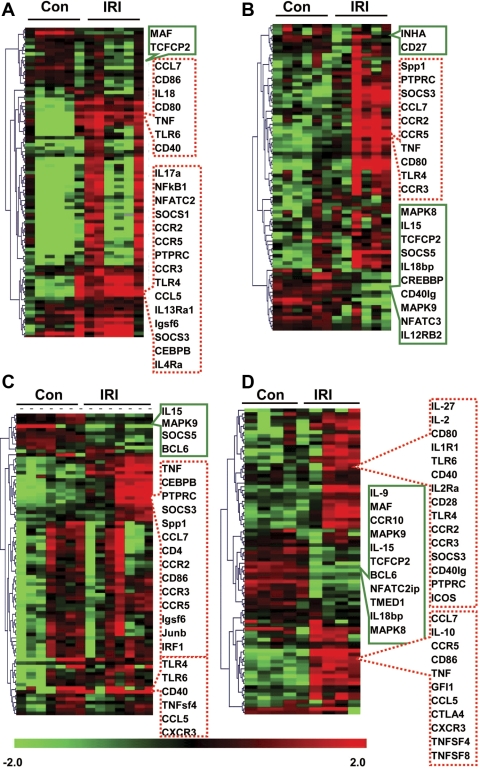
Marked transcriptional changes in multiple genes occurred in CD3+ T cells infiltrating into the kidney after IRI, which persisted up to 28 days after IRI (Table 1). Of 89 genes studied, there were significant changes in 24, 22, 24, and 37 at 6 h, 3 days, 10 days, and 28 days after IRI. The numbers of upregulated genes were higher than downregulated genes except for day 3 after IRI (upregulated genes: 22, 10, 20, and 26, and downregulated genes: 2, 12, 4, and 11 at 6 h, 3 days, 10 days, and 28 days after IRI, respectively). The hierarchical clustering analysis revealed several clusters of genes, those that were markedly up- or downregulated in IRI kidneys compared with control nonischemic kidneys at each time point, which showed that IRI induced significant genetic responses in kidney T cells (Fig. 1).
Fig. 1.
Hierarchical clustering of genes affected by ischemia-reperfusion injury (IRI) in T cell population. In the study, 24, 22, 24, and 37 genes showed significant changes at 6 h, day 3, day 10, and day 28, respectively. Genes significantly affected by IRI after 45 min of ischemia were combined and clustered using MeV software. Fold-change values (log2) were calculated by subtracting the average of their corresponding controls (n = 5–8) from individual gene expression values of each biological replicate. The clustering was conducted based on the gene expression pattern rather than amplitude using uncentered Pearson correlation and applying an average linkage algorithm. A: 6 h data. B: day 3 data. C: day 10 data. D: day 28 data. Boxes with green solid lines, downregulated genes; boxes with red dotted lines, upregulated genes; Con, contralateral kidney; IRI, ischemic kidney.
Genes that were continuously affected by IRI are associated with chemokine and cytokine signaling and the costimulatory pathway.
Table 2 lists the genes that demonstrated significant changes at more than three time points. A total of 15 genes were considered commonly expressed genes. Twelve of them were upregulated, and the other three were downregulated. Pubmetrix analysis for gene lists from CD3+ T cells after IRI identified that 12 and 14 genes among 15 candidate genes had been associated with renal IRI and kidney inflammation, respectively, according to the current PubMed database. All of them were well known for their role related to immune cell trafficking. The commonly expressed genes were categorized into three groups. One group consisted of chemokine ligands and receptors. CCL7 and CCL5, and their receptors CCR2, CCR3, and CCR5 were highly upregulated throughout all time points. Other genes were associated with cytokine signaling and the costimulatory pathway for antigen presentation. Whereas genes related to cytokine signaling showed various expression between time points, expression of genes in the costimulatory pathway peaked at 6 h after IRI, subsequently decreased, and then increased on day 28 after IRI.
Table 2.
List of candidate genes
| Gene Title | Pubmetrix |
|||
|---|---|---|---|---|
| Gene Symbol | Kidney IRI | Renal inflammation | Immune cell trafficking | |
| Genes associated with chemokines | ||||
| C-C motif chemokine ligand 7 | CCL7 | 0 | 4 | 14 |
| C-C motif chemokine receptor 5 | CCR5 | 1 | 26 | 165 |
| C-C motif chemokine receptor 2 | CCR2 | 3 | 28 | 91 |
| C-C motif chemokine ligand 5 | CCL5 | 7 | 62 | 115 |
| C-C motif chemokine receptor 3 | CCR3 | 0 | 4 | 49 |
| Genes related to cytokine signaling | ||||
| Tumor necrosis factor-α | TNF-α | 169 | 965 | 253 |
| Suppressor of cytokine signaling 3 | SOCS3 | 3 | 6 | 1 |
| Toll-like receptor 4 | TLR4 | 7 | 51 | 30 |
| Protein tyrosine phosphatase Rc | PTPRC | 4 | 10 | 58 |
| IL-15 | IL-15 | 1 | 8 | 8 |
| Costimulatory molecules | ||||
| CD80 | CD80 | 11 | 11 | 36 |
| CD86 | CD86 | 6 | 8 | 46 |
| CD40 | CD40 | 1 | 29 | 55 |
| Transcript factor CP2 | Tfcp2 | 0 | 0 | 1 |
| MAPK9 | MAPK9 | 3 | 6 | 2 |
Genes which showed significant changes (fold-change >2.0 or <−2.0 and P < 0.05) at more than 3 time points studied) were listed as candidate genes. Pubmetrix analysis demonstrated that 12, 14, and 15 genes among 15 candidate genes were related to kidney IRI, renal inflammation, and immune cell trafficking, respectively, in the current PubMed database. (Genes in italics were downregulated.)
Global functional analysis.
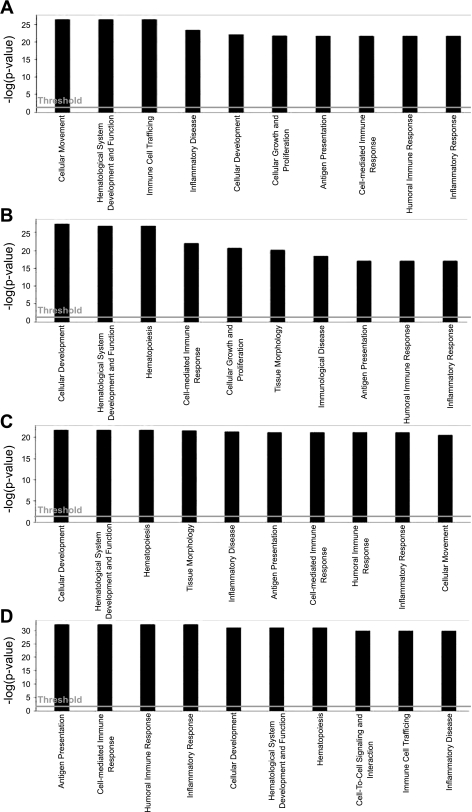
An Ingenuity pathway analysis demonstrated 10 different types of molecular and cellular functions associated with genes which showed significant changes from each time point (Fig. 2). Inflammatory responses related to antigen presentation, cell mediated-, and humoral immune response were identified as common biological functions where candidate genes affected by IRI at all time points were engaged. In the data from 6 h, cellular movement (P < 0.00005) and immune cell trafficking (P < 0.00004) were shown to have the highest significance involving 14 genes from our primary candidate list (Table 1). On day 3, cellular development (P < 0.00006, 19 genes were involved) and cell-mediated immunity (P < 0.00005, 18 genes were involved) were the most significant functions. On day 10, the genes were highly linked with cellular development (P < 0.000006) and tissue morphology (P < 0.000009) with 21 and 22 genes, respectively. On day 28, the significant functional processes were a shift to antigen presentation (P < 0.000004) and cell-mediated response (P < 0.000004), related to 33 and 35 genes, respectively.
Fig. 2.
Global functional pathway with significantly up- and downregulated genes at each time point. The significance value of each function is a measure of how likely a group of our candidate genes is involved in a represented (x-axes) function. The significance is expressed as a P value that is calculated using the right-tailed Fisher's exact test and represented as negative log value (y-axis). The threshold line represents significant P value 0.05 (1.3 in −log units). A: 6 h. B: day 3. C: day 10. D: day 28.
Biological functional network.
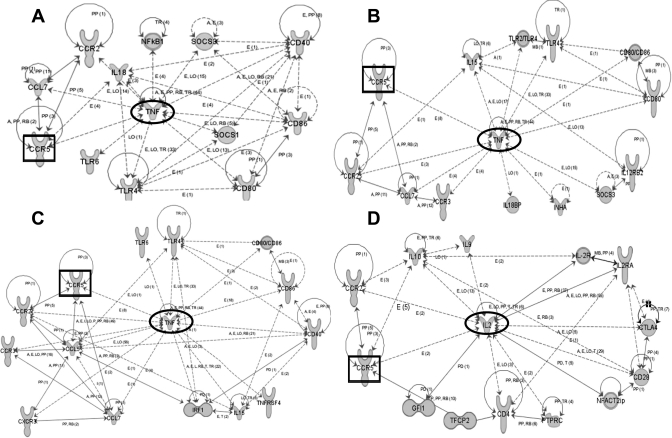
Ingenuity pathway analysis was then used to build functional networks between the candidate genes at each time point. To identify effector genes which work actively through interaction with other genes during whole time points, we conducted a genomic network analysis based on 24, 22, 24, and 37 activated transcripts at each time point (Fig. 3). The network score for each time point was >26, and the network score means the probability that a network would be assembled by chance, which is statistically significant (P < 0.001), where a level = >3. The top network derived from target genes at 6 h was related to cellular movement and immune cell trafficking, and TNF was centered with 13 other genes (Fig. 3A). On day 3, TNF was surrounded by 11 genes, which were related to immunological disease and cellular movement (Fig. 3B). The network for day 10 contained genes associated with hematological development and cellular movement, and TNF also occupied a central position connected with 13 genes (Fig. 3C). The network derived from the data for day 28 contains 14 genes centered around IL-2, which was associated with cellular growth and proliferation and antigen presentation (Fig. 3D). CCR5 was the only component that participated in networks for a significant functional pathway at all time points.
Fig. 3.
Ingenuity-pathway network among significantly changed genes of kidney-infiltrating T cells after IRI. The relationship is presented as follows: solid lines (binding), solid arrows (direct activation), or broken arrows (indirect activation). Arrows point to the element on which an action is performed. A box represents the most common gene showed at all of time points (CCR5), and a circle represents the genes which have highest relationship with other genes (center of the network). Each network is involved in certain biological functions. A: 6 h, cell movement and immune cell trafficking. B: day 3, immunological disease and cell movement. C: day 10, hematological development and cell movement. D: day 28, cell growth/proliferation and antigen presentation.
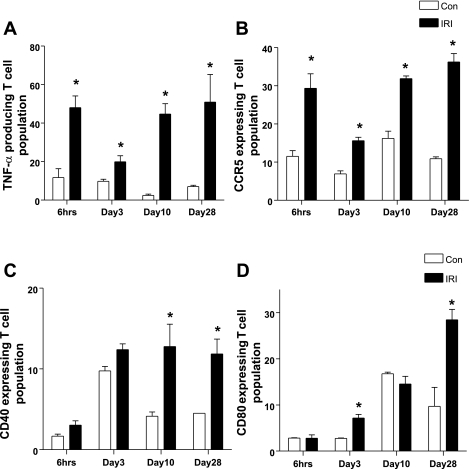
Validation of protein expression of candidate genes.
We assessed protein expression of selected candidate genes by flow cytometry. TNF-α and CCR5 expression were increased in T lymphocytes of IRI kidney at all time points, which correlated very well with PCR findings. CD40 expression was higher on days 10 and 28, and CD80 expression was higher on days 3 and 28 in IRI kidneys compared with contralateral kidneys, which is consistent with gene expression data, although upregulation of both genes at 6 h did not coincide with protein expression (Fig. 4).
Fig. 4.
Validation of protein expression of candidate genes on kidney infiltrating T cell by flow cytometry. Protein production of TNF-α (A) and CCR5 (B) were increased in T cells from ischemic kidney at all of time points. CD40 (C) and CD80 (D) expression on kidney-infiltrating T cells were increased in later time points. *P < 0.05 vs. contralateral kidney.
Anti-CCR5 antibody attenuated renal dysfunction following IRI.
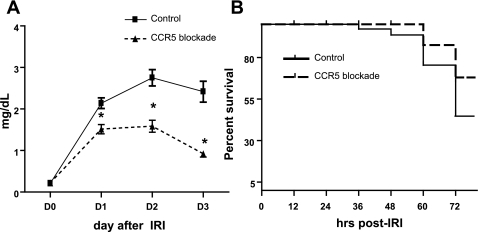
Renal dysfunction following IRI was evaluated with daily serum creatinine measured until day 3 after bilateral IRI (Fig. 5A). Mice treated with a CCR5-blocking antibody had lower serum creatinine compared with the isotype control antibody group from day 1 to day 3 after IRI (control vs. blocking antibody; day 1: 2.12 ± 0.13 vs. 1.53 ± 0.11, day 2: 2.62 ± 0.20 vs. 1.67 ± 0.14, and day 3: 2.24 ± 0.25 vs. 0.92 ± 0.08 mg/dl, P < 0.05). Survival on day 3 after IRI in the blocking antibody group trended higher, but was not statistically significant (control vs. blocking antibody 61 vs. 75%, P = 0.06) (Fig. 5B).
Fig. 5.
Effects of CCR5 blockade on renal function (A) and survival (B). All mice were treated with either an antibody against CCR5 or a control antibody before undergoing kidney ischemia followed by reperfusion. The blood samples were taken at indicated time points, and serum creatinine concentration was measured as a marker of renal function. All mice were followed up for mortality to 72 h after surgery, and mice survival was analyzed. A: serum creatinine level was significantly lower at all of time points in mice with CCR5 blockade compared with mice treated with control antibody. B: survival rate after kidney IRI trended higher in CCR5-blocked mice than in control mice by 72 h, but it was not significant (P = 0.06). *P < 0.05 vs. isotype control antibody group; n = 16–18 per group.
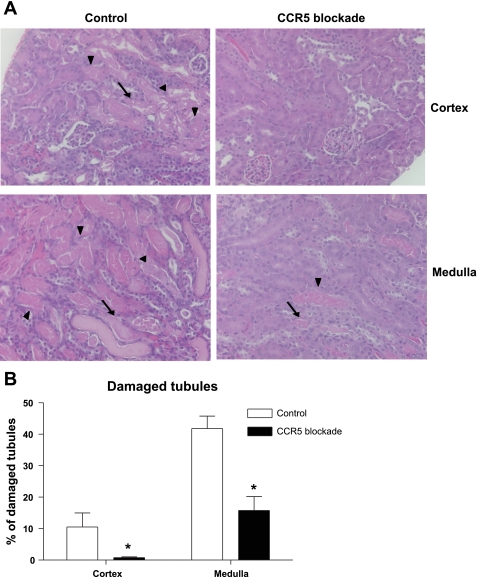
Anti-CCR5 antibody reduced renal tubular injury following IRI.
CCR5-blocking antibody-treated mice had reduced tubular injury and necrotic debris. There were significant differences in the percentage of damaged tubules consistently in both cortex and outer medulla (control vs. blocking antibody: 8.4 ± 3.8 vs. 1.1 ± 0.28 in cortex, and 41.8 ± 4.0 vs. 18.8 ± 4.6 in medulla, respectively, Fig. 6).
Fig. 6.
Histological assessment of renal tubular damage. Upon euthanasia, the kidneys were harvested and stained with hematoxylin and eosin for histological examination. The degree of renal tubular damage was blindly scored by a renal pathologist. A and B: representative photomicrographs of the ischemic kidney from mice treated with CCR5 or control antibody. On day 3, postischemic kidneys of CCR5-blocking antibody-treated mice showed less severe tubular damage compared with control both in cortex and medulla. C: kidney tubular damage score. Arrows indicate necrotic tubules, and arrowheads indicate necrotic debris. Magnification ×200. *P < 0.05 vs. isotype control antibody group; n = 10/group.
Anti-CCR5 antibody attenuated T cell activation following IRI.
We investigated a subpopulation of T cells, B cells, NKT cells, NK cells, macrophage, neutrophil, and dendritic cells of the postischemic kidneys in both treatment groups (Table 3). Because the cell counts of KMNCs were not different between groups, the percent population also reflected absolute proportions. The population of total CD3+ T cells, and their subtypes, CD4+ and CD8+ T cells, were not different between groups. However, the percentage of activated T cells with CD69 expression was lower in the CCR5-blocking antibody group compared with control (control vs. blocking antibody; 22.4 ± 3.8 vs. 13.9 ± 1.0% population on day 3 after IRI, P < 0.05). Blocking antibody treatment had little effect on B cells, activation and differentiation of B cells, NKT cells, activated NKT cells, NK cells, macrophages, neutrophils, and dendritic cells.
Table 3.
Population of T cells, B cells, NK T cells, NK cells, macrophages, neutrophils, and dendritic cells in postischemic kidneys of control (isotype antibody) and CCR5-blocking antibody-treated groups on day 3 after IRI
| Control | CCR5 Blockade | |
|---|---|---|
| Cell counts, ml | 3.0 ± 0.9 × 105 | 3.1 ± 0.7 × 105 |
| % Population | ||
| Total T cells (CD3+) | 40.5 ± 2.1 | 40.9 ± 4.3 |
| Activated T cells (CD3+CD69+) | 22.4 ± 3.8 | 13.9 ± 1.0* |
| CD4 T cells (CD3+CD4+) | 64.2 ± 3.0 | 66.5 ± 3.4 |
| Activated CD4 T cells (CD3+CD4+ CD69+) | 19.3 ± 4.1 | 12.5 ± 1.4 |
| CD8 T cells (CD3+CD8+) | 27.4 ± 3.5 | 27.4 ± 1.5 |
| Activated CD8 T cells (CD3+CD8+ CD69+) | 28.8 ± 4.3 | 20.1 ± 2.2 |
| B cells (CD19+) | 24.6 ± 6.0 | 30.4 ± 1.5 |
| Activated B cells (CD19+CD69+) | 2.6 ± 0.3 | 2.8 ± 0.5 |
| Differentiated B cells (CD19+MHCII+) | 16.6 ± 2.3 | 22.6 ± 2.4 |
| NKT cells (CD3+ NK1.1+) | 1.5 ± 0.15 | 1.8 ± 0.17 |
| Activated NKT cells (CD3+ NK1.1+ CD69+) | 64.2 ± 7.2 | 53.6 ± 3.7 |
| NK cells (CD3− NK1.1+) | 9.2 ± 0.9 | 10.0 ± 0.8 |
| Macrophages (F4/80highCD11clow) | 18.5 ± 1.6 | 15.1 ± 2.0 |
| Neutrophils (Mac-1highLy-6Ghigh) | 11.0 ± 1.7 | 6.7 ± 1.9 |
| Dendritic cells (CD11c+ cells) | 4.3 ± 0.9 | 5.1 ± 0.3 |
Values are means ± SE.
P < 0.05 compared with control.
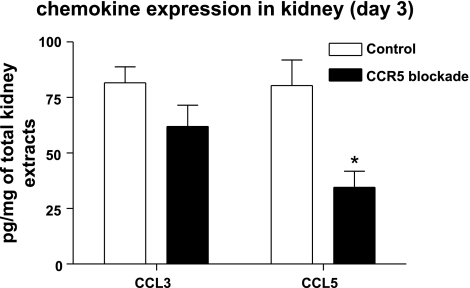
Anti-CCR5 antibody decreased CCL5 expression.
Chemokine ligands related to CCR5, such as CCL3 and CCL5, were measured by ELISA in kidney tissue. CCL3 and CCL5 levels were increased after IRI. Blocking antibody treatment significantly attenuated CCL5 expression in kidneys (Fig. 7).
Fig. 7.
Effect of CCR5 blockade on renal CCL3 and CCL5. The protein levels of CCL3 and CCL5 were measured in whole ischemic kidneys from mice with CCR5 blockade or control antibody at days 0 and 3. CCL3 and CCL5 levels were increased after IRI compared with those at day 0. The CCL5 level in postischemic kidneys was significantly less increased in the CCR5 blockade group than that in the control group. *P < 0.05 vs.. D3 isotype control antibody group; n = 8/group.
DISCUSSION
We have demonstrated that the small population of CD3+ T cells infiltrating into the kidney after IRI can undergo molecular profiling, which then led to the identification of a novel mechanistic pathway in this disease. Robust changes were found in chemokines, cytokines, and costimulatory molecules in kidney T cells after IRI. CCR5 was one of genes most highly increased in kidney-infiltrating T cells after IRI. Antibody blockade to CCR5 attenuated functional and histological changes in the kidney after IRI and led to decreased activation of kidney-infiltrating T cells.
A number of studies have demonstrated the important role for T cells in kidney IRI as in other nonimmunological renal diseases such as hypertension and diabetes (6, 16, 24). Rapid trafficking of T lymphocytes into the kidney was observed as early as 1 h after IRI (4, 31). The pathogenic role of T cells in kidney IRI has been revealed by renal protection after IRI in athymic nu/nu mice (11), peripheral T cell depletion (7), and inactivation of T cell chemoattractant (47). CD4+ T cells, particularly IFN-γ producing, are important mediators of IRI (11, 15). T cell receptor (TCR) diversity and activation participated in kidney IRI (38–39). Th1 and Th2 T cell subsets have different effects on IRI (43, 49), and recently a small T cell population, regulatory T cells, has been found to confer protection from kidney IRI (19, 25). Moreover, increased numbers of activated effector-memory T cells were found in the postischemic kidneys as late as 6 wk after IRI, suggesting that T cells are also involved in long-term changes after IRI and could be responding to a newly exposed neoantigen (5). There is preliminary evidence in humans as well that T cells mediate IRI (9, 27). Despite many studies demonstrating the role of T cells in experimental kidney IRI, the underlying mechanisms are largely unknown
Gene expression profiling can be useful in providing mechanistic insight and identifying novel biomarkers for kidney IRI (20, 37). While a previous study has analyzed the transcriptome of the pure T cell population using cultured T cells in a rejection model (21), low numbers of kidney-infiltrating T cells during IRI have limited discovery in this setting, and defining transcriptional characteristics of T cells infiltrating into the kidney after IRI has been challenging. There have been recent advances in methods that effectively elute mononuclear cells from the kidney and isolate pure CD3+ T cells. To circumvent the problem of a small number of cells, we started with a technique which we have previously used to elute mononuclear cells from kidney using Percoll gradient methods (4). We then isolated CD3+ T cells by magnetic bead separation from kidney mononuclear cells and extracted RNA from the pure T cell population. RNA purity was checked by three different methods for confirming the quality of samples. We then applied an array-based QRT-PCR to study multiple genes with a small amount of RNA at the same time and identify specific biological pathways of interest. Quantitative PCR array analysis enables detection of multiple transcriptional changes from very small amounts of RNA (23). Combining these techniques, we were able to measure transcriptional changes in diverse candidate genes from purified CD3+ T cells infiltrating into the kidney after IRI.
IRI led to robust transcriptional activities in T cells as early as 6 h after IRI, and changes in gene expression persisted even 4 wk after IRI. Supervised hierarchical clustering identified several clusters linked with up- or downregulated genes in ischemic kidneys at each time point, which showed significant differences compared with T cells in the noninjured kidney. Fifteen genes that showed significant changes in expression at more than three time points were stratified into three groups: chemokines, cytokines, and costimulatory molecules. The functional analysis tools linked our gene expression to pathways associated with various immune responses. Immune cell trafficking and cellular movement gene changes were prominent at 6 h and day 3, and there was a change in the pattern with an increase in cellular development products related to immune responses on day 10. By day 28, there was a shift in T cell gene expression toward cellular and humoral immune responses involved with antigen presentation.
CCR5 was repeatedly among the highest upregulated genes at all time points, as well as the only one candidate gene that demonstrated significant interaction within key biological functional networks at all time points studied. This suggested that CCR5 could be playing an important role in IRI. Moreover, tools were available to define the exact function of CCR5 in IRI with blocking antibodies. CCR5 is a cellular receptor for RANTES, macrophage inflammatory protein-1 in T lymphocytes, macrophage, and dendritic cells (33). We first validated the increased CCR5 gene expression in T cells after IRI at the protein level by flow cytometry. CCR5 expression was also shown in macrophages and dendritic cells. However, it was not significantly different between ischemic and contralateral kidneys (data not shown). We hypothesized that CCR5 on T lymphocytes was a mediator of kidney IRI. Antibody blockade of CCR5 attenuated kidney IRI, although the effect on survival was not significant. Moreover, CCR5 blockade led to decreased T cell activation in the kidney after IRI. These data are consistent with previous reports that blockade of CCR5 attenuated intestinal and cardiac IRI in mice (2, 10). In addition, clinical studies on the role of CCR5 in allograft rejection linked its inactive allele carriers to longer graft survivals (22). An inactive CCR5 phenotype was also correlated with lower mortality in dialysis patients (30). Measurement of chemokines showed that CCL5 was decreased with antibody blockade of CCR5 compared with isotype control. CCL5 is a well-known chemoattractant and one of the chemokines related to tissue injury in renal IRI. Reduced T cell activation after IRI in CCR5 blockade might explain the decrease in CCL5 and attenuation of renal IRI. The exact role and mechanisms by which CCR5 is involved in IRI requires further study.
In addition to CCR5, other T cell candidate genes were revealed by our transcriptional discovery work. CCL7, a ligand for CCR1, -2, and -3, was the highest expressing gene. CCL7 is known to modulate lymphocyte activation and maturation (1) and was also found to be increased in stroke (48). Chemoattractant activities of CCL7 and CCL5, a well-known chemokine in kidney IRI (13), might be enhanced by the upregulation of target receptors such as CCR3 and CCR5. SOCS3 was another prominent T cell gene we found. This molecule was shown to confer cardioprotection in ischemic injury (51), whereas SOCS3 overexpression led to increase in oxidative stress (32) and suppression of regulatory T cells (28), which could worsen IRI. Another key gene, TNF-α, showed constant elevated expression in T lymphocytes after IRI, and it could be associated with poor renal outcome (13). TLR4 was also elevated in trafficking T cells, can mediate IRI (3, 35, 50), and increased TLR4 expression in T cells induced recruitment of memory T cells after burn injury (14). CD80, CD86, and CD40 were altered in kidney-trafficking T cells and are well-known costimulatory molecules which strengthen interaction between T lymphocytes and antigen-presenting cells in IRI (36, 41, 45). Future studies can explore the role of these molecules in the T cell response to kidney IRI.
Although our PCR profiling interrogated only 89 transcripts in kidney T cells, we focused on important lymphocyte pathways. Moreover, array-based QRT-PCR is a very accurate and effective way of investigating transcriptome with small amounts of RNA. This technique does not require mRNA validation at the transcript level like microarray chips, and thus one can proceed directly to protein level validation. We focused on select highest-expressing and biologically significant targets and found that protein expression on T cells was consistent with our PCR array data.
In summary, we established a technique which enabled discovery-oriented genetic profiling for a pure single T cell population infiltrating into the kidney after IRI and found robust transcriptional activities in kidney T cells. Among genes showing changes in expression associated with chemokine and cytokine signaling and costimulatory pathway for antigen presentation, CCR5 was one of the highest, and the only one with significant interactions for major biological process throughout all time points studied. Antibody blockade of CCR5 in mice protected renal function after IRI. Analysis of transcriptional activities in kidney T cells after IRI can help to elucidate mechanisms of IRI mediated by T cells and lead to future candidate diagnostic and therapeutic strategies.
GRANTS
G. J. Ko was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (E00029). H. Rabb was supported by the National Institutes of Health, National Kidney Foundation, and by a research gift from Rogelio Miro of Panama.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: G.J.K. and H.R. provided conception and design of research; G.J.K., D.L., H.R.J., E.H., T.W., C.C., and M.L. performed experiments; G.J.K., M.L., L.C.R., D.N.G., and H.R. analyzed data; G.J.K., L.C.R., D.N.G., and H.R. interpreted results of experiments; G.J.K. prepared figures; G.J.K. and H.R. drafted manuscript; G.J.K., D.N.G., and H.R. edited and revised manuscript; G.J.K. and H.R. approved final version of manuscript.
REFERENCES
- 1. Adarichev VA, Vermes C, Hanyecz A, Ludanyi K, Tunyogi-Csapo M, Finnegan A, Mikecz K, Glant TT. Antigen-induced differential gene expression in lymphocytes and gene expression profile in synovium prior to the onset of arthritis. Autoimmunity 39: 663–673, 2006 [DOI] [PubMed] [Google Scholar]
- 2. Akahori T, Sho M, Kashizuka H, Nomi T, Kanehiro H, Nakajima Y. A novel CCR5/CXCR3 antagonist protects intestinal ischemia/reperfusion injury. Transplant Proc 38: 3366–3368, 2006 [DOI] [PubMed] [Google Scholar]
- 3. Andrade CF, Kaneda H, Der S, Tsang M, Lodyga M, Chimisso Dos Santos C, Keshavjee S, Liu M. Toll-like receptor and cytokine gene expression in the early phase of human lung transplantation. J Heart Lung Transplant 25: 1317–1323, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Ascon DB, Lopez-Briones S, Liu M, Ascon M, Savransky V, Colvin RB, Soloski MJ, Rabb H. Phenotypic and functional characterization of kidney-infiltrating lymphocytes in renal ischemia reperfusion injury. J Immunol 177: 3380–3387, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Ascon M, Ascon DB, Liu M, Cheadle C, Sarkar C, Racusen L, Hassoun HT, Rabb H. Renal ischemia-reperfusion leads to long term infiltration of activated and effector-memory T lymphocytes. Kidney Int 75: 526–535, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Awad AS, Huang L, Ye H, Duong ET, Bolton WK, Linden J, Okusa MD. Adenosine A2A receptor activation attenuates inflammation and injury in diabetic nephropathy. Am J Physiol Renal Physiol 290: F828–F837, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Awad AS, Ye H, Huang L, Li L, Foss FW, Jr, Macdonald T, Lynch KR, Okusa MD. Selective sphingosine 1-phosphate 1 receptor activation reduces ischemia-reperfusion injury in mouse kidney. Am J Physiol Renal Physiol 290: F1516–F1524, 2006 [DOI] [PubMed] [Google Scholar]
- 8. Barrier A, Olaya N, Chiappini F, Roser F, Scatton O, Artus C, Franc B, Dudoit S, Flahault A, Debuire B, Azoulay D, Lemoine A. Ischemic preconditioning modulates the expression of several genes, leading to the overproduction of IL-1Ra, iNOS, and Bcl-2 in a human model of liver ischemia-reperfusion. FASEB J 19: 1617–1626, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Bogetti D, Sankary HN, Jarzembowski TM, Manzelli A, Knight PS, Thielke J, Chejfec G, Cotler S, Oberholzer J, Testa G, Benedetti E. Thymoglobulin induction protects liver allografts from ischemia/reperfusion injury. Clin Transplant 19: 507–511, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Braunersreuther V, Pellieux C, Pelli G, Burger F, Steffens S, Montessuit C, Weber C, Proudfoot A, Mach F, Arnaud C. Chemokine CCL5/RANTES inhibition reduces myocardial reperfusion injury in atherosclerotic mice. J Mol Cell Cardiol 48: 789–798, 2010 [DOI] [PubMed] [Google Scholar]
- 11. Burne MJ, Daniels F, El Ghandour A, Mauiyyedi S, Colvin RB, O'Donnell MP, Rabb H. Identification of the CD4(+) T cell as a major pathogenic factor in ischemic acute renal failure. J Clin Invest 108: 1283–1290, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burne-Taney MJ, Rabb H. The role of adhesion molecules and T cells in ischemic renal injury. Curr Opin Nephrol Hypertens 12: 85–90, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Burne-Taney MJ, Yokota N, Rabb H. Persistent renal and extrarenal immune changes after severe ischemic injury. Kidney Int 67: 1002–1009, 2005 [DOI] [PubMed] [Google Scholar]
- 14. Cairns B, Maile R, Barnes CM, Frelinger JA, Meyer AA. Increased Toll-like receptor 4 expression on T cells may be a mechanism for enhanced T cell response late after burn injury. J Trauma 61: 293–298, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Day YJ, Huang L, Ye H, Li L, Linden J, Okusa MD. Renal ischemia-reperfusion injury and adenosine 2A receptor-mediated tissue protection: the role of CD4+ T cells and IFN-gamma. J Immunol 176: 3108–3114, 2006 [DOI] [PubMed] [Google Scholar]
- 16. De Miguel C, Guo C, Lund H, Feng D, Mattson DL. Infiltrating T lymphocytes in the kidney increase oxidative stress and participate in the development of hypertension and renal disease. Am J Physiol Renal Physiol 300: F734–F742, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Perrot M, Young K, Imai Y, Liu M, Waddell TK, Fischer S, Zhang L, Keshavjee S. Recipient T cells mediate reperfusion injury after lung transplantation in the rat. J Immunol 171: 4995–5002, 2003 [DOI] [PubMed] [Google Scholar]
- 18. Fiorina P, Ansari MJ, Jurewicz M, Barry M, Ricchiuti V, Smith RN, Shea S, Means TK, Auchincloss H, Jr, Luster AD, Sayegh MH, Abdi R. Role of CXC chemokine receptor 3 pathway in renal ischemic injury. J Am Soc Nephrol 17: 716–723, 2006 [DOI] [PubMed] [Google Scholar]
- 19. Gandolfo MT, Jang HR, Bagnasco SM, Ko GJ, Agreda P, Satpute SR, Crow MT, King LS, Rabb H. Foxp3+ regulatory T cells participate in repair of ischemic acute kidney injury. Kidney Int 76: 717–729, 2009 [DOI] [PubMed] [Google Scholar]
- 20. Grigoryev DN, Liu M, Hassoun HT, Cheadle C, Barnes KC, Rabb H. The local and systemic inflammatory transcriptome after acute kidney injury. J Am Soc Nephrol 19: 547–558, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hidalgo LG, Einecke G, Allanach K, Halloran PF. The transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4+ CTL, CD8+ CTL and NK cells. Am J Transplant 8: 627–636, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Hummel M, Bara C, Hirt S, Haverich A, Hetzer R. Prevalence of CCR5Delta32 polymorphism in long-term survivors of heart transplantation. Transpl Immunol 17: 223–226, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Jacob N, Yang H, Pricop L, Liu Y, Gao X, Zheng SG, Wang J, Gao HX, Putterman C, Koss MN, Stohl W, Jacob CO. Accelerated pathological and clinical nephritis in systemic lupus erythematosus-prone New Zealand Mixed 2328 mice doubly deficient in TNF receptor 1 and TNF receptor 2 via a Th17-associated pathway. J Immunol 182: 2532–2541, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim SY, Park KH, Gul R, Jang KY, Kim UH. Role of kidney ADP-ribosyl cyclase in diabetic nephropathy. Am J Physiol Renal Physiol 296: F291–F297, 2009 [DOI] [PubMed] [Google Scholar]
- 25. Kinsey GR, Sharma R, Huang L, Li L, Vergis AL, Ye H, Ju ST, Okusa MD. Regulatory T cells suppress innate immunity in kidney ischemia-reperfusion injury. J Am Soc Nephrol 20: 1744–1753, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 27. Loverre A, Divella C, Castellano G, Tataranni T, Zaza G, Rossini M, Ditonno P, Battaglia M, Palazzo S, Gigante M, Ranieri E, Schena FP, Grandaliano G. T helper 1, 2 and 17 cell subsets in renal transplant patients with delayed graft function. Transpl Int 24: 233–242, 2011 [DOI] [PubMed] [Google Scholar]
- 28. Matsumura Y, Kobayashi T, Ichiyama K, Yoshida R, Hashimoto M, Takimoto T, Tanaka K, Chinen T, Shichita T, Wyss-Coray T, Sato K, Yoshimura A. Selective expansion of foxp3-positive regulatory T cells and immunosuppression by suppressors of cytokine signaling 3-deficient dendritic cells. J Immunol 179: 2170–2179, 2007 [DOI] [PubMed] [Google Scholar]
- 29. Mishra J, Ma Q, Prada A, Mitsnefes M, Zahedi K, Yang J, Barasch J, Devarajan P. Identification of neutrophil gelatinase-associated lipocalin as a novel early urinary biomarker for ischemic renal injury. J Am Soc Nephrol 14: 2534–2543, 2003 [DOI] [PubMed] [Google Scholar]
- 30. Muntinghe FL, Verduijn M, Zuurman MW, Grootendorst DC, Carrero JJ, Qureshi AR, Luttropp K, Nordfors L, Lindholm B, Brandenburg V, Schalling M, Stenvinkel P, Boeschoten EW, Krediet RT, Navis G, Dekker FW. CCR5 deletion protects against inflammation-associated mortality in dialysis patients. J Am Soc Nephrol 20: 1641–1649, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noiri E, Doi K, Inagi R, Nangaku M, Fujita T. Contribution of T lymphocytes to rat renal ischemia/reperfusion injury. Clin Exp Nephrol 13: 25–32, 2009 [DOI] [PubMed] [Google Scholar]
- 32. Numata K, Kubo M, Watanabe H, Takagi K, Mizuta H, Okada S, Kunkel SL, Ito T, Matsukawa A. Overexpression of suppressor of cytokine signaling-3 in T cells exacerbates acetaminophen-induced hepatotoxicity. J Immunol 178: 3777–3785, 2007 [DOI] [PubMed] [Google Scholar]
- 33. Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal 16: 1201–1210, 2004 [DOI] [PubMed] [Google Scholar]
- 34. Perico N, Cattaneo D, Sayegh MH, Remuzzi G. Delayed graft function in kidney transplantation. Lancet 364: 1814–1827, 2004 [DOI] [PubMed] [Google Scholar]
- 35. Pulskens WP, Teske GJ, Butter LM, Roelofs JJ, van der Poll T, Florquin S, Leemans JC. Toll-like receptor-4 coordinates the innate immune response of the kidney to renal ischemia/reperfusion injury. PLoS One 3: e3596, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rabb H. The T cell as a bridge between innate and adaptive immune systems: implications for the kidney. Kidney Int 61: 1935–1946, 2002 [DOI] [PubMed] [Google Scholar]
- 37. Ritter T, Kupiec-Weglinski JW. Gene therapy for the prevention of ischemia/reperfusion injury in organ transplantation. Curr Gene Ther 5: 101–109, 2005 [DOI] [PubMed] [Google Scholar]
- 38. Satpute SR, Park JM, Jang HR, Agreda P, Liu M, Gandolfo MT, Racusen L, Rabb H. The role for T cell repertoire/antigen-specific interactions in experimental kidney ischemia reperfusion injury. J Immunol 183: 984–992, 2009 [DOI] [PubMed] [Google Scholar]
- 39. Savransky V, Molls RR, Burne-Taney M, Chien CC, Racusen L, Rabb H. Role of the T-cell receptor in kidney ischemia-reperfusion injury. Kidney Int 69: 233–238, 2006 [DOI] [PubMed] [Google Scholar]
- 40. Seifarth C, Mack M, Steinlicht S, Hahn EG, Lohmann T. Transient chemokine receptor blockade does not prevent, but may accelerate type 1 diabetes in prediabetic NOD mice. Horm Metab Res 38: 167–171, 2006 [DOI] [PubMed] [Google Scholar]
- 41. Shen X, Wang Y, Gao F, Ren F, Busuttil RW, Kupiec-Weglinski JW, Zhai Y. CD4 T cells promote tissue inflammation via CD40 signaling without de novo activation in a murine model of liver ischemia/reperfusion injury. Hepatology 50: 1537–1546, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shen XD, Gao F, Ke B, Zhai Y, Lassman CR, Tsuchihashi S, Farmer DG, Busuttil RW, Kupiec-Weglinski JW. Inflammatory responses in a new mouse model of prolonged hepatic cold ischemia followed by arterialized orthotopic liver transplantation. Liver Transpl 11: 1273–1281, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Shen XD, Ke B, Zhai Y, Gao F, Anselmo D, Lassman CR, Busuttil RW, Kupiec-Weglinski JW. Stat4 and Stat6 signaling in hepatic ischemia/reperfusion injury in mice: HO-1 dependence of Stat4 disruption-mediated cytoprotection. Hepatology 37: 296–303, 2003 [DOI] [PubMed] [Google Scholar]
- 44. Star RA. Treatment of acute renal failure. Kidney Int 54: 1817–1831, 1998 [DOI] [PubMed] [Google Scholar]
- 45. Takada M, Chandraker A, Nadeau KC, Sayegh MH, Tilney NL. The role of the B7 costimulatory pathway in experimental cold ischemia/reperfusion injury. J Clin Invest 100: 1199–1203, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thakar CV, Zahedi K, Revelo MP, Wang Z, Burnham CE, Barone S, Bevans S, Lentsch AB, Rabb H, Soleimani M. Identification of thrombospondin 1 (TSP-1) as a novel mediator of cell injury in kidney ischemia. J Clin Invest 115: 3451–3459, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S, Diao H, Guan Q, Cruikshank WW, Delovitch TL, Jevnikar AM, Du C. Decreased renal ischemia-reperfusion injury by IL-16 inactivation. Kidney Int 73: 318–326, 2008 [DOI] [PubMed] [Google Scholar]
- 48. Wang X, Li X, Yaish-Ohad S, Sarau HM, Barone FC, Feuerstein GZ. Molecular cloning and expression of the rat monocyte chemotactic protein-3 gene: a possible role in stroke. Brain Res Mol Brain Res 71: 304–312, 1999 [DOI] [PubMed] [Google Scholar]
- 49. Yokota N, Burne-Taney M, Racusen L, Rabb H. Contrasting roles for STAT4 and STAT6 signal transduction pathways in murine renal ischemia-reperfusion injury. Am J Physiol Renal Physiol 285: F319–F325, 2003 [DOI] [PubMed] [Google Scholar]
- 50. Zhai Y, Qiao B, Shen XD, Gao F, Busuttil RW, Cheng G, Platt JL, Volk HD, Kupiec-Weglinski JW. Evidence for the pivotal role of endogenous toll-like receptor 4 ligands in liver ischemia and reperfusion injury. Transplantation 85: 1016–1022, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Zhou M, Wang YL. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol Biol Cell 19: 318–326, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]