Abstract
The Fanconi anemia (FA) pathway participates in interstrand cross-link (ICL) repair and the maintenance of genomic stability. The FA core complex consists of eight FA proteins and two Fanconi anemia-associated proteins (FAAP24 and FAAP100). The FA core complex has ubiquitin ligase activity responsible for monoubiquitination of the FANCI-FANCD2 (ID) complex, which in turn initiates a cascade of biochemical events that allow processing and removal of cross-linked DNA and thereby promotes cell survival following DNA damage. Here, we report the identification of a unique component of the FA core complex, namely, FAAP20, which contains a RAD18-like ubiquitin-binding zinc-finger domain. Our data suggest that FAAP20 promotes the functional integrity of the FA core complex via its direct interaction with the FA gene product, FANCA. Indeed, somatic knockout cells devoid of FAAP20 displayed the hallmarks of FA cells, including hypersensitivity to DNA cross-linking agents, chromosome aberrations, and reduced FANCD2 monoubiquitination. Taking these data together, our study indicates that FAAP20 is an important player involved in the FA pathway.
Keywords: mitomycin C, DNA repair, foci
Fanconi anemia (FA) is a rare recessive genetic disorder characterized by bone marrow failure, congenital developmental defects, and cancer predisposition (1–4). Cellular features of FA include chromosomal instability and hypersensitivity to cross-linking agents (5). Fifteen FA complementation genes have been identified so far. These genes form several complexes to orchestrate interstrand cross-linking (ICL) repair. The FA core complex is composed of eight of the FA gene products (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM), in addition to FAAP24 and FAAP100 (6, 7), and acts as an E3 ligase to ubiquitinate FANCI/FANCD2 (I/D2) complex (8–11). The monoubiquitinated FANCI/FANCD2 complex interacts with Fanconi anemia-associated nuclease 1 (FAN1), which has exonuclease and endonuclease activity that may unhook the ICL, facilitate translesion synthesis, and promote downstream homologous recombination (HR) repair (12–15).
Besides the FA core complex and FANCI/FANCD2, there are several other FA proteins that likely act downstream of FANCI/FANCD2 and participate in HR repair. These proteins include BRCA2 (FANCD1) and PALB2 (FANCN), both of which are essential for HR repair (16–18). Another downstream FA protein, BACH1 (FANCJ), is also a bona fide double-strand break repair factor (19). BRCA2/FANCD1, PALB2/FANCN, and BACH1/FANCJ are all recruited to ICL sites (20), indicating that they are directly involved in ICL repair. More recently, mutations in two other genes, RAD51C/FANCO and SLX4/FANCP, were identified in patients with FA phenotypes (21–23), suggesting that there may be additional FA genes responsible for this disease.
Not all FA proteins function in a linear pathway involved in ICL repair. Many of the downstream FA proteins are involved in HR repair and associated with breast cancer susceptibility (16–19, 24, 25). These proteins all have functions besides ICL repair. More recently, another DNA repair protein, RAD18, which is best known for its role in UV lesion bypass, has also been shown to participate in the activation of the FA pathway via its ability to promote proliferating cell nuclear antigen (PCNA) monoubiquitination (26). It remains to be resolved how PCNA monoubiquitination is linked with the activation of the FA pathway.
Among all of the FA genes, mutations in FANCA (∼60%), FANCC (∼14%), and FANCG (∼10%) account for over 80% of the mutations identified in patients (27). However, FANCA, FANCC, and FANCG are orphan proteins that do not share extensive sequence homology with other proteins. Thus, it is still unknown how these proteins function in the FA pathway. We reason that the identification of new FA-associated proteins may help us understand how these orphan proteins participate in DNA repair. In this study, we report the identification of C1orf86 isoform2 as a previously undescribed FANCA-interacting protein (Fanconi anemia-associated protein 20 kDa, hereafter referred as FAAP20). Genetic inactivation of FAAP20 revealed many features of FA cells, highlighting that FAAP20 is a key component of the FA core complex and participates in ICL repair.
Results
FAAP20 Is a Unique Component of the FA Core Complex.
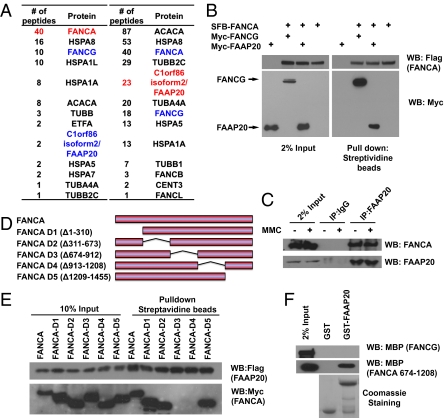
We performed tandem affinity purification (TAP) using FANCA as bait to identify FANCA-associated proteins. After excluding general contaminants, such as heat-shock proteins and ribosomal proteins, we identified FAAP20 as a potential FANCA-binding partner (Fig. 1A). FAAP20 (LOC1999990 isoform 2) encodes a 20-kDa protein with unknown function (gene name: C1orf86 isoform 2; accession number: NP_872339.2). To confirm its association with FANCA, we performed a reverse purification using FAAP20 as bait and showed that FANCA and FANCG copurified with FAAP20 (Fig. 1A), indicating that FAAP20 is a potential component of the FA core complex.
Fig. 1.
FAAP20 is a FANCA-binding protein. (A) 293T cells stably expressed SFB-FANCA or SFB-FAAP20, respectively, were subjected to TAP and mass spectrometry analysis. Red indicates the bait protein and blue indicates the known or putative-associated proteins. Number of peptides recovered from mass spectrometry analysis is also presented. (B) 293T cells were transfected with constructs encoding SFB-FANCA along with constructs encoding Myc-FANCG or Myc-FAAP20. Coprecipitation and immunoblotting were carried out as indicated. (C) Lysates prepared from control or MMC-treated 293T cells were incubated with protein A agarose beads conjugated with rabbit IgG or anti-FAAP20 antibodies. Western blotting was performed using indicated antibodies. (D) Schematic illustration of wild-type and deletion mutants of FANCA used in this study. (E) 293T cells were transfected with constructs encoding SFB-FAAP20 along with constructs encoding Myc-tagged wild-type or deletion mutants of FAAP20. Precipitation and immunoblotting were conducted as indicated. (F) Pull-down assays were performed using bacterially expressed and purified GST-FAAP20 and MBP-fused FANCA (residues 674–1032) or FANCG. Immunoblotting were conducted using anti-MBP antibody.
FAAP20 Binds Directly to FANCA.
To verify that FAAP20 interacts with FANCA, we coexpressed triple-tagged (SFB-tag: S-protein tag, FLAG epitope, tag and streptavidin-binding peptide tag) FANCA with Myc-tagged FAAP20 or FANCG. As expected, we observed a robust binding of SFB-tagged FANCA with Myc-tagged FANCG. In addition, we found a strong interaction between FANCA and FAAP20 (Fig. 1B), suggesting that they exist in the same complex. Moreover, we showed that endogenous FANCA coimmunoprecipitated with endogenous FAAP20 and this interaction occurs independently of mitomycin C (MMC) treatment (Fig. 1C).
We generated a series of internal deletion mutants of FANCA (FANCA-D1 to FANCA-D5) (Fig. 1D) and observed that two of them, FANCA-D3 and FANCA-D4, failed to interact with FAAP20 (Fig. 1E), indicating that FAPP20 binds to the middle region of FANCA. In addition, we performed a pull-down assay using bacterial expressed GST-fused FAAP20 and maltose-binding protein (MBP)-fused FANCA (aa674-1208) or MBP-fused FANCG. GST-FAAP20 pulled down MBP-fused FANCA (residues 674–1208), but not MBP-fused FANCG (Fig. 1F), suggesting that FAAP20 binds directly to FANCA, but not to FANCG.
We then used a series of FAAP20 internal-deletion mutants to map a FANCA-binding region on FAAP20 (Fig. 2A). Although wild-type FAAP20 coprecipitated with FANCA, two N-terminal deletion mutants of FAAP20 (FAAP20-D1 and FAAP20-D2) failed to do so (Fig. 2B). To further narrow down the residues on FAAP20 that are responsible for FAAP20-FANCA interaction, we aligned the human FAAP20 sequence with those of FAAP20 from other species and noted three highly conserved motifs at the N terminus of FAAP20 (residues 40–45, residues 76–81, and residues 83–87). Thus, we constructed three alanine substitution mutants, 6A1 (40WAELLR/AAAAAA45), 6A2 (76EVFTVG/AAAAAA81), and 6A3 (83 KTFSWT/AAAAAA87), to disrupt each of these conserved motifs (Fig. 2C), respectively, and examined their ability to interact with FANCA. The 6A1 mutant completely abolished the interaction between FAAP20 and FANCA (Fig. 2D), suggesting that this N-terminal motif within FAAP20 is necessary for FAAP20-FANCA interaction.
Fig. 2.
A conserved N-terminal region of FAAP20 is required for its interaction with FANCA. (A) Illustration of deletion mutants of FAAP20 used in this study. (B and D) 293T cells were transfected with plasmids encoding SFB-FANCA along with those encoding Myc-tagged wild-type or mutants of FAAP20. Precipitation was conducted using streptavidin beads and immunoblotting was performed using anti-Flag or anti-Myc antibodies as indicated. (C) Alignment of the N terminus of FAAP20 from different species. The conserved amino acids are shaded in black. Three mutants of FAAP20 with six alanine substitutions were generated (6A1, 6A2, and 6A3).
FAAP20 Contains an Evolutionarily Conserved RAD18-like Ubiquitin-Binding Zinc-Finger Domain.
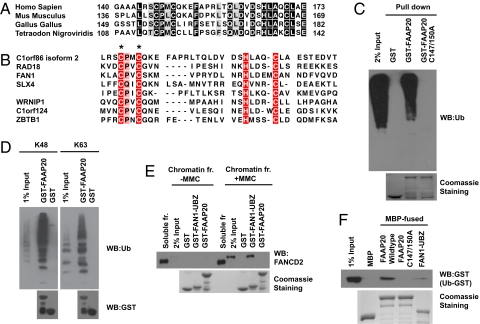
We noticed that FAAP20 contains an evolutionarily conserved ubiquitin-binding zinc-finger (UBZ) 4-type domain that belongs to the RAD18 family of zinc-finger (ZNF) domains (Fig. 3A). This domain is also present in other DNA damage-repair proteins, such as FAN1, RAD18, and SLX4/FANCP, which are involved in the FA pathway (Fig. 3B). Prompted by the ability of these zinc-finger domains in recognizing ubiquitin, we examined whether FAAP20 would also bind to ubiquitin through its UBZ domain. Indeed, GST-FAAP20 was able to pull down endogenous ubiquitin chains, whereas GST alone or the GST-FAAP20 C147/150A mutant failed to do so (Fig. 3C). FAAP20 interacted with both K48- and K63-linked ubiquitin chains without any notable preference (Fig. 3D). Given that monoubiquitinated FANCD2 is important for the function of the FA pathway (9), we tested but did not observe any appreciable binding of FAAP20 with monoubiquitinated FANCD2 in vitro, whereas the FAN1-UBZ (residues 1–100) domain was able to do so (Fig. 3E). We also examined whether or not FAAP20 would bind to monoubiquitin. In this regard, we performed an in vitro pull-down assay using bacterial-expressed carboxyl terminal-tagged ubiquitin (ub-GST). We observed that MBP-FAAP20 and MBP-FAN1-UBZ interacted with ub-GST, but MBP or MBP-FAAP20-C147/150A did not (Fig. 3F). Thus, although FAAP20 contains a ubiquitin-binding domain, the physiological partners of this UBZ domain remain unknown.
Fig. 3.
FAAP20 contains a RAD18-like UBZ domain at its C terminus. (A) Alignment of the RAD18-like UBZ domain of FAAP20 from different species. Identical residues are shaded in black. (B) Alignment of FAAP20 UBZ domain with other RAD18-like UBZ domains. The conserved Cys and His residues that define the two dyads of the ubiquitin-binding ZNF domain are shaded in red. The asterisks denote the conserved Cys147 and Cys150 residues mutated in the the FAAP20 C147/150A mutant. (C) FAAP20 binds to ubiquitin via its RAD18-like UBZ domain. GST pull-down experiments were carried out using 293T lysates and GST, GST-FAAP20, or GST-FAAP20 C147/150A mutant. Immunoblotting was conducted using anti-Ub antibody. (D) In vitro pull-down experiments were performed using K48- or K63-linked ubiquitin chains and GST-fusion proteins, as indicated. (E) Chromatin lysates were prepared from control or MMC-treated cells. In vitro pull-down experiments were performed using indicated GST-fusion proteins and Immunoblotting was conducted using anti-FANCD2 antibody. (F) FAAP20 binds to monoubiquitin. In vitro pull-down assays were performed using ub-GST and immobilized MBP-fusion proteins, as indicated.
FAAP20 Is Required for ICL Repair.
To further substantiate the role of FAAP20 in the FA pathway, we used the eChIP assay with a defined ICL (20) to test whether FAAP20 localize to ICL sites (Fig. 4A). Indeed, we observed a fourfold enrichment of FAAP20 at cross-linked substrates (Fig. 4A), suggesting that FAAP20 is recruited to ICLs in vivo. As a positive control, FANCD2 was also enriched at ICL substrates.
Fig. 4.
FAAP20 deficiency sensitizes cells to ICL damage and leads to genomic instability. (A) Schematic representation of the plasmid substrates used in the eChIP assay. The presence of psoralen-ICL and PCR primer locations are indicated. Relative enrichment of FAAP20 at ICLs was calculated by normalizing comparative concentration from real-time PCR of each sample against that of its input. Error bars represent SD from three independent experiments. CTL, control; XL, cross-linked. (B) Clonogenic survival assay of HCT116 cells, FANCL-deficient cells, and FAAP20-deficient cells following MMC treatment. (C) Whole-cell extracts were prepared from HCT116 cells, FAAP20-deficient cells, or FANCL-deficient cells mock-treated or treated with MMC for 24 h. Western blotting was conducted using indicated antibodies. (D) HCT116 cells, FAAP20-deficient cells, and FANCL-deficient cells were mock-treated or treated with 1 μM MMC for 24 h. Immunostaining was performed using anti-FANCD2 antibody and cells were counterstained with DAPI, as indicated. (Magnification: 100×.) (E) Quantification results were the average of two independent experiments and were presented as mean ± SEM. More than 300 cells were counted in each experiment. (F) HCT116 cells or FAAP20-deficient cells were exposed to a low dose of MMC and then treated with colcemid. A representative micrograph shows radial chromosome formation and chromosome breaks marked by arrows that were observed in FAAP20-deficient cells. (Magnification: 100×.) (G) Quantification of chromosome aberration were the average of two independent experiments using wild-type, FAAP20−/− cells, and FANCL−/− cells. The data were presented as mean ± SEM. (H) HCT116 cells or FAAP20-deficient cells were mock-treated or treated with 50 nM MMC. Cell-cycle distributions were analyzed by FACS and presented as percentages of cells in G1, S, or G2/M phases.
To study the function of FAAP20 in the FA pathway and ICL repair, we transiently knocked down FAAP20 in U2OS using siRNA and examined FANCD2 monoubiquitnation, as well as FANCD2 foci formation following MMC treatment (Fig. S1A). U2OS cells with FAAP20 down-regulation showed a reduction of FANCD2 monoubiquitination and FANCD2 foci formation (Fig. S1). However, monoubiquitnation of FANCD2 was not absent in FAAP20 knockdown cells. This finding could be because of incomplete knockdown of FAAP20 by siRNA. Alternatively, this finding may suggest that FAAP20 is not as important as other FA core components.
To test whether FAAP20 is essential for the activation of the FA pathway, we decided to generate a FAAP20-deficient cell line in HCT116 colon carcinomas and a control FANCL-deficient HCT116-derivative cell line. After targeting both alleles with virus vectors containing homology arms of exon 2 and 3 of FAAP20 and exon 2 and 3 of FANCL, we screened for clones with the correctly targeted alleles by PCR analysis (Fig. S2) and confirmed the absence of FAAP20 or FANCL in these cell lines by Western blotting (Fig. 4C).
Because the FA pathway is important for ICL repair, depletion of any of the FA proteins leads to hypersensitivity to DNA cross-linking agents. As expected, we observed that both FAAP20- and FANCL-deficient cells exhibited enhanced sensitivity to MMC. It is worth noting that FAAP20-deficient cells showed lower sensitivity than FANCL-deficient cells (Fig. 4B).
The FA core complex acts as an ubiquitin ligase that monoubiquitinates the FANCI/FANCD2 complex during S phase and also upon exposure to cross-linking agents. Although only FANCL has intrinsic E3 ligase activity, depletion of any component of the FA core complex compromises FANCI/FAND2 monoubiquitination (1, 8, 28). Thus, monoubiquitination of FANCD2 has been used as a surrogate marker for the integrity or the activation of the FA pathway. Because FAAP20 is a FANCA-associated protein, we reasoned that FAAP20-deficient cells might display defect in FANCD2 ubiquitination. Western blotting analysis revealed the presence of both unmodified and monoubiquitinated forms of FANCD2 in parental HCT116 cells, and an increase of monoubiquitinated FANCI and FANCD2 after MMC treatment (Fig. 4C). In contrast, ubiquitination of FANCI and FANCD2 was largely abrogated in FAAP20- and FANCL-deficient cells (Fig. 4C), suggesting that like FANCL, FAAP20 is also a component of the FA core complex and contributes to the activation of the FA pathway following MMC treatment. Interestingly, we observed a reduction in FANCA protein level in FAAP20-deficient cells (Fig. 4C), indicating that as a FANCA-binding protein, FAAP20 may stabilize FANCA in the cell. In addition to reduced FANCD2 monoubiquitnation, FAAP20-deficient cells also displayed defective FANCD2 foci formation (Fig. 4 D and E). Moreover, increased G2/M accumulation and genomic instability, including radial chromosome formation and chromosome breaks, are also the hallmarks of FA cells (29). As a matter of fact, we observed all of these phenotypes in FAAP20-deficient cells (Fig. 4 G and H). These data further validated that FAAP20 is a component of the FA pathway.
Binding to FANCA Is Required for FAAP20 Function in the FA Pathway.
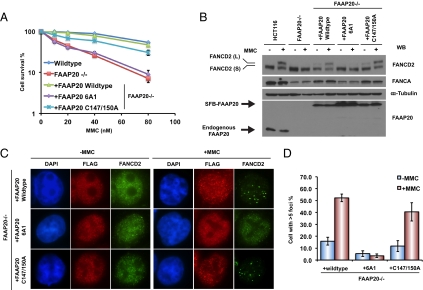
We reconstituted FAAP20-deficient cells with wild-type FAAP20 and observed that the FANCA level, MMC hypersensivity, and MMC-induced FANCD2 monoubiquitination and foci formation were all restored (Fig. 5). These data strongly suggest that the defects observed in FAAP20-deficient cells are a result of the loss of FAAP20 in these cells.
Fig. 5.
FANCA-binding is required for FAAP20 function in the FA pathway. (A) Clonogenic survival assay of HCT116 cells, FAAP20-deficient cells, and FAAP20-deficient cells reconstituted with wild-type FAAP20, the 6A1, or the C147/150A mutant of FAAP20. (B) Whole-cell extracts were prepared from HCT116 cells, FAAP20-deficient cells, and FAAP20-deficient cells complemented with indicated constructs with or without MMC treatment. Western blotting was conducted using indicated antibodies. (C) FAAP20-deficient cells reconstituted with indicated SFB-tagged wild-type or mutant FAAP20 were mock-treated or treated with MMC for 24 h. Immunostaining was performed using anti-Flag and anti-FANCD2 antibodies. Cells were counterstained with DAPI as indicated. (Magnification: 100×.) (D) Quantification results were the average of two independent experiments and were presented as mean ± SEM. More than 100 cells were counted in each experiment.
To understand whether the FANCA-binding activity and the UBZ domain of FAAP20 are important for its function in ICL repair, we also reconstituted the FAAP20-deficient cells with FAAP20-6A1 and FAAP20-C147/150A mutants, respectively. As expected, FAAP20-6A1 could not restore FANCA level and thus this mutant of FAAP20 could not rescue MMC sensitivity or MMC-induced FANCD2 monoubiquitination and foci formation in FAAP20-deficient cells (Fig. 5). On the other hand, the C147/150A mutant of FAAP20 rescued all of the above defects observed in FAAP20-deficient cells (Fig. 5). Taken together, these data indicate that FANCA-binding, but not its UBZ domain, is critical for FAAP20 function in ICL repair.
Discussion
In this study, we identified FAAP20 as a unique component of the FA core complex. FAAP20 binds directly to FANCA and stabilizes FANCA in the cell. The FANCA binding is essential for its function in the FA pathway, which is at least one mechanism of how it functions in the FA core complex.
The FA core complex comprises several subcomplexes, including FANCL-FANCB-FAAP100 (4), FANCC-FANCE-FANCF (30), FANCA-FANCG (31, 32), and FANCM-FAAP24 (33). Although it is known that the loss of any component in the FA core complex would lead to similar defects in FANCD2 ubiquitination and MMC sensitivity, it is not yet clear how these subcomplexes function together. An early study indicates that FANCA and FANCG stabilize each other and promote the nuclear localization of the FA core complex (34). This finding is similar to the situation in the present study. FANCA and FANCG consistently copurified with FAAP20 (Fig. 1A), suggesting that these three components likely form a stable subcomplex. The destabilization of FANCA in FAAP20-deficient cells could, in part, contribute to the defects observed in these cells. It is noteworthy to point out that unlike other FA cells, residual FANCD2 monoubiquitination was detectable in FAAP20-deficient cells. This phenotype is similar to that observed in FANCMΔ2/Δ2 mouse embyrionic fibroblasts (35). The significance of this observation is currently unclear.
FAAP20 contains a RAD18-like ZNF domain that binds to ubiquitin. At least three RAD18 UBZ domain-containing proteins, RAD18, FAN1, and SLX4/FANCP, are known to play critical roles in the FA pathway (12–15, 21, 22, 26, 36, 37). In particular, the UBZ domain of FAN1 binds directly to monoubiquitinated FANCD2 and thus recruits FAN1 to ICL sites to carry out its function in ICL repair (12–15). However, in the case of FAAP20, we showed that the UBZ domain of FAAP20 is not critical for FANCD2 monoubiquitination and MMC sensitivity. Further study is required to elucidate the cellular function of this highly conserved UBZ domain of FAAP20.
In conclusion, our data suggest that FAAP20 interacts with FANCA and participates in the regulation of the FA pathway. It is likely that via stabilizing FANCA, FAAP20 modulates the ubiquitin ligase activity of the FA core complex, which in turn regulates the FANCI/FANCD2 monoubiquitination following DNA damage. Up to now, no patient was found having mutation with several genes encoding FA pathway-related proteins (including FAN1, FAAP100, and FAAP24). Because the cellular phenotypes of FAAP20-deficient cells are rather mild compared with FANCL-deficient cells, it is possible that patients with FAAP20 mutation would display mild FA phenotypes and thus be difficult to diagnose. Knocking out the FAAP20 gene in mice may provide some clues to the function of this protein in mammals and may help the diagnosis of these patients.
Materials and Methods
Plasmids.
FAAP20, FANCG, FANCA, and FAN1 cDNAs were purchased from Open Biosystems. The cDNAs were cloned into the pDONR201 vector using Gateway cloning technology (Invitrogen). All of the deletions and point mutations were generated by site-directed mutagenesis using standard protocols. The corresponding entry vectors were transferred into a Gateway-compatible destination vector harboring an N-terminal triple-tag (S-protein, Flag, and a streptavidin-binding peptide), HA-Flag epitope tag, or a myc epitope tag for expression in mammalian cells, and GST or MBP tag for expression in bacteria, respectively.
TAP of Protein Complexes.
TAP was performed as previously described (38). Briefly, 293T cells were transfected with constructs encoding SFB-tagged FANCA or FAAP20 and selected with media containing puromycin (2 μg/mL). Cell lines stably expressing these tagged proteins were confirmed by Western blotting and immunofluorescence staining. For TAP, cells were lysed in NETN buffer (20 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM MgCl2) for 20 min in 4 °C. The crude lysates were cleared by centrifugation at 18,407 × g (Eppendorf 5424, Hamburg, Germany) at 4 °C for 30 min and rocked with streptavidin-conjugated beads (Amersham) for 2 h at 4 °C. The immunocomplexes were washed with NETN three times and eluted with 2 mg/mL biotin. The eluent was then incubated with S-protein Agarose beads (Novagen) for 2 h at 4 °C. The beads were then washed three times. The protein mixtures were eluted and analyzed by the Taplin Mass Spectrometry Facility at Harvard Medical School (Boston, MA).
Antibodies.
The primary antibodies used in this study were as follows: polyclonal anti-C1orf86 isoform 2 (FAAP20) antibody (Sigma-Aldrich; HPA038829); anti-myc antibody (Santa Cruz Biotechnology; sc-40); anti-FLAG antibody (Sigma-Aldrich; F1804); polyclonal anti-FANCA and anti-FANCI antibodies (Bethyl Laboratories; A301-980A and A301-254A); monoclonal anti-FANCD2 antibody (Santa Cruz Biotechnology; sc-20022); polyclonal anti-FANCD2 antibody (Novus Biologicals; NB100-182); polyclonal anti-MBP antibody (Millipore; 05–912); monoclonal anti-Ub antibody (Millipore; 04–263); monoclonal anti-GST (Santa Cruz; SC-138); polyclonal anti-FANCL antibodies were a generous gift from Weidong Wang (National institute on Aging, National Institutes of Health, Baltimore, MD).
Cell Cultures and Transfection.
Human embryonic kidney 293T cells and human colorectal cancer HCT116 cells were cultured in RPMI 1640 and DMEM, respectively, supplemented with (vol/vol) FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin, and maintained in 5% CO2 at 37 °C. Plasmid and siRNA transfection was performed using Lipofectamine 2000 and oligofectamineb (Invitrogen), respectively, according to the manufacturer's instructions. The coding strand for control siRNA was UCCAGUGAAUCCUUGAGGUUU and that for FAAP20 siRNA was UCCGAAAGCACAGAAGACGUUU. All siRNA were purchased from Dharmacon.
Immunoprecipitation, GST Pull-Down, and Western Blotting Analysis.
Cells were lysed in NETN buffer containing protease inhibitors. For immunoprecipitation of endogenous protein complexes, cell extracts were incubated with protein-A beads and antibody against FAAP20 for 2 h at 4 °C. For precipitation of SFB-tagged proteins or pull-down experiments, cell extracts were incubated with either streptavidin beads or GST-fusion proteins immobilized on glutathione beads for 2 h at 4 °C. For in vitro binding assay, ub-GST were eluted with glutathione and then incubated with beads coated with bacterial expressed MBP, MBP-FAAP20, MBP-FAAP20 C147/150A, or MBP FAN1-1-100. The beads were washed with NETN buffer and proteins were eluted by boiling in 1× Laemmli buffer. Samples were resolved by SDS/PAGE, transferred to polyvinylidene difluoride membrane, and immunoblotted with antibodies as indicated.
Immunofluorescence Staining.
Cells cultured on coverslips were washed in PBS, fixed in 3% paraformaldehyde for 15 min and then permeabilized in 0.5% triton solution for 5 min at room temperature. Samples were incubated with primary antibodies for 30 min, washed, and incubated with secondary antibodies for 30 min. Samples were then counterstained with DAPI and mounted on the glass slides with an antifade solution and visualized using a Nikon Eclipse 90i fluorescence microscope.
Somatic Knockout of FAAP20 and FANCL.
For the generation of somatic knockout cells, adeno-associated virus-based strategy was used as previously described (39). The targeting adeno-associated viruses were packaged in 293T cells by transfecting 3 μg of the targeting vector, pHelper, and pRC plasmids. Viruses were harvested at 72 h after transfection. Human colon cancer cell line HCT116 was infected for 48 h and selected with geneticin for 20 d. The geneticin-resistant clones were then screened using genomic PCR with primers derived from the neomycin-resistant gene and the upstream region of the left homologous arm or the downstream region of the right homologous arm. After the first allele was targeted, the neomycin-resistant gene was excised using viruses expressing Cre-recombinase. Second targeting was performed using the same approach.
Clonogenic Assay.
Cells were seeded at a density of 700 cells onto 6-cm dishes in triplicate. Twenty-four hours after seeding, the cells were treated with the indicated concentrations of MMC for 24 h. Cells were then washed free of drugs and incubated in fresh medium for another 10–14 d. The cells were then fixed and stained with 0.5% crystal violet in 20% ethanol. Colonies containing more than 50 cells were counted.
MMC-Induced Radial Chromosome Analysis.
Cells were plated in 10-cm dishes and treated with 0.063 μM MMC for 48 h. After treatment, cells were exposed to colcemid for 8 h, swollen using 0.075 M KCl, and fixed with 3:1 methanol:acetic acid. Slides were stained with Giemsa and 50 metaphase spreads were scored for radials in two independent experiments.
eChIP Assay.
Control substrates or substrates containing a defined cross-link were introduced into HEK293T cells stably expressing SFB-tagged FAAP20. ChIP was carried out as previously described (20).
Cell Cycle Analysis.
Cells were exposed to 50 nM MMC and were allowed to grow for 24 h. Cells were trypsinized and fixed in 70% ethanol overnight. Cells were then washed in PBS, nuclei were stained with propidium iodide (4 μg/mL), treated with RNase (2 μg/mL) at room temperature for 30 min, and analyzed in a flowcytometer using FACS Flow Jo software.
Supplementary Material
Acknowledgments
We thank our colleagues in the J.C. laboratory for insightful discussions and technical assistance. This work was supported in part by the Cancer Prevention Research Institute of Texas, Multi-Investigator Award, Grant RP110465-P2 (to J.C.). J.C. is a recipient of Era of Hope Scholar Award W81XWH-05-1-0470 from the Department of Defense and a member of the MD Anderson Cancer Center (CA016672).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118720109/-/DCSupplemental.
References
- 1.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy RD, D'Andrea AD. The Fanconi anemia/BRCA pathway: New faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 3.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling C, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Gurtan AM, D'Andrea AD. Dedicated to the core: Understanding the Fanconi anemia complex. DNA Repair (Amst) 2006;5:1119–1125. doi: 10.1016/j.dnarep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 10.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 11.Dorsman JC, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 14.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 17.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 18.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 19.Litman R, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Shen X, et al. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoepker C, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 22.Crossan GP, et al. Sanger Mouse Genetics Project Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaz F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 24.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 25.Levran O, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 26.Geng L, Huntoon CJ, Karnitz LM. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol. 2010;191:249–257. doi: 10.1083/jcb.201005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: Recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 28.Grompe M, van de Vrugt H. The Fanconi family adds a fraternal twin. Dev Cell. 2007;12:661–662. doi: 10.1016/j.devcel.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Hanlon Newell AE, et al. Loss of homologous recombination or non-homologous end-joining leads to radial formation following DNA interstrand crosslink damage. Cytogenet Genome Res. 2008;121:174–180. doi: 10.1159/000138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Léveillé F, et al. The Fanconi anemia gene product FANCF is a flexible adaptor protein. J Biol Chem. 2004;279:39421–39430. doi: 10.1074/jbc.M407034200. [DOI] [PubMed] [Google Scholar]
- 31.Reuter T, Herterich S, Bernhard O, Hoehn H, Gross HJ. Strong FANCA/FANCG but weak FANCA/FANCC interaction in the yeast 2-hybrid system. Blood. 2000;95:719–720. [PubMed] [Google Scholar]
- 32.Garcia-Higuera I, Kuang Y, Näf D, Wasik J, D'Andrea AD. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol. 1999;19:4866–4873. doi: 10.1128/mcb.19.7.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciccia A, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Higuera I, Kuang Y, Denham J, D'Andrea AD. The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 2000;96:3224–3230. [PubMed] [Google Scholar]
- 35.Bakker ST, et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet. 2009;18:3484–3495. doi: 10.1093/hmg/ddp297. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto KN, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci USA. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood. 2011;117:5078–5087. doi: 10.1182/blood-2010-10-311761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung JW, et al. SET nuclear oncogene associates with microcephalin/MCPH1 and regulates chromosome condensation. J Biol Chem. 2011;286:21393–21400. doi: 10.1074/jbc.M110.208793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods. 2008;5:163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.