Abstract
Messenger RNA (mRNA) localization and regulated translation can spatially restrict gene expression to each of the thousands of synaptic compartments formed by a single neuron. Although cis-acting RNA elements have been shown to direct localization of mRNAs from the soma into neuronal processes, less is known about signals that target transcripts specifically to synapses. In Aplysia sensory-motor neuronal cultures, synapse formation rapidly redistributes the mRNA encoding the peptide neurotransmitter sensorin from neuritic shafts into synapses. We find that the export of sensorin mRNA from soma to neurite and the localization to synapse are controlled by distinct signals. The 3′ UTR is sufficient for export into distal neurites, whereas the 5′ UTR is required for concentration of reporter mRNA at synapses. We have identified a 66-nt element in the 5′ UTR of sensorin that is necessary and sufficient for synaptic mRNA localization. Mutational and chemical probing analyses are consistent with a role for secondary structure in this process.
Keywords: FISH, RNA stem loop, RNA transport, SHAPE analysis, synaptogenesis
Messenger RNA (mRNA) localization and regulated translation provide a means of spatially restricting gene expression within distinct subcellular compartments. In the brain, local protein synthesis is critical to the development and experience-driven refinement of neural circuits, playing roles in axon guidance, synaptogenesis, and synaptic plasticity (1, 2). A large but select population of transcripts localizes to axons and dendrites (3–8), indicating that local translation subserves diverse cell biological functions. Where studied, the localization of mRNAs to axons or dendrites has been shown to depend on cis-acting localization elements (LEs) usually found in the 3′ UTR, although occasionally present in the coding sequence or 5′ UTR (1, 2, 9). These cis-acting mRNA LEs recruit specific transacting RNA binding proteins, and the resulting messenger ribonucleoproteins are packaged into RNA transport granules that interact with molecular motors to be delivered to their final subcellular destination (10–12).
In situ hybridization studies in neurons indicate that localized mRNAs in neurons are targeted to distinct subcellular compartments and domains within neuronal processes. For example, MAP2 mRNA concentrates within proximal dendrites, whereas calcium-calmodulin dependent protein kinase IIα (CaMKIIα) mRNA extends to distal dendrites (13). mRNA localization also seems to be dynamically regulated during development and with activity. In mature neurons, β-actin mRNA localizes to dendrites, and its concentration to distal dendrites is stimulated by depolarization (14). Stimuli that activate NMDA or neurotrophic receptor tyrosine kinase 2 (TrkB) receptors drive specific BDNF mRNA isoforms into distal dendrites of hippocampal neurons (15). High-frequency stimulation of perforant path projections to the dentate gyrus has been shown to direct localization of the mRNA encoding the immediate–early gene Arc selectively and specifically to activated dendritic lamina (16) and to drive localization of preexisting CaMKIIα mRNA into synaptosome fractions (17). Together, these findings indicate that specific transcripts undergo constitutive as well as developmental- and activity-dependent localization to distinct subcellular compartments.
Although a handful of cis-acting RNA LEs, ranging in size from five to several hundreds nucleotides, have been shown to mediate constitutive localization of specific mRNAs into distal processes of neurons (18), little is known about cis-acting elements that target mRNAs to more restricted subcellular compartments, such as synapses, or that mediate activity-dependent redistribution of mRNAs within neuronal processes. To identify such cis-acting LEs, we generated and expressed chimeric reporters to study the localization of the mRNA encoding the Aplysia sensory neuron (SN)-specific peptide transmitter, sensorin (19). Release of sensorin from the SN is required for both synapse formation and long-term facilitation (19, 20). The localization of sensorin mRNA is regulated by synapse formation, such that it is diffusely localized in neurites of isolated SNs (which do not form synapses) but concentrates at synapses in SNs paired with target motor neurons (MNs) (19). Synapse formation does not alter the localization of other neuritically localized mRNAs, including those encoding α-tubulin and β-thymosin (4), suggesting that sequences within the sensorin mRNA specify its synaptic localization. Dissection of the mechanisms underlying sensorin mRNA localization thus provides not only a means of identifying cis-acting LEs involved in the export of mRNA from the soma to distal neuronal processes and in the localization of mRNA specifically to synapses, but also LEs that dynamically mediate changes in mRNA localization in response to synapse formation.
We previously demonstrated that the full-length 5′ and 3′ UTRs of sensorin are sufficient for synaptic localization of reporter mRNAs and for stimulus-induced translational regulation of the reporter at synapses (21). Although the 3′ UTR was sufficient for localization of reporter mRNA to distal neurites, the 5′ UTR was required for synaptic localization of the reporter (21). To define the minimal LE for synaptic targeting, we have now characterized a series of deletion and point mutations in the 5′ UTR, focusing on stem-loop structures. Our studies identify a 66-nt-long stem-loop cis-element in 5′ UTR, just upstream of the translational start site, that functions as a synaptic LE (sLE).
Results
To define the RNA sequences that mediate localization of sensorin mRNA to neurites and to synapses, we fused the 5′ UTR, coding sequence, and/or the 3′ UTR of sensorin to the coding sequence of fluorescent protein dendra2 (Fig. 1A). We microinjected each construct into cultured Aplysia SNs, fixed the cultures 48 h later, and processed them for FISH with antisense dendra2 riboprobes to detect reporter mRNA. To differentiate between sequences mediating neuritic localization and sequences mediating synaptic localization, we performed experiments in isolated SNs that do not form chemical synapses (19, 22) and in SNs forming glutamatergic synapses with target MNs.
Fig. 1.
The 3′ UTR of sensorin is sufficient to target reporter mRNA into distal neurites. pNEX vectors encoding translational reporters were microinjected into isolated Aplysia SNs in culture (DIV 2). Neurons were fixed (DIV 4) and processed for FISH with dendra antisense riboprobes. (A) Cartoons of reporter constructs in pNEX3 expression vector. (B) Representative images of reporter (dendra2) mRNA FISH in isolated SNs. (B1–B6) Neurolucida tracing of each SN; (B1′–B6′) FISH (detected with dendra antisense riboprobes for B1′–B5′ and with sensorin antisense riboprobes for B6′). The FISH signal only extends to distal neurites when the 3′ UTR of sensorin is present; distal localization is enhanced by 5′ UTR. (Scale bar, 100 μm.) (C) Quantification of the distribution of reporter mRNA within sensory neurites. Neurites were linearized and divided into proximal, middle, and distal segments. The percentage of total FISH signal in distal segments is shown (see also Fig. S1). ***P < 0.0001, Kruskal-Wallis one-way analysis of variance followed by Dunnett's multiple comparison test.
As shown in Fig. 1B, when no sensorin sequences were included, dendra2 reporter mRNA was restricted to the soma and proximal neurites of isolated SNs. Addition of either the 5′ UTR or the coding sequence of sensorin to dendra2 did not alter this pattern of distribution. In contrast, the sensorin 3′ UTR distributed dendra2 reporter mRNA into distal neurites, and addition of 5′ UTR increased the distal distribution. To quantify the localization, we measured fluorescence pixel intensity along neurite length and measured the percentage of FISH signal in the proximal, middle, and distal thirds of the neurites. As shown in Fig. 1C, in the absence of any sensorin sequence, or with the sensorin 5′ UTR or coding sequence alone, little reporter RNA was present in the distal third (9% ± 1%, 12% ± 1%, and 14% ± 4%, respectively). In contrast, addition of the 3′ UTR significantly increased the FISH signal in the distal third (to 26% ± 3%), and when both 5′ and 3′ UTRs were included, the percentage of reporter RNA in the distal third rose to 45% ± 3%. By comparison, FISH for endogenous sensorin mRNA in parallel sets of cultures revealed that 34% ± 2% of signal was present in the distal third (Fig. 1C and Fig. S1). Note that the mean pixel intensity in the cell body did not differ significantly between reporter constructs (Fig. S1), indicating that differences in distal localization were not due to changes in expression levels. Collectively, our results show that the sensorin 3′ UTR is sufficient to promote mRNA localization from the soma into distal processes. The sensorin 5′ UTR does not promote distal localization on its own but enhances distal localization of 3′ UTR-containing reporters.
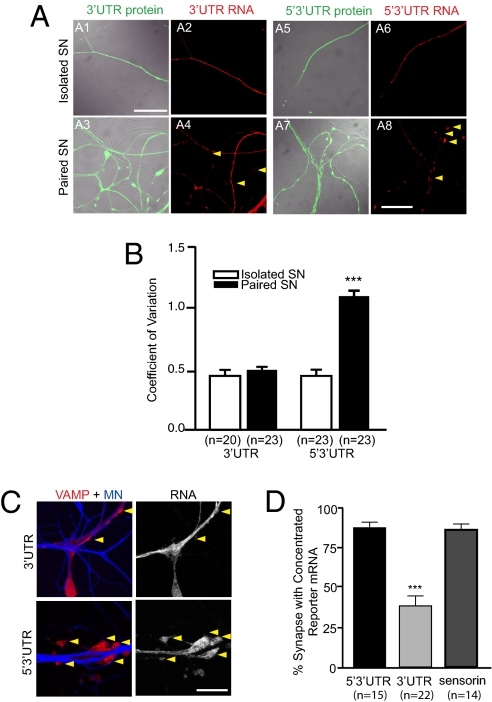
We next asked which sequences were required for synaptic localization by expressing the reporters shown in Fig. 1A in SNs that were paired with MNs. As shown in Fig. 2A, although the reporter with the 3′ UTR of sensorin localized to distal sensory neurites, it did not concentrate at synapses. In contrast, the reporter with both the 5′ and the 3′ UTR of sensorin not only localized to distal neurites but also concentrated at SN–MN synapses. We quantified this redistribution by comparing the coefficient of variation (CV) of the FISH signal in isolated SNs and in SN–MN cocultures (Fig. 2B). As previously reported for endogenous sensorin mRNA (19), we found that the CV was significantly greater when the 5′3′ UTR reporter construct was expressed in SN–MN cocultures than in isolated SNs (0.46 ± 0.04 in SN vs. 1.07 ± 0.09 in SN–MN; P < 0.001, unpaired Student t test), whereas the CV of 3′ UTR reporter was not significantly different in cocultures compared with isolated SNs (0.46 ± 0.04 in SN vs. 0.51 ± 0.04 in SN–MN). The CV of cytoplasmic diffusible dendra reporter proteins from the same neurites did not differ between SN and SN–MN, showing that the different CV of reporter mRNA was not due to local volumetric variation (5′3′ UTR reporter protein: 0.53 ± 0.05 in SN vs. 0.52 ± 0.03 in SN–MN; 3′ UTR-reporter protein: 0.41 ± 0.02 in SN vs. 0.56 ± 0.03 in SN–MN).
Fig. 2.
The 5′ UTR of sensorin is required for localizing reporter mRNA to synapse. Expression vectors encoding dendra2 reporters with either the 3′ UTR or both the 5′ and 3′ UTRs (5′3′ UTR) of sensorin were microinjected into Aplysia SN (DIV 2) cultured in isolation (isolated SN) or with MNs. Neurons were fixed (DIV 4) and processed for FISH using dendra2 antisense riboprobes. (A) Representative photomicrographs of dendra reporter protein (green, A1, A3, A5, A7) and mRNA (red, A2, A4, A6, A8) distribution in isolated and paired SNs. (Scale bar, 50 μm.) (B) Quantification of the change in distribution of reporter mRNA by measuring the CV (SD/mean) of the FISH signal. ***P < 0.0001, unpaired Student t test. (C) The 5′3′ UTR reporter or the 3′ UTR reporter was coexpressed with the presynaptic marker VAMP-mCherry in Aplysia SNs paired with target MNs on DIV 2. On DIV 4, the MN was injected with the volume filling dye Alexa Fluor 647 (blue), and images of VAMP-mCherry and blue Alexa Fluor were acquired, followed by fixation and FISH with dendra antisense riboprobes. Left: Merged VAMP/MN images of a coculture with SN overexpressing 5′3′ UTR or 3′ UTR reporter and VAMP-mCherry (VAMP-mCherry in red, MN in blue); Right: FISH signal for reporter mRNA. (Scale bar, 100 μm.) (D) Quantification of the percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA (error bars, SEM). ***P < 0.0001, one-way ANOVA followed by Dunnett's multiple comparison test. See also ref. 21 and Fig. S2.
To confirm that the sites of reporter mRNA concentration represented synapses, we expressed the presynaptic marker vesicle-associated membrane protein (VAMP)-mCherry in the SN and labeled the postsynaptic MN with Alexa Fluor 647 (Fig. 2C). We defined synapses as SN varicosities of >2 μm diameter containing a concentration of VAMP-mCherry and contacting the Alexa Fluor (blue)-labeled MN. To measure synaptic localization, we quantified the percentage of VAMP-mCherry–positive varicosities adjacent to MNs that contained clusters of reporter mRNA. We limited our data analysis to neurons in which reporter mRNA was abundant in adjacent neurites (mean fluorescent RNA in situ intensity >40 in an 8-bit image). As shown in Fig. 2D, 87% ± 3% of synapses colocalized with 5′3′ UTR reporter mRNA concentration, whereas only 38% ± 7% also contained 3′ UTR reporter mRNA concentration (see also ref. 21) (Fig. S2). Together, these data indicate that the dynamic redistribution of sensorin mRNA from neuritic shafts to synapses upon synapse formation is mediated by signals in the 5′ UTR.
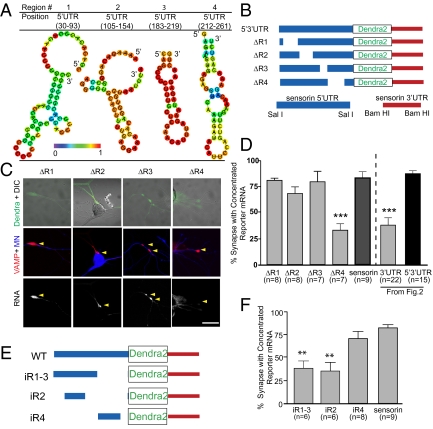
Studies of mRNA localization in other cell types have revealed that LEs often consist of stem-loop structures (1, 18, 23). To define the minimal sequences within the sensorin 5′ UTR that mediate localization to synapses, we focused on stem-loop secondary structures predicted by structural motif discovery programs. Twenty stem-loop structures were predicted, present within four distinct regions of the 5′ UTR (Fig. 3A and Fig. S3) (23–25). We deleted each region from the 5′3′ UTR dendra reporter to generate reporters with intact 3′ UTR and 5′ UTRs lacking one of the four regions predicted to contain local stem-loop structures (ΔR1–4; Fig. 3B).
Fig. 3.
A region directly upstream of the sensorin translation start site, in the 5′ UTR, is necessary and sufficient for synaptic localization of the reporter mRNA. (A) Representative graphic depictions of potential stem-loop structures in the 5′ UTR of sensorin predicted by RNAfold (35); color scale denotes base pair probabilities. (B) Cartoon representation of deletions made to the reporter construct. (C) Representative images of deletion constructs ΔR1–4 expressed in SNs synaptically connected with MNs. Top: Photomicrograph of Dendra protein (SN, green) merged with DIC image. Middle: Synapses marked as VAMP-mCherry clusters (red) contacting the MN (blue, Alexa 647). Bottom: FISH images showing clustering of reporter mRNA. (D) Quantification of synaptic localization of reporter mRNAs as the percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA. (E) Cartoon of insertion constructs (iR1–4) in which the designated regions of the sensorin 5′ UTR were inserted into a control pNEX 5′ UTR to test their ability to localize the reporter mRNA to synapses. The mutants were coexpressed in SNs paired with target MNs at DIV 2, and neurons were fixed on DIV 4 and processed for FISH. (F) Percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA.
We expressed these reporters together with VAMP-mCherry in SNs paired with MNs and quantified synaptic localization. As shown in Fig. 3 C and D, ΔR1 and ΔR3 retained synaptic localization comparable to the intact 5′3′ UTR reporter (81% ± 2% for ΔR1 and 80% ± 9% for ΔR3, compared with 87% ± 3% for intact 5′ UTR). The synaptic localization of ΔR2 was not as high as that of the intact 5′3′ UTR reporter or endogenous sensorin mRNA but was significantly greater than a 3′ UTR-dendra reporter lacking the 5′ UTR (68% ± 7% compared with 38% ± 7%). In contrast, the synaptic localization of ΔR4 mRNA was completely abolished and did not differ significantly from that of the 3′ UTR reporter (33% ± 6% compared with 38% ± 7%). RNA FISH intensity in the soma for each mutant was used as an indicator for overall RNA expression level. There was no correlation between RNA FISH intensity and synaptic localization, indicating that synaptic localization of mRNA did not depend on mRNA expression level (Fig. S2).
To test whether R4 was sufficient for localizing distally localized reporter mRNA to synapses, we inserted R4 (iR4, 66 nt) into the dendra reporter, downstream of the Rous sarcoma virus (RSV) promoter and upstream of the translation initiation site (iR4; Fig. 3E). This reporter showed synaptic localization that was not significantly different from endogenous sensorin mRNA (Fig. 3F; 70% ± 7% and 83% ± 3%, respectively), indicating that the 66-nt R4 sequence is sufficient for localizing RNA to synapses (Fig. 3F). By comparison, insertion of a sequence including R1, R2, and part of R3 (iR1–3) or R2 (iR2) was not sufficient to mediate synaptic mRNA localization (iR1–3: 37% ± 9% and iR2: 35% ± 10%; Fig. 3 E and F). Because R4 was necessary and sufficient for synaptic RNA localization, we call it a synaptic LE (sLE).
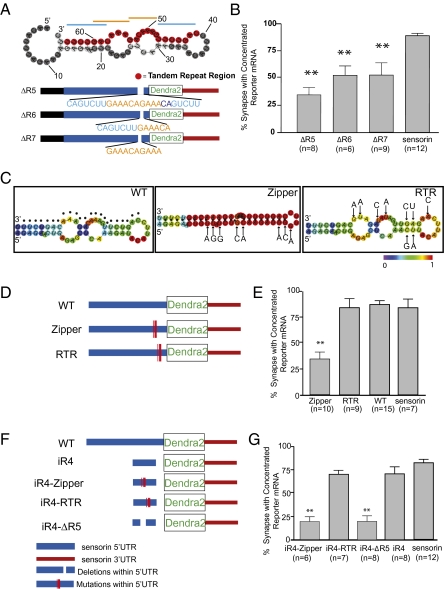
We next set out to further define the sLE. Closer inspection of the primary sequence of this structure revealed a 24-nt-long sequence that contains two repeated 7mers (5′-CAGTCTTGAAACAGAAACAGTCTT-3′, with two “CAGTCTT”s at the ends and two “GAAACAG”s in the center; Fig. 4A). Repeated hexamers or heptamers have been identified in LEs in various RNAs, often sites of recognition by RNA binding proteins. We thus set out to test whether this 24-nt tandem repeat element constituted the minimal synaptic LE. When we deleted either the entire 24-nt double-7mer-repeating sequence from 5′3′ UTR-dendra reporter (ΔR5), just one set of the two heptamers (ΔR6), or 10 nt of the center repeat (ΔR7), the synaptic concentration of the reporter mRNA was strongly reduced (Fig. 4 A and B; 34% ± 6%, 52% ± 9%, and 53% ± 11%, respectively). The fact that loss of a single repeat reduced localization suggested to us that synaptic RNA targeting depended on the secondary structure rather than the primary sequence of the sLE.
Fig. 4.
Stem-loop structure (66 nt) localizes reporter mRNA to synapses. (A) Predicted secondary structure of the region corresponding to R4 [RNAfold (35); highlighted in red is the 24-nt primary sequence containing a double-tandem 7mer-repeat (indicated with blue/orange lines)]. Mutants were generated in which the entire 24-nt (ΔR5), the first 13-nt set of repeat elements (ΔR6), or 10 nt of the center repeat (ΔR7) were deleted from the 5′3′ UTR reporter construct. (B) Mutants were coexpressed with VAMP-mCherry in SNs paired with target MNs, and the percentage of synapses containing reporter RNA was measured. **P < 0.01, one-way ANOVA followed by Dunnett's multiple comparison test compared with 3′ UTR reporter or endogenous sensorin. See also Figs. S2, S3, and S5. (C) Predicted secondary structures [RNAfold (35)] of WT (dots denote tandem repeat region described in Fig. 3); zipper construct, 8-point mutations were introduced to collapse the predicted secondary structure without disrupting the tandem repeat sequence; RTR construct, 9 mutations were introduced to disrupt the primary sequence of the tandem repeat region, while retaining predicted secondary structure. (D) Cartoon showing the location within the sensorin 5′ UTR of the mutations in the zipper and RTR constructs. The mutants were coexpressed in SNs paired with target MNs at DIV 2, and neurons were fixed on DIV 4 and processed for FISH. (E) Percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA. (F) Cartoon of mutant insertion reporter constructs, including WT iR4 (66 nt), iR4-RTR (iR4 with mutations shown in RTR in C), iR4-Zipper (iR4 with mutations shown in zipper in C), or iR4-ΔR5 (iR4 lacking the 24-nt repeat element). The mutants were coexpressed in SNs paired with target MNs at DIV 2, and neurons were fixed on DIV 4 and processed for FISH. (G) Percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA. **P < 0.001, one-way ANOVA followed by Dunnett's multiple comparison test.
To specifically test the role of secondary structure in mediating synaptic RNA localization, we generated and analyzed two additional reporters in which we introduced (i) eight mutations to “collapse” the two internal loops into a long stem (Fig. 4 C and D, “Zipper”); and (ii) nine mutations to maintain the predicted secondary structure while disrupting the primary sequence of the tandem repeats (Fig. 4 C and D, “RTR” for “remove tandem repeats”). The synaptic localization of the Zipper mutant was completely abolished (Fig. 4E; 34% ± 7% of VAMP-mCherry sites containing reporter RNA). In contrast, the synaptic localization of the RTR mutant was intact (Fig. 4E; 84% ± 8% of VAMP-mCherry sites containing reporter RNA). Moreover, insertion of the RTR 66-nt sequence into the dendra reporter, downstream of the RSV promoter and upstream of the translation initiation site (iR4-RTR), was sufficient to mediate synaptic localization of reporter RNA, whereas insertion of the Zipper 66-nt sequence (iR4-Zipper) was not (iR4-RTR: 70% ± 4%; iR4-Zipper: 20% ± 3%; Fig. 4 F and G). Together, these data are consistent with a critical role for the secondary structure of the sLE in mediating synaptic RNA localization.
Because the half-life of an RNA could affect the ability of that transcript to concentrate at synapses, we measured the stability of a subset of constructs by expressing them in isolated SNs, severing the soma and then fixing the remaining neurites at 0 or 48 h after soma removal. FISH was performed and mean pixel intensity measured. Our data show that all reporter RNAs were stable over a 48-h period (Fig. S4).
To further analyze the secondary structure of the sLE, we used selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) (26, 27). SHAPE takes advantage of the difference in reactivity between base-paired and unpaired nucleotides to the electrophile N-methylisatoic anhydride (NMIA). Modification of unpaired RNA at the 2’ hydroxyl group with NMIA blocks primer extension by reverse transcriptase, which can be detected by sequencing gels with single-nucleotide resolution.
We used SHAPE to test the structure of two sequences that localize to the synapse, R4 and RTR, and one sequence that does not, Zipper (Fig. 5). Consistent with algorithm-based prediction, Zipper collapses the two loops into a long stem while maintaining the tandem repeat elements. In contrast, the reactivity of RTR is remarkably similar to the wild-type sequence R4. Incorporation of the SHAPE data (ΔGs) into a secondary structure prediction program (28) decreased the minimal free energy (ΔG) of R4 from −7.2 to −23.1, of RTR from −7.7 to −21.3, and of Zipper from −20.5 to −74.1 kcal/mol (Fig. 5). Chemical analysis by SHAPE thus supports the predicted in silico secondary structure of the identified synaptic LE.
Fig. 5.
SHAPE analysis of the 66-nt element and mutants. Upper: SHAPE reactivities, determined from the sequencing gels in Fig. S6. Residues with intensities between 0 and 20%, 20% and 50%, and 50% and 100% are labeled gray, yellow, and red, respectively. Bars show the amounts of modification at each position relative to the most highly modified nucleotide. Numbers denote nucleotide position. Lower: SHAPE modification intensities mapped onto the predicted secondary structures; structures were generated using RNAfold (35). Residues with intensities between 0 and 20%, 20% and 50%, and 50% and 100% are shown in gray, yellow, and red, respectively. Both ΔG and ΔGs (predicted with SHAPE data) were generated using RNAstructure (28). Numbers denote nucleotide position.
Discussion
Our studies identify a cis-acting LE that mediates localization of mRNA to neuronal synapses. Our data support a multistep mRNA localization mechanism within neurons, in which specific cis-acting LEs mediate localization from the soma to the neuronal process, and other cis-acting LEs mediate further targeting to synapses (29). This type of multistep mechanism for localization to distinct subcellular compartments has previously been reported for myelin basic protein mRNA localization in oligodendrocytes (29) and protein kinase Mzeta in neurons (30). Although our data demonstrate that the 5′ UTR is necessary for synaptic localization, they do not rule out the possibility that synaptic localization involves combined actions of the 5′ and 3′ UTRs.
Bioinformatic analyses of the 5′ UTR of sensorin does not reveal homology at the primary sequence level with any known LEs. Although it is possible that the identified 66-nt sequence is unique, we believe that it is more likely that the conservation is at the level of secondary structure or 3D structure because mutations that maintain secondary structure localize reporter RNA to synapses despite disrupting the primary sequence (Fig. 4 A–C). As RNA secondary structure alignment programs are developed and optimized (31, 32), it will be interesting to use the R4 stem-loop structure to search for similar structures in other mRNAs and to then determine whether these mRNAs are synaptically localized.
The large size of cultured Aplysia sensory-motor neurons facilitates the study of mRNA subcellular localization in neurons. The ability to compare mRNA localization in neurons that do and do not form chemical synapses provides a means of detecting dynamic changes in mRNA localization that occur upon synapse formation. Together, this level of spatial resolution and control over stimulation (synapse formation) permits determination of where and when mRNAs localize in neurons. As such, our studies extend studies in mammalian neurons that have described cis-acting LEs for mRNA localization and/or dynamic changes in mRNA localization, in which analysis has been restricted to the level of localization to proximal and distal dendrites, rather than to specific compartments within the dendrite (e.g., refs. 16 and 33).
Our data show that mRNAs are remarkably stable within isolated neuronal processes. As shown in Fig. S4, we do not detect any decrease in FISH signal for endogenous sensorin or for any of the reporter RNAs in neurites that have been severed from their cell bodies after 48 h. Although it has been proposed that localized mRNAs are transported in structures in which they are protected from degradation (10), these studies specifically monitor RNA stability in isolated neuronal processes.
Our discovery of a cis-acting RNA sLE is a first step toward dissecting the mechanisms whereby RNAs localize to distinct subcellular compartments within neurons. It opens the door to identifying the specific RNA binding proteins, cytoskeletal elements, molecular motors, and transport structures that function to regulate gene expression with exquisite spatial and temporal control within neural synapses and circuits.
Materials and Methods
Reporter Constructs.
pNEX-dendra2, sensorin 3′ UTR pNEX-dendra2, sensorin 5′ UTR pNEX dendra2, sensorin 5′3′ UTR-dendra2, and mCherry-VAMP constructs were generated as previously described (21).
Aplysia Neuronal Cultures, Microinjection, Electrophysiology, and Stimulations.
Aplysia SN–MN cultures were prepared as previously described (34). Reporter plasmids were microinjected into SNs 24–36 h after plating. Synaptic connectivity was assayed by measuring excitatory post-synaptic potential (EPSP) amplitude between SN and target MN, as previously described (21).
Live Cell Imaging.
Confocal images were acquired on a Zeiss Pascal scanning laser microscope. Green dendra2 protein was excited with a 488-nm Argon laser (at 2.5 mW). To detect VAMP-mCherry–positive SN varicosities in contact with Alexa Fluor 647-labeled MNs, pNEX vectors encoding VAMP-mCherry were microinjected into SNs 24–36 h after plating, and the MN was microinjected with Dextran, Alexa Fluor 647 (Invitrogen) 60 min before imaging on day in vitro (DIV) 4. Neuronal morphology was traced from differential interference contrast (DIC) images using Neurolucida trace software (MBF Bioscience).
FISH.
Cells were fixed with 4% (wt/vol) PFA/30% (wt/vol) sucrose in PBS and processed for FISH as previously described (21). Sense riboprobe did not produce any background signal; antisense riboprobe did not produce signal in MNs [which do not express sensorin (21)]. Because dendra2 protein fluorescence does not persist after processing of samples for FISH, we manually aligned RNA images to live cell images on the basis of the morphology of SN and MN. We limited our data analysis to neurons in which reporter mRNA was abundant (e.g., mean fluorescent RNA in situ intensity above 40 in an 8-bit image) in adjacent shafts.
Statistical Analysis.
GraphPad Prism software was used for all statistical analysis as specified in figure legends. Kruskal-Wallis test followed by Dunnett's multiple comparison tests were used when data distribution did not follow Gaussian distribution.
SHAPE Analysis.
SHAPE was used to determine paired and unpaired regions within predicted secondary structures (26, 27). Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Braslow, K. Cadenas, M. Lee, and H. Huang for assistance with “blind” image analysis; D. Gunning for technical assistance; University of California, Los Angeles academic technology services for statistical consulting; and D. Black, S. L. Zipursky, and members of the Martin laboratory for helpful discussions. This work was supported by National Institutes of Health Grants R01NS045324 (to K.C.M.) and 5T32NS007449 (to E.J.M.), and Grants-in-Aid #23870016 (to D.O.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116269109/-/DCSupplemental.
References
- 1.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27:1065–1077. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- 4.Moccia R, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon MM, Choi SH, Jamieson CAM, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki T, Tian QB, Kuromitsu J, Kawai T, Endo S. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci Res. 2007;57:61–85. doi: 10.1016/j.neures.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Zivraj KH, et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian M, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci. 2006;26:7139–7142. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 13.Steward O, Wallace CS. mRNA distribution within dendrites: Relationship to afferent innervation. J Neurobiol. 1995;26:447–449. doi: 10.1002/neu.480260316. [DOI] [PubMed] [Google Scholar]
- 14.Tiruchinapalli DM, et al. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 17.Håvik B, Røkke H, Bårdsen K, Davanger S, Bramham CR. Bursts of high-frequency stimulation trigger rapid delivery of pre-existing alpha-CaMKII mRNA to synapses: A mechanism in dendritic protein synthesis during long-term potentiation in adult awake rats. Eur J Neurosci. 2003;17:2679–2689. doi: 10.1046/j.1460-9568.2003.02712.x. [DOI] [PubMed] [Google Scholar]
- 18.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 20.Hu JY, Glickman L, Wu F, Schacher S. Serotonin regulates the secretion and autocrine action of a neuropeptide to activate MAPK required for long-term facilitation in Aplysia. Neuron. 2004;43:373–385. doi: 10.1016/j.neuron.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Wang DO, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glanzman DL, Kandel ER, Schacher S. Identified target motor neuron regulates neurite outgrowth and synapse formation of aplysia sensory neurons in vitro. Neuron. 1989;3:441–450. doi: 10.1016/0896-6273(89)90203-1. [DOI] [PubMed] [Google Scholar]
- 23.Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc Natl Acad Sci USA. 2008;105:14885–14890. doi: 10.1073/pnas.0803169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton RS, Davis I. Identifying and searching for conserved RNA localisation signals. Methods Mol Biol. 2011;714:447–466. doi: 10.1007/978-1-61779-005-8_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 28.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ainger K, et al. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muslimov IA, et al. Dendritic transport and localization of protein kinase Mzeta mRNA: Implications for molecular memory consolidation. J Biol Chem. 2004;279:52613–52622. doi: 10.1074/jbc.M409240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamada M, Sato K, Asai K. Improving the accuracy of predicting secondary structure for aligned RNA sequences. Nucleic Acids Res. 2011;39:393–402. doi: 10.1093/nar/gkq792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taly J-F, et al. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat Protoc. 2011;6:1669–1682. doi: 10.1038/nprot.2011.393. [DOI] [PubMed] [Google Scholar]
- 33.Muslimov IA, Iacoangeli A, Brosius J, Tiedge H. Spatial codes in dendritic BC1 RNA. J Cell Biol. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Wang DO, Martin KC. Preparation of Aplysia sensory-motor neuronal cell cultures. J Vis Exp. 2009 doi: 10.3791/1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.